Introduction
A number of genetic and epigenetic factors have been
regarded as cancer pathogen (1). A
previous study of cancer epigenetics offered clues as to how
DNA-modification, including DNA methylation, histone variants or
miRNA varation, may be involved in the pathogenesis of certain
types of cancer (2). Histone
modification causes changes in the chromatin, and subsequently this
chromatin unfolds and remodels, altering the control of gene
transcription by affecting the approaching areas of the
transcription start site (3). In
addition, changes in chromatin influence the progression of cancer,
including the occurrence, metastasis, invasion and prognosis
(4). The molecular mechanisms of
histone variation affect gene expression, which is associated with
tumor progression and differentiation (5).
Histone variants have been regarded to have a
crucial role in the regulation of nuclear activation regulation by
changing chromatin structure (6,7).
However, the underlying mechanisms of this process are yet to be
determined. Two predominant histone variants, H3 and H2A, have been
established as participants in tissue-restricted protein expression
(8). Similar to its counterpart,
the H2A isoform MacroH2A has a crucial role in the transformation
of these variants. MacroH2A is the only histone with a tripartite
structure of the N-terminal histone-fold, which contains an
unstructured linker domain and a unique C-terminal Macro domain
(9,10). This unique structure makes MacroH2A
one of the most frequent alterations among all the histone variants
(11,12). Furthermore, MacroH2A has been shown
to have an important role in tumor pathogenesis and development via
the modification of chromatin condensation and regulation of gene
expression (13). Through gene
knockdown, it has been determined that MacroH2A promotes cancer
development via upregulation of the cyclin-dependent protein kinase
8 (CDK8) signaling pathway. It has been established that CDK8
belongs to the CDK family and is involved in the regulation of the
cell cycle (14,15). The results of these studies
indicate that MacroH2A affects tumor progression through changes to
the cell cycle process. However, until now, the effects of MacroH2A
on cell cycle regulatory genes have not been well illustrated.
In normal cells, the cell cycle processes of cell
division and proliferation are strictly regulated. The key
mediators of the cell cycle, CDKs and cyclins, regulate the
transition of the cell cycle (16–18).
During this process, dysfunctional expression of CDKs and/or
cyclins, induced by abnormal metabolic activity or environmental
factors, may trigger arrest or delay checkpoints in cell division
(19,20). Furthermore, CDKs and cyclins have
important roles in stabilizing the genome in the S phase (21,22).
As a result, abnormal expression of CDKs and cyclins may induce the
disorganization of cell proliferation, which results in the
dysfunction of cell division (23,24).
The aim of the present study was to investigate the effects of
MacroH2A on U2-OS osteosarcoma cells, by inducing MacroH2A
overexpression, interference, overexpression rescue and
interference rescue, and subsequently detecting the cell cycle
progression using flow cytometry. Furthermore, the regulation of
cyclin D and CDK expression levels by MacroH2A was
investigated.
Materials and methods
Cell culture
The U2-OS acute osteosarcoma cell line was purchased
from the American Type Culture Collection (Manassas, VA, USA). The
cells were maintained in Dulbecco’s modified Eagle’s medium
(Gibco-BRL, Grand Island, NY, USA), supplemented with 10% fetal
bovine serum (FBS; Invitrogen Life Technologies, Carlsbad, CA, USA)
at 37°C in a humidified incubator containing 5% CO2.
Following cell culture, the cell viability was analyzed by trypan
blue exclusion. The cells were grown on glass cover slips for
detection of cell morphology and were observed using an inverted
microscope (Leica DMI3000B, Wetzlar, Germany).
Vector design and transfection
To examine the biophysical properties of the U2-OS
cell line following knockdown and overexpression of MacroH2A,
interference and overexpression vectors were created. The MacroH2A
shRNA recombinant expression plasmid was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA; sc-62576-SH), and the
MacroH2A recombinant expression plasmid was produced in-house as
follows: the open reading frame fragment was obtained from OriGene
Technologies (SC110068; Rockville, MD, USA) and cloned into the
plasmid between the XhoI and BamHI sites of a pcDNA3.1(t) plasmid
(Invitrogen Life Technologies) to obtain the pcDNA3.1(t)-MacroH2A
recombinant plasmid. Once the plasmids has been obtained, the cells
were transfected with pcDNA3.1(t)-MacroH2A and/or MacroH2A shRNA
Plasmid using Lipofectamine™ 2000 (Invitrogen Life Technologies)
according to the manufacturer’ instructions. The cells were
collected for analysis following a 24-h incubation period at
37°C.
Following culture, the cells were divided into five
groups (with five parallel experiments performed for each group),
including a control (non-treated) group, a MacroH2A interference
group (transfected with 1 μg MacroH2A shRNA Plasmid), a MacroH2A
overexpression group (1 μg pcDNA3.1(t)-MacroH2A Plasmid
transfection), a MacroH2A interference rescue group (transfected
with 1 μg MacroH2A shRNA Plasmid transfection for 12 h followed by
transfection with 1 μg pcDNA3.1(t)-MacroH2A Plasmid for 12 h) and a
MacroH2A overexpression rescue group (transfected with 1 μg
pcDNA3.1(t)-MacroH2A for 12 h followed by transfection with 1 μg
MacroH2A shRNA Plasmid for 12 h).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To measure the change in the expression of mRNA
transcripts in the treatment groups, RT-qPCR was performed. Total
RNA was isolated from the cultured cells using TRIzol®
reagent (Invitrogen Life Technologies) according to the
manufacturer’s instructions. A total of 1 μg total RNA was
transcribed to cDNA using a Takara cDNA Library Construction kit
(Takara Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer’s instruction. The transcription levels of MacroH2A,
IGF-1, cyclin D1, cyclin D3, cyclin A, CDK4, CCNA2, CDC25A, CDK and
CDK8 were quantified with an ABI 7500 realtime PCR system (Applied
Biosystems, Inc., Foster City, CA, USA). The following primers were
used: Forward: 5′-CGTTCAAGTACCGGATCAGC-3′ and reverse:
5′-CAACTGCCAGCAAGATGTGT-3′ for MacroH2A, forward:
5′-CTGGAGATGTATTGCGCACC-3′ and reverse: 5′-AGACTTGCTTCTGTCCCCTC-3′
for IGF1, forward: 5′-CCGTCCATGCGGAAGATC-3′ and reverse:
5′-GAAGACCTCCTCCTCGCACT-3′ for Cyclin D1, forward:
5′-AACTGACCGGGAGATCAAGG-3′ and reverse: 5′-TGGCTTCAGATCTCGGTGAA-3′
for Cyclin D3, forward: 5′-AGTGTGAGAGTCCCCAATGG-3′ and reverse:
5′-CCTTGATCTCCCGGTCAGTT-3′ for CDK4, forward:
5′-TGTTTCAGCTTCTCCGAGGT-3′ and reverse: 5′-AGACTTCGGGTGCTCTGTAC-3′
for CDK6, forward: 5′-TGGTCACGTCTACAAAGCCA-3′ and reverse:
5′-AGCCACACCTTCCTATCAGC-3′ for CDK8, and forward:
5′-AACAGCGACACCCATCCTC-3′ and reverse:
5′-CATACCAGGAAATGAGCTTGACAA-3′ for GAPDH. These primers for these
genes were designed by Applied Biosystems, Inc., and span at least
one intron/exon border to eliminate genomic DNA amplification.
GAPDH was used as an endogenous control. qPCR was performed as
follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 sec
and 60°C for 45 sec, using the TaqMan® Universal PCR
Master Mix (Applied Biosystems, Inc.). The 2−ΔΔCt method
was used to analyze relative mRNA expression.
Western blot analysis
Following incubation, the cells from each group were
collected by centrifugation at 200 × g for 5 min at 4°C and washed
three times with ice-cold phosphate-buffered saline (PBS;
Invitrogen Life Technologies). The cell pellets were resuspended in
radioimmunoprecipitation assay lysis buffer (Pierce, Rockford, IL,
USA) and proteins were obtained from the lysate by centrifugation.
Proteins were separated by 10% SDS-PAGE and transferred to
polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA).
Once blocked with 4% non-fat milk, the membranes were incubated
with the following antibodies, purchased from Abcam (Cambridge, MA,
USA): monoclonal anti-MacroH2A (ab83782), anti-cyclin D1 (ab7958),
anti-cyclin D3 (ab112034), anti-CDK4 (ab7955), anti-CDK6 (ab151247)
and anti-CDK8 (ab123940) and anti-GAPDH (ab9485). Subsequently,
horseradish peroxidase-conjugated anti-mouse IgG secondary antibody
was added to the membranes and incubated for 1 h at room
temperature. The protein bands were detected using enhanced
chemiluminescent substrate (ECL plus, GE Healthcare Life Sciences,
Pittsburgh, PA, USA). Images were acquired using a Fujifilm FLA
5,000 image reader (Fujifilm, Valhalla, NY, USA).
Immunofluorescence
U2-OS osteosarcoma cells (4×104) were
grown on sterile glass cover slips and fixed for 30 min in 4%
paraformaldehyde solution (Sigma-Aldrich St. Louis, MO, USA) in
phosphate buffer. Following incubation with blocking reagent (5%
FBS in PBS) for 30 min, the glasses were washed with PBS and
incubated with primary antibodies overnight at 4°C. The following
primary antibodies were used: MacroH2A rabbit polyclonal, 1:1,000
dilution, 40 kDa (ab83782); Cyclin D1 rabbit polyclonal, 1:1,000
dilution, 33 kDa (ab7958); Cyclin D3 rabbit polyclonal, 1:1,000
dilution, 33kDa (ab112034); CDK4 rabbit polyclonal, 1:500 dilution,
34kDa (ab7955), CDK6 rabbit polyclonal, 1:1500 dilution, 37kDa
(ab151247); CDK8 rabbit polyclonal, 1:500 dilution, 53kDa
(ab1123940), GAPDH rabbit polyclonal, 1:5,000 dilution, 37kDa
(ab9485), all obtained from Abcam. The immunofluorescence assays
were performed by incubating cover slips with fluorescein
isothiocyanate-conjugated secondary antibodies (F5262;
Sigma-Aldrich). A conventional fluorescence microscope (Carl Zeiss,
Oberkochen, Germany) was used for visualization.
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD). The statistical significance of the results was analyzed
using the Student’s t-test or one-way analysis of variance test.
P<0.05 was considered to indicate a statistically significant
difference. All analyses were performed using SPSS version 13.0
(SPSS Inc., Chicago, IL, USA).
Results
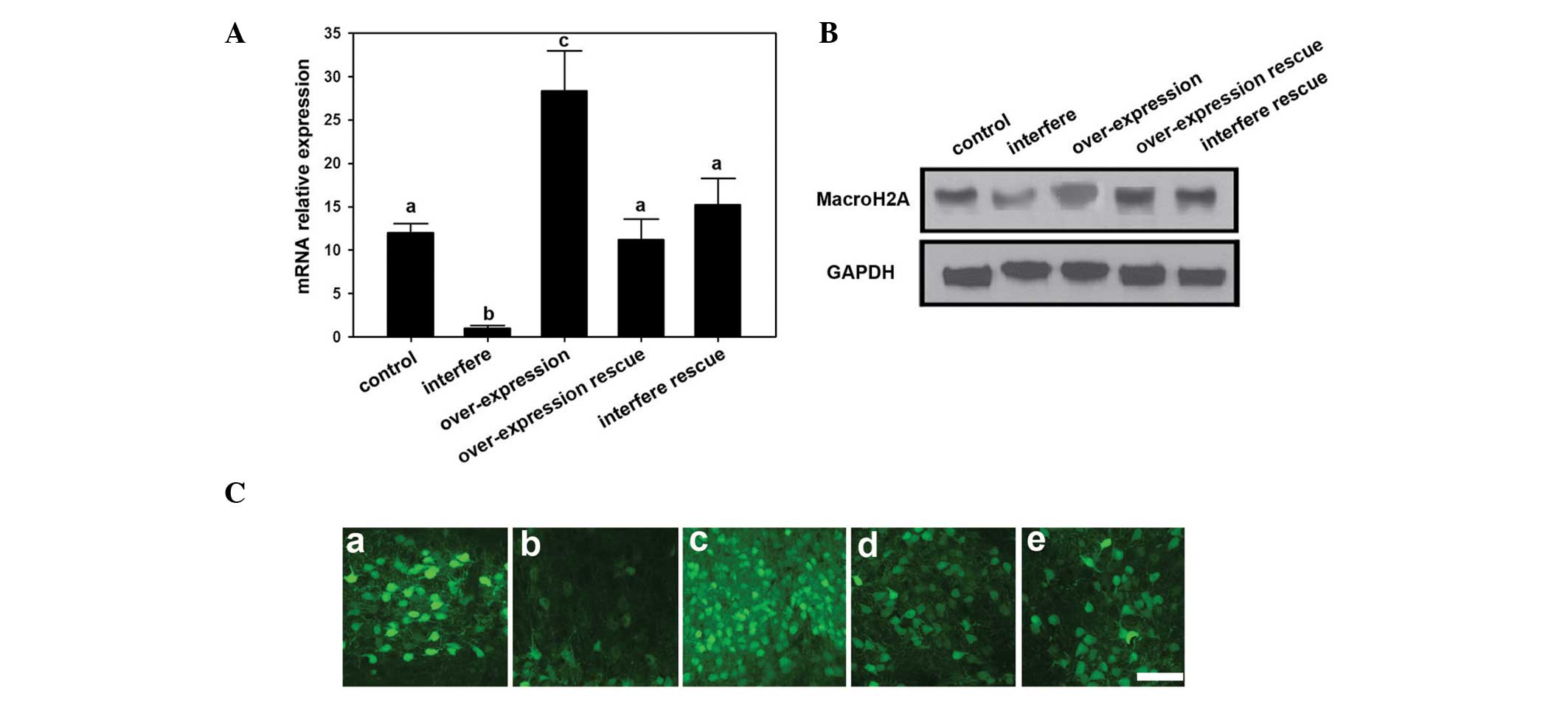
Overexpression and transfer of MacroH2A
in the U2-OS osteosarcoma cell line
To detect the effect of MacroH2A on osteosarcoma
cells, interference and overexpression MacroH2A vectors were
constructed and transfected into osteosarcoma cells using
Lipofectamine™ 2000. The mRNA expression level was analyzed by
RT-qPCR. Among all of the groups, the highest MacroH2A mRNA
expression level was in the MacroH2A overexpression group, while
the control, MacroH2A overexpression rescue and MacroH2A
interference rescue groups showed median expression. The MacroH2A
interference group had the lowest MacroH2A mRNA expression levels
(Fig. 1A). Furthermore, the
protein expression was detected by western blot. Accordingly, the
interference group had the lowest MacroH2A protein expression level
compared with those of the other groups. The overexpression group
had the highest MacroH2A protein density signals among all the
groups. Through the use of immunofluorescence analyses, the protein
expression levels were shown to have similar expression
differentiation to that observed by western blotting.
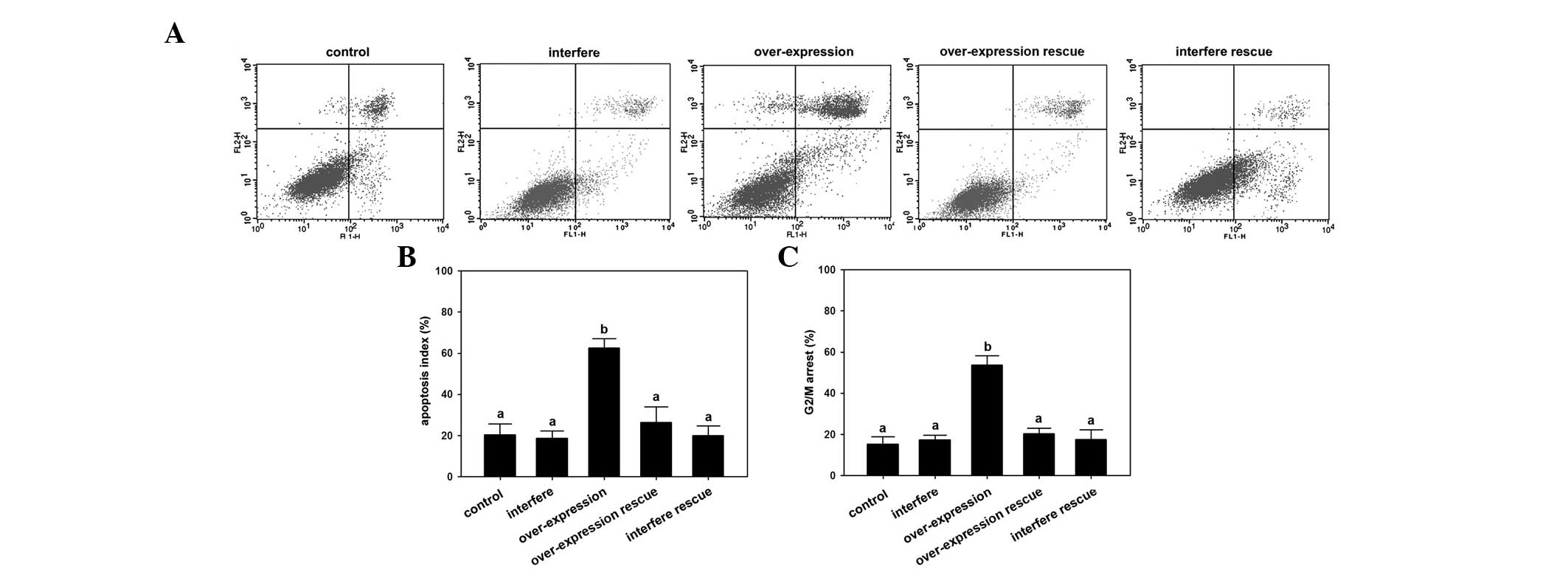
Regulation of osteosarcoma cell line
progression by MacroH2A
To elucidate the effects of MacroH2A on cellular
regulation, the cell cycle progression and cell apoptosis levels
were detected by flow cytometry. The results revealed that the
highest apoptosis rate occurred in the overexpression group
compared with those of the other groups (P<0.05; Fig. 2A and B). Furthermore, the
overexpression group showed a significantly higher G2/M
arrest rate compared with the other groups of U2-OS osteosarcoma
cells (P<0.05; Fig. 2C).
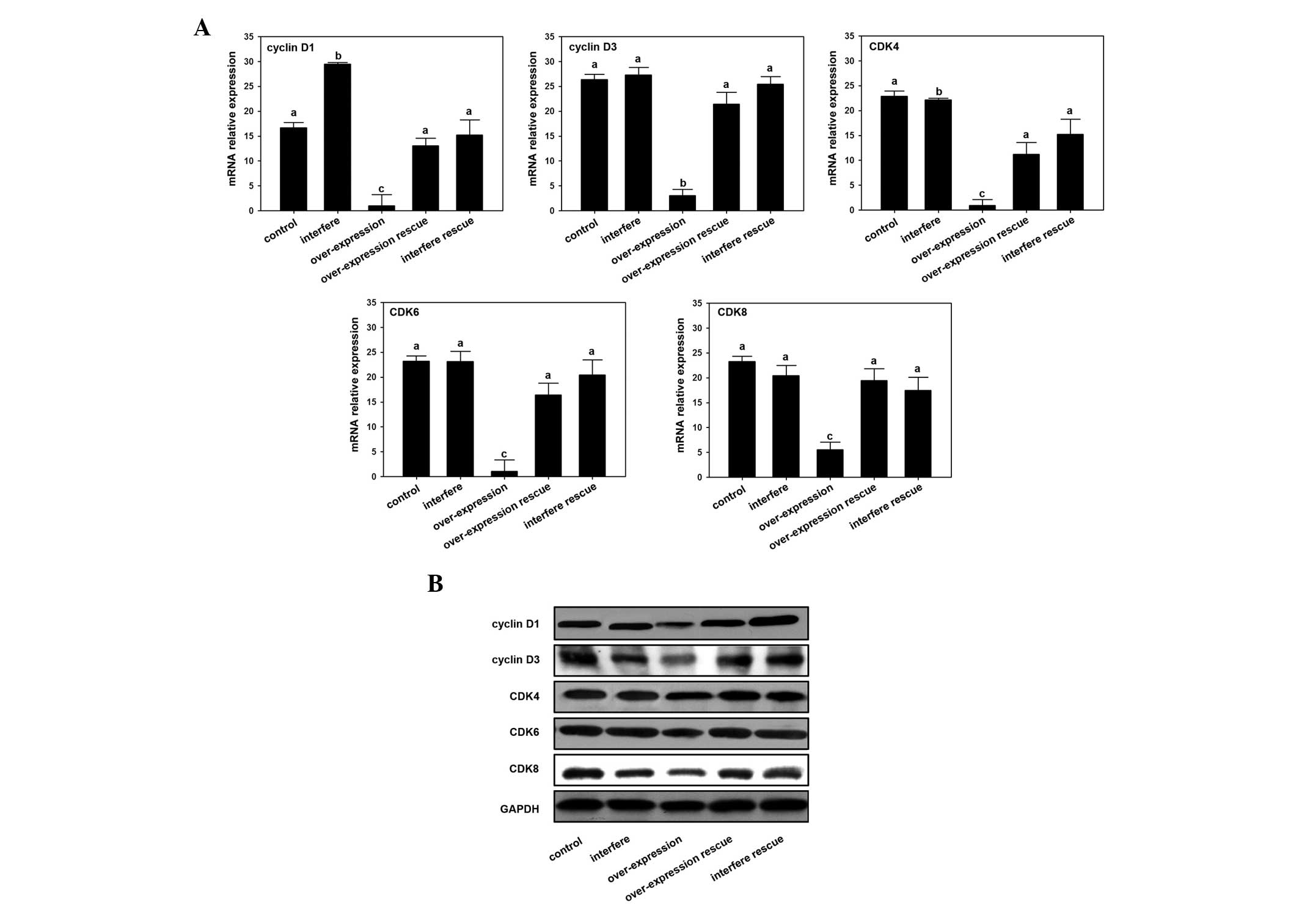
MacroH2A regulates cyclin D and CDK
expression in osteosarcoma cells
Through assaying the expression of cyclin D and CDK
following interference or overexpression of MacroH2A, the mechanism
of MacroH2A-induced osteosarcoma cell line inhibition was
demonstrated. The results showed that expression levels of cyclin
D1 and cyclin D2 were downregulated by the overexpression of
MacroH2A mRNA and protein (Fig.
3). The expression level of cyclin D1 was highest in the
MacroH2A interference group compared with those of the other
groups, while the expression level of cyclin D2 in the MacroH2A
interference group showed no significant difference compared with
the control and two rescue groups (Fig. 3). In addition, the level of
expression of CDK mRNA in the MacroH2A overexpression group was
significantly reduced compared with the other groups. The results
of western blotting also revealed that levels of the CDK4, CDK6 and
CDK8 proteins in the MacroH2A overexpression group were decreased
compared with the other groups (Fig.
3).
Discussion
The present study demonstrated the effect of
MacroH2A on the progression of the U2-OS osteosarcoma cell line
through the interference and/or overexpression of MacroH2A and
subsequent rescue experiments. The results revealed that the
treatments were efficient at artificially regulating the MacroH2A
expression, and that MacroH2A suppressed the progression of U2-OS
cells. Similar results have previously been reported in other types
of cancers, including testicular, ovarian, lung, bladder, cervical,
breast, colorectal and endometrial cancers (25–30).
In these types of cancer, a low expression level of MacroH2A was
found, which may be associated with cancer initiation and
development. A study on melanoma indicated that the MacroH2A
expression levels are strongly negative correlated with tumor
development (13). Dardenne et
al (31) illustrated that the
alternative splicing of MacroH2A also promoted tumor progression in
breast cancer cells.
Malignant tumor cells are characterized by
metastasis and invasion, which is based on cell cycle progression.
It has been established that MacroH2A has a central role in this
process. In addition, MacroH2A maintains genomic stabilization
during the replication of the genome by preventing DNA damage
(9). A lack of MacroH2A may
upregulate CDK8 expression levels, which contributes to cell
apoptosis by blocking premature mitotic entry (13). The results of the current study
indicated that the overexpression of MacroH2A triggers apoptosis in
U2-OS osteosarcoma cells. Thus, the low expression of MacroH2A in
tumor cells seems to be a protective effect against apoptosis.
Kapoor et al (13)
demonstrated that the upregulation of MacroH2A expression
suppressed the progression of melanoma cells by controlling the
expression of the cell cycle factor CDK8. In addition, the current
study demonstrated that following the overexpression of MacroH2A,
the apoptosis rate of U2-OS osteosarcoma cells was increased
significantly, while no such changes were observed in the other
groups. In addition, the arrest stages during these cell cycles
were analyzed. The results revealed that the MacroH2A
overexpression group showed a significantly higher stagnancy at the
G2/M stage, which indicates that the overexpression of
MacroH2A in U2-OS osteosarcoma cells causes G2/M phase
arrest. These results suggest that MacroH2A may trigger apoptosis
by controlling the cell cycle in U2-OS osteosarcoma cells. In
addition, these changes show that MacroH2A expression blocks the
process by which the divided cells pass through the S-phase into
the G2-phase during mitosis. In summary, these results
demonstrate that during mitosis, an increased expression of
MacroH2A arrests osteosarcoma cells at the functional
G2/M stage, which blocks the cell cycle transition into
mitosis, leading to cell apoptosis and preventing the metastasis
and invasion tumor cells. These results provide insight into
possible novel therapeutic strategies for the treatment of
osteosarcoma. MacroH2A-induced inhibition of U2-OS osteosarcoma
cells may be caused by its regulation of cell cycle factors. Thus,
the current study subsequently addressed the regulation of cell
cycle factors by MacroH2A.
To understand the molecular mechanisms behind the
MacroH2A-induced apoptosis of osteosarcoma cells and the cell
cycle, the expression levels of a number of cell cycle factors was
investigated, including cyclin D1, cyclin D3, CDK4, CDK6 and CDK8
gene. The results showed that all five genes had a significantly
lower expression level in the MacroH2A overexpression group
compared with those of the other groups. Cyclin D1 has been
reported to be upregulated in a number of types of cancer, and is
thus regarded as an oncogene (32,33).
Furthermore, cyclin D1 may affect molecular process in cancer
progression, including translocation, amplification and
stabilization of mRNA. Additionally, cyclin D3 is a key factor in
the accumulation and progression of cancer cells (34). In the present study, it was
revealed that by upregulating MacroH2A expression, the expression
of cyclin D1 and cyclin D3 could be suppressed. The CDK family,
including CDK2, CDK3, CDK4, CDK5, CDK6, CDK7 and CDK8, also has a
crucial role in the cell cycle (35). Among these family members, CDK4,
CDK6 and CDK8 are the most critical in the cell cycle, controlling
DNA synthesis at the beginning of the cell cycle and progressing
cells from the G1-phase into the S-phase (36). Several types of cancer have been
determined to have a higher expression level of CDKs. This
attribute may be a potential target in the cancer therapeutic
strategy. In the current study, it was demonstrated that MacroH2A
downregulates the expression of CDK4, CDK6 and CDK8 in U2-OS
osteosarcoma cells. Hence, we propose that by upregulating the
levels of MacroH2A in osteosarcoma cells, the expression of cyclin
D and CDK may be suppressed, hence inhibiting the progression of
the osteosarcoma cells.
In conclusion, the results of the present study
indicate that the overexpression of MacroH2A induces apoptosis in
U2-OS osteosarcoma cells and arrests the cells at the
G2/M-phase boundary of the cell cycle. In addition, it
was determined that a high expression level of MacroH2A suppresses
cell cycle progression. The underlying molecular mechanism involves
the downregulation of cyclin D1, cyclin D3, CDK4, CDK6 and CDK8
genes by MacroH2A overexpression. Therefore, the expression of
MacroH2A maintains a regular cell cycle by controlling the cyclin D
and CDKs genes. This study has elucidated the association between
chromatin structure modification and the cell cycle in osteosarcoma
cells, which may offer a novel insight into potential treatment
strategies for osteosarcoma.
References
|
1
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kulaeva OI, Gaykalova DA and Studitsky VM:
Transcription through chromatin by RNA polymerase II: histone
displacement and exchange. Mutat Res. 618:116–129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neely KE and Workman JL: The complexity of
chromatin remodeling and its links to cancer. Biochim Biophys Acta.
1603:19–29. 2002.PubMed/NCBI
|
|
5
|
Hake S, Xiao A and Allis C: Linking the
epigenetic ‘language’ of covalent histone modifications to cancer.
Br J Cancer. 90:761–769. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peterson CL and Laniel MA: Histones and
histone modifications. Curr Biol. 14:R546–R551. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Redon C, Pilch D, Rogakou E, et al:
Histone H2a variants H2AX and H2AZ. Curr Opin Genet Dev.
12:162–169. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pehrson JR and Fried VA: MacroH2A, a core
histone containing a large nonhistone region. Science.
257:1398–1400. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kustatscher G, Hothorn M, Pugieux C,
Scheffzek K and Ladurner AG: Splicing regulates NAD metabolite
binding to histone macroH2A. Nat Struct Mol Biol. 12:624–625. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brooks WH: X chromosome inactivation and
autoimmunity. Clin Rev Allergy Immunol. 39:20–29. 2010. View Article : Google Scholar
|
|
12
|
Chadwick BP and Willard HF: Histone H2A
variants and the inactive X chromosome: identification of a second
macroH2A variant. Hum Mol Genet. 10:1101–1113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kapoor A, Goldberg MS, Cumberland LK, et
al: The histone variant macroH2A suppresses melanoma progression
through regulation of CDK8. Nature. 468:1105–1109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rickert P, Seghezzi W, Shanahan F, Cho H
and Lees E: Cyclin C/CDK8 is a novel CTD kinase associated with RNA
polymerase II. Oncogene. 12:2631–2640. 1996.PubMed/NCBI
|
|
15
|
Donner AJ, Szostek S, Hoover JM and
Espinosa JM: CDK8 is a stimulus-specific positive coregulator of
p53 target genes. Mol Cell. 27:121–133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sánchez I and Dynlacht BD: New insights
into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol.
16:311–321. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Satyanarayana A and Kaldis P: Mammalian
cell-cycle regulation: several Cdks, numerous cyclins and diverse
compensatory mechanisms. Oncogene. 28:2925–2939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fisher RP: CDKs and cyclins in
transition(s). Curr Opin Genet Dev. 7:32–38. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andreassi MG: DNA damage, vascular
senescence and atherosclerosis. J Mol Med. 86:1033–1043. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morgan DO: Cyclin-dependent kinases:
engines, clocks, and microprocessors. Annu Rev Cell Dev Biol.
13:261–291. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tyson JJ, Csikasz-Nagy A and Novak B: The
dynamics of cell cycle regulation. Bioessays. 24:1095–1109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sherr CJ and Roberts JM: CDK inhibitors:
positive and negative regulators of G1-phase progression. Genes
Dev. 13:1501–1512. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taylor WR and Stark GR: Regulation of the
G2/M transition by p53. Oncogene. 20:1803–1815. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sporn JC, Kustatscher G, Hothorn T, et al:
Histone macroH2A isoforms predict the risk of lung cancer
recurrence. Oncogene. 28:3423–3428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Novikov L, Park JW, Chen H, et al:
QKI-mediated alternative splicing of the histone variant MacroH2A1
regulates cancer cell proliferation. Mol Cell Biol. 31:4244–4255.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haugstetter A, Loddenkemper C, Lenze D, et
al: Cellular senescence predicts treatment outcome in metastasised
colorectal cancer. Br J Cancer. 103:505–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buschbeck M and Di Croce L: Approaching
the molecular and physiological function of macroH2A variants.
Epigenetics. 5:118–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang R and Adams PD: Heterochromatin and
its relationship to cell senescence and cancer therapy. Cell Cycle.
6:784–789. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Collado M, Gil J, Efeyan A, et al: Tumour
biology: senescence in premalignant tumours. Nature. 436:642. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dardenne E, Pierredon S, Driouch K, et al:
Splicing switch of an epigenetic regulator by RNA helicases
promotes tumor-cell invasiveness. Nat Struct Mol Biol.
19:1139–1146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shtutman M, Zhurinsky J, Simcha I, et al:
The Cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway.
Proc Natl Acad Sci USA. 96:5522–5527. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu M, Wang C, Li Z, Sakamaki T and Pestell
RG: Minireview: Cyclin D1: normal and abnormal functions.
Endocrinology. 145:5439–5447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hunter T and Pines J: Cyclins and cancer
II: Cyclin D and CDK inhibitors come of age. Cell. 79:573–582.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gray N, Détivaud L, Doerig C and Meijer L:
ATP-site directed inhibitors of Cyclin-dependent kinases. Curr Med
Chem. 6:859–876. 1999.PubMed/NCBI
|
|
36
|
Lapenna S and Giordano A: Cell cycle
kinases as therapeutic targets for cancer. Nat Rev Drug Discov.
8:547–566. 2009. View
Article : Google Scholar : PubMed/NCBI
|