Introduction
Osteosarcomas (OS) are among the most frequently
occurring secondary malignancies in childhood cancer (1). OS most often originates in the
metaphyses of long bones in adolescents and young adults (2). During the past 30 years, an optimal
treatment strategy for OS has been developed, which consists of
multi-agent chemotherapy and aggressive surgical resection of all
sites of disease involvement (3).
However, ~80% of patients with localized OS develop metastatic
disease following surgical resection (4). Patients with primary metastatic OS
are a heterogeneous group, and a five-year event-free survival rate
of up to 75% is reported for patients presenting with unilateral
lung metastases (5). Furthermore
<20% of patients with high-grade osteosarcoma are clinically
diagnosed with metastatic disease at the initial diagnosis, and
long-term survival rates of patients with metastatic OS range
between 10–40% (6,7). Therefore, due to the high rate of
systemic spread, complete recovery following surgical treatment
alone is rare (8).
Recently, microarray analysis (9) has been widely used in screening for
differentially expressed genes (DEGs), in order to identify
potential genes that may be investigated for the treatment of
various human diseases (10,11).
Previous studies have focused on the microarray analysis of OS. A
previous study used genome-wide cDNA microarrays to investigate the
transcriptome profile of the human Saos-2 and U-2 OS cell lines.
Genes associated with focal adhesion were shown to be
differentially regulated in the two cell lines (12). Luo et al (13), identified a total of 1,836 DEGs in
OS. In addition, a previous study detected a total of 35 aberrantly
expressed genes in three cell lines of OS (IOR/OS9, IOR/OS10, and
IOR/OS15), eight were upregulated and 27 were downregulated, as
compared with the expression levels in osteoblasts (14). Another DEG analysis yielded 75
upregulated genes and 97 downregulated genes in osteoblastic, as
compared with non-osteoblastic OS samples (15). Based on comparative genomic
hybridization, chromosomal imbalances have also been shown to exist
in OS (16). Therefore, DNA
microarray analysis may be considered an effective approach for the
identification of genes associated with OS, and may provide
potential treatment strategies for OS.
The present study used a microarray analysis to
identify the DEGs between metastatic and non-metastatic OS samples,
using the BioConductor package in R language. Functional annotation
and pathway enrichment analyses of the DEGs were conducted using
the MAS system and the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway, respectively. In addition, the co-expression of
DEGs was analyzed using Pearson’s correlation coefficient.
Protein-protein interaction (PPI) networks of the co-expressed
genes were constructed using Cytoscape and the submodules of the
network were selected for by MCODE. The results of the present
study may provide increased understanding of the underlying
molecular mechanisms of metastatic OS, and identify novel
therapeutic targets for its diagnosis and treatment.
Materials and methods
Samples
The gene expression profile GSE9508 (17) was downloaded from the public
functional genomics database Gene Expression Omnibus (GEO,
http://www.ncbi.nlm.nih.gov/geo/). A
total of 23 specimens, which included 11 human metastatic OS
samples, seven non-metastatic OS samples and five normal samples,
were obtained. The GSE9508 expression profile is based on the
GPL6076 Whole Human Genome Oligo Microarray G4112A platform
(Agilent Technologies, Inc., Santa Clara, CA, USA).
Data pretreatment and analysis of
DEGs
Using the BioConductor package (http://bioconductor.org/) and Gene Spring software
(Silicon Genetics, Redwood City, CA, USA) in R language, the
probe-level data were converted into expression values. The
expression values of all of the probes in each sample were reduced
to a single value by determining the average expression value.
Missing data were imputed and quantile normalization for complete
data was performed, as previously described (18), using the preprocessCore package in
R language (19). When numerous
probes were mapped to one gene, the mid-value of the data was
defined as the expression level of the gene. However, when numerous
genes were mapped by one probe, this probe was considered to lack
specificity, and was removed from the analysis.
The normal samples were classed as the controls and
the normalized data were analyzed using a t-test (20,21)
implemented in LIMMA package (22). The differential genes in the
metastatic and non-metastatic OS samples were considered to be
those with a P-value <0.05. The differential genes that were
uniquely expressed in the metastatic OS samples, as compared with
the non-metastatic OS samples, were considered to be DEGs.
Functional enrichment and pathway
enrichment analyses
Gene Ontology (GO) (23) and KEGG pathway (24) enrichment analyses of DEGs were
conducted using the Molecule Annotation System 3.0 (MAS, http://bioinfo.capitalbio.com/mas3/). GO
functional enrichment analysis encompasses three categories:
Biological process, cellular component and molecular function, with
a cutoff criteria of gene count >2 and P<0.01. The KEGG
pathway analysis was performed using the same method and parameter
settings (count >2 and P<0.01), to identify the pathways that
the DEGs are involved in.
Co-expression analysis of the DEGs
The co-expression of DEGs in the metastatic OS
samples was also analyzed. Based on the previously obtained DEG,
the co-expression of numerous pairs of DEGs in metastatic OS was
identified by Pearson’s correlation coefficient (PCC) (5,25,26).
A pair of DEGs with PCC ≥0.98 and P<0.01, were identified as
significantly co-expressed genes, under specific conditions.
Construction of a co-expression network
and identification of functional modules
The human PPI network was downloaded from the Human
Protein Reference Database (http://www.hprd.org/) (27). The specific interactional patterns
of DEGs in metastatic OS were extracted according to the human PPI
network, and the dysfunctional protein networks associated with
metastatic OS were constructed by integrating the specific
interactional patterns of DEGs and the gene co-expression
information. The genes which belonged to the top 10% (nodal points
≥ 6) were screened for further analysis.
The topological characteristics of the co-expression
network were examined using Cytoscape MCODE (28), which is a clustering algorithm used
for directed or undirected graphs. The MCODE algorithm includes the
following steps: Vertex-weighting, complex prediction and optional
post-processing (29). The
weighting scheme defined a measure of local density for a vertex’s
neighborhood. Complexes with a high vertex weight were then used as
seed and the complex neighbor vertices were checked to determine
whether they were a part of the complex. Post-processing was
performed with a minimum degree of 3. The submodules were then
obtained from the regulatory network.
Results
Screening DEGs in metastatic OS
Pretreatment and standardization of the data from
all of the available samples was conducted, and the DEGs of
non-metastatic and metastatic OS samples were compared with the
normal samples. A total of 965 characteristic genes were then
identified as DEGs in metastatic, as compared with non-metastatic,
OS samples (P<0.05).
GO clustering and KEGG pathway analysis
of the DEGs
To explore the function of the DEGs in metastatic
OS, the DEGs were mapped to the GO database using the MAS system.
The results were analyzed based on three categories: Biological
process, cellular component and molecular function. The DEGs
clustered according to biological process are shown in Table I, and include regulation of
transcription, DNA-dependent (P=3.11E-82), signal transduction
(P=2.14E-45) and cell adhesion (P=3.65E-32). These results indicate
that the majority of DEGs were associated with transcription,
signal transduction and cell adhesion. The DEGs clustered according
to cellular component were shown to predominantly be associated
with the nucleus (P=9.23E-250), cytoplasm (P=2.59E-177) and
integral to the membrane (P=4.41E-126) (Table II), thus suggesting that the
cellular components in which the DEGs were expressed, was
relatively comprehensive. The clustering of DEGs according to
molecular function is shown in Table
III, and included binding activities, such as protein binding
(P=3.43E-232), zinc ion binding (P=2.23E-94) and nucleotide binding
(P=2.03E-88).
 | Table IGene ontology (GO) functional
enrichment of differentially expressed genes according to
biological process, in metastatic osteosarcoma. |
Table I
Gene ontology (GO) functional
enrichment of differentially expressed genes according to
biological process, in metastatic osteosarcoma.
| GO accession
number | Count | Adjusted
P-value | Description |
|---|
| GO:0006355 | 100 | 3.11E-82 | Regulation of
transcription, DNA-dependent |
| GO:0006350 | 81 | 3.09E-56 | Transcription |
| GO:0007165 | 81 | 2.14E-45 | Signal
transduction |
| GO:0055114 | 37 | 7.82E-36 | Oxidation
reduction |
| GO:0007155 | 37 | 3.65E-32 | Cell adhesion |
| GO:0007275 | 52 | 3.34E-28 | Development |
| GO:0015031 | 25 | 6.24E-18 | Protein
transport |
| GO:0044419 | 24 | 6.51E-18 | Protein amino acid
phosphorylation |
| GO:0006468 | 18 | 2.36E-17 | Interspecies
interaction between organisms |
| GO:0000122 | 15 | 1.51E-16 | Negative regulation
of transcription from RNA polymerase II promoter |
 | Table IIGene ontology (GO) functional
enrichment of differentially expressed genes according to cellular
component, in metastatic osteosarcoma. |
Table II
Gene ontology (GO) functional
enrichment of differentially expressed genes according to cellular
component, in metastatic osteosarcoma.
| GO accession
number | Count | Adjusted
P-value | Description |
|---|
| GO:0005634 | 265 | 9.23E-250 | Nucleus |
| GO:0005737 | 236 | 2.59E-177 | Cytoplasm |
| GO:0016021 | 174 | 4.41E-126 | Integral to
membrane |
| GO:0016020 | 191 | 1.56E-123 | Membrane |
| GO:0005886 | 125 | 1.84E-92 | Plasma
membrane |
| GO:0005576 | 90 | 1.24E-74 | Extracellular
region |
| GO:0005829 | 57 | 4.75E-55 | Cytosol |
| GO:0005783 | 50 | 1.76E-46 | Endoplasmic
reticulum |
| GO:0005739 | 52 | 3.91E-46 | Mitochondrion |
| GO:0005794 | 43 | 1.16E-38 | Golgi
apparatus |
 | Table IIIGene ontology (GO) functional
enrichment of differentially expressed genes according to molecular
function, in metastatic ostersarcoma. |
Table III
Gene ontology (GO) functional
enrichment of differentially expressed genes according to molecular
function, in metastatic ostersarcoma.
| GO accession
number | Count | Adjusted
P-value | Description |
|---|
| GO:0005515 | 283 | 3.43E-232 | Protein
binding |
| GO:0008270 | 109 | 2.23E-94 | Zinc ion
binding |
| GO:0000166 | 100 | 2.03E-88 | Nucleotide
binding |
| GO:0046872 | 112 | 4.23E-72 | Metal ion
binding |
| GO:0005524 | 72 | 2.55E-66 | ATP binding |
| GO:0003677 | 70 | 2.03E-47 | DNA binding |
| GO:0003700 | 48 | 5.97E-43 | Transcription
factor activity |
| GO:0016740 | 57 | 1.43E-41 | Transferase
activity |
| GO:0005509 | 44 | 3.33E-38 | Calcium ion
binding |
| GO:0003723 | 37 | 1.10E-33 | RNA binding |
To further explore the detailed changes to
biological pathways in metastatic OS, a KEGG pathway enrichment
analysis was conducted to select the altered pathways. The top 10
pathways were identified and are listed in Table IV. The most significantly enriched
pathways were associated with the ribosome (P=1.03E-07), axon
guidance (P=1.37E-07), cytokine-cytokine receptor interaction
(P=3.93E-07) and focal adhesion (P=8.58E-06), which was also
enriched in the DEGs clustered according to biological process.
 | Table IVKyoto Encyclopaedia of Genes and
Genomes (KEGG) pathways of differentially expressed genes in
metastatic osteosarcoma. |
Table IV
Kyoto Encyclopaedia of Genes and
Genomes (KEGG) pathways of differentially expressed genes in
metastatic osteosarcoma.
| KEGG | Count | Adjusted
P-value | Description |
|---|
| KEGG_PATHWAY | 10 | 3.36E-08 | Pathogenic
Escherichia coli infection-EHEC |
| KEGG_PATHWAY | 10 | 3.36E-08 | Pathogenic
Escherichia coli infection-EPEC |
| KEGG_PATHWAY | 12 | 1.03E-07 | Ribosome |
| KEGG_PATHWAY | 13 | 1.37E-07 | Axon guidance |
| KEGG_PATHWAY | 17 | 3.93E-07 | Cytokine-cytokine
receptor interaction |
| KEGG_PATHWAY | 8 | 8.28E-06 | Adipocytokine
signaling pathway |
| KEGG_PATHWAY | 13 | 8.58E-06 | Focal adhesion |
| KEGG_PATHWAY | 10 | 2.34E-05 | Tight junction |
| KEGG_PATHWAY | 12 | 5.07E-05 | Regulation of actin
cytoskeleton |
| KEGG_PATHWAY | 13 | 5.28E-05 | Neuroactive
ligand-receptor interaction |
PPI network and submodules of DEGs
A gene co-expression analysis was performed in
metastatic OS, based on the PCC values. Among the 965 DEGs, a total
of 182 gene pairs were identified as co-expressed genes pairs with
PCC>0.98 and P<0.01.
In addition, the interaction patterns of DEGs in
metastatic OS were obtained by mapping them to the human PPI
network. The dysfunctional network associated with metastatic OS
was then constructed, by integrating the interaction data of the
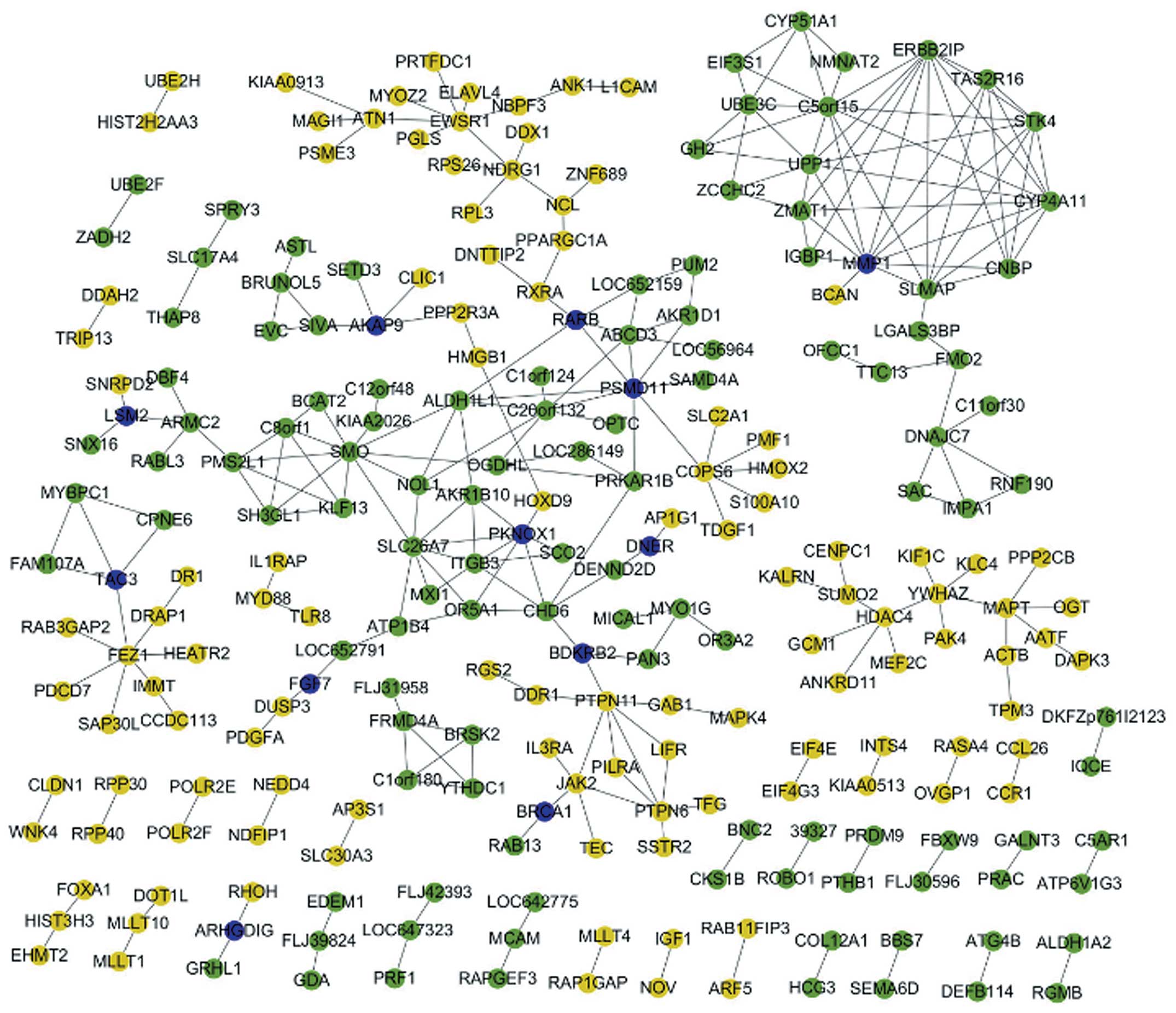
DEGs and the associations of co-expressed genes (Fig. 1). This network included the
interaction of DEGs in metastatic OS samples. In the network, hub
nodes included matrix metalloproteinase 1 (MMP1), smoothened (SMO),
ewing sarcoma breakpoint region 1 (EWSR1), unnamed protein product
1, sarcolemmal membrane-associated protein (SLMAP), fasciculation
and elongation protein ζ-1 (FEZ1).
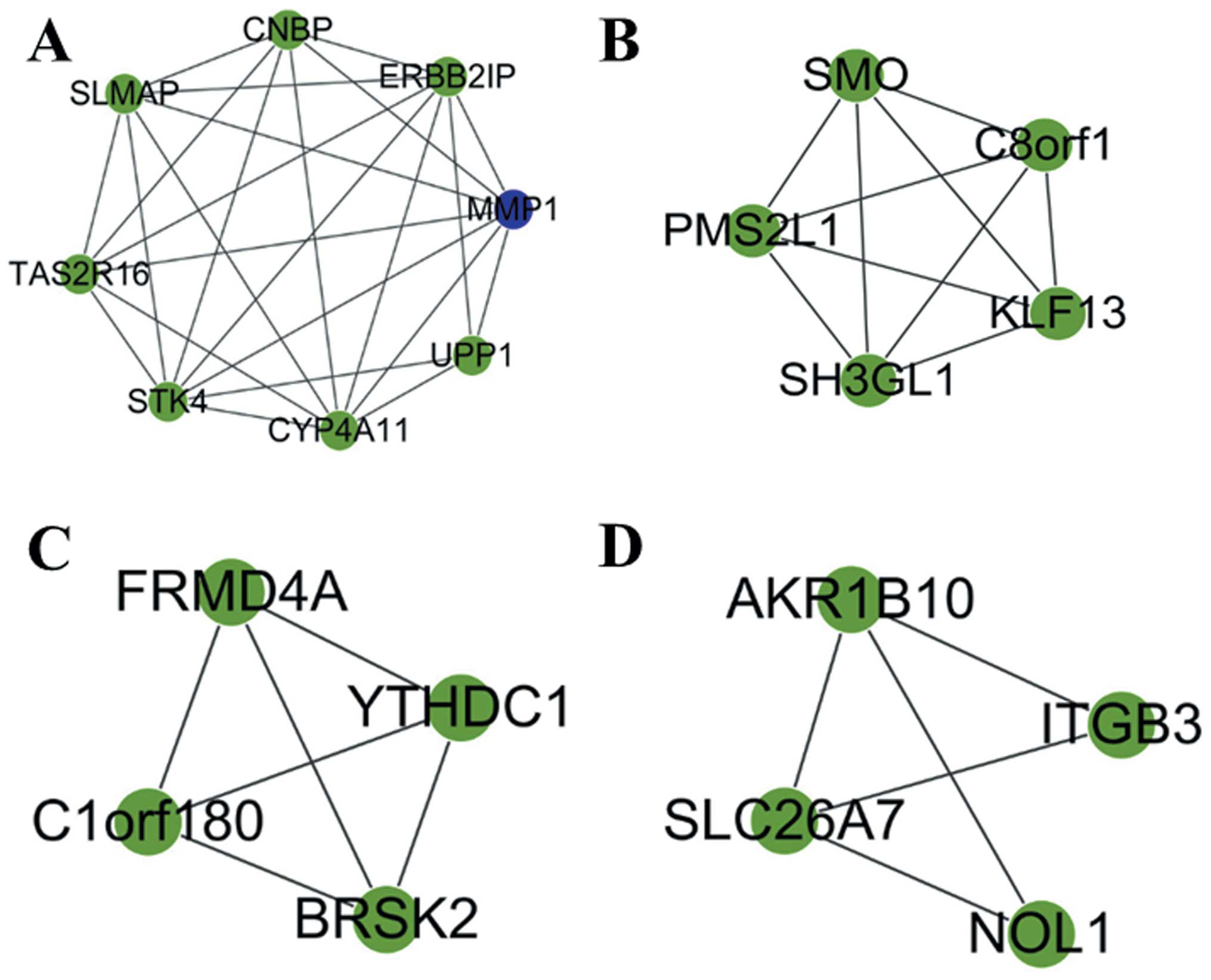
Four functional submodules, which were dysfunctional
in the process of metastatic OS, were then selected using Cytoscape
(Fig. 2). MMP1, SMO, brain
selective kinase 2 (BRSK2), aldo-keto reductase family 1 member B10
(AKRIB10), as well as other genes were included within these four
submodules.
Discussion
OS is one of the most frequent secondary cancers
that occurs following childhood malignancy, particularly metastatic
OS. Due to the low recovery rates and the high incidence of
metastatic OS, it is necessary to explore the molecular mechanisms
of OS, in order to identify an effective prevention and treatment
strategy.
In the present study, 965 DEGs between metastatic
and non-metastatic OS samples were identified by comparing their
gene expression profiles with those of normal samples. The DEGs
were then subjected to functional annotation, based on three
categories: Biological process, cellular component and molecular
function. A KEGG pathway analysis was also performed. The ribosome
and axon guidance were identified as the most significantly altered
pathways. Biogenesis and translational control are essential
cellular processes associated with ribosomes, which are governed at
numerous levels. Numerous tumor suppressors and proto-oncogenes
have previously been shown to either affect the formation of the
mature ribosome or regulate the activity of translation factors
(30). Therefore, the DEGs
identified in the present study may have important effects on
metastatic OS, through the ribosome pathway.
Axons are guided along specific pathways by
attractive and repulsive cues in the extracellular environment
(31). The results of the present
study suggest that axon guidance may be an important pathway in
metastatic OS. Furthermore, the cytokine-cytokine receptor
interaction pathway has previously been shown to be present in
astrocytomas, and the autoregulation of interleukin-1 and
cytokine-receptor interactions was shown to exist in primary human
astrocytoma cells (32). In the
present study, focal adhesion was identified in the GO biological
process analysis of DEGs and the KEGG pathway. Focal adhesion was
previously identified as a prominent determinant in cancer
initiation, progression and metastasis (33–36).
The present study constructed a PPI network of the
co-expressed genes, which was then screened for submodules. Genes,
such as MMP1, SMO, EWSR1 and FEZ1, were selected as the hub nodes,
suggesting that these genes are associated with the crucial
function of the whole network. MMP1 not only existed in the PPI
network, but was also identified as one of the co-expressed genes
in metastatic OS. The functions of MMP1 have been reported in
previous studies. It has been identified as a candidate marker that
may be useful for identification of breast lesions, which can
develop into cancer (37). MMP1
has also been shown to possess important functions in OS primary
tumors and OS metastasis to the lung, which is the predominant site
of OS metastasis (38). There has
also been a correlation reported between MMP expression and the
oncological outcome of OS patients, thus suggesting the prognostic
significance of MMPs in OS (39).
Furthermore, the regulation of MMP gene expression has vital roles
in tumor invasion (40). Hence,
MMP1 may have a vital role in metastatic OS.
SMO is a distant relative of the G protein-coupled
receptors. It has been shown to mediate the Hedgehog signaling
pathway during embryonic development and may initiate ligand
independent pathway activation in tumorigenesis (41). The inactivation of SMO has been
suggested as a potential target for the treatment of patients with
OS (42). EWSR1 represents one of
the most commonly involved genes in sarcoma translocations
(43), and in a study regarding a
fusion transcript in osteogenic sarcoma, it was shown to be closely
associated with the molecular mechanisms of small cell osteogenic
and Ewing sarcomas (44). EWSR1
gene rearrangements have also been identified in soft tissue
myoepithelial tumors (45). FEZ1
is a tumor suppressor gene that maps to chromosome 8p22, which is a
frequently deleted chromosomal region in numerous human
malignancies (46). A previous
study showed that it was associated with p53 (47), it may be suggested that the
abnormal expression of FEZ1 may inhibit the normal function of p53,
resulting in the occurrence and metastasis of cancer. Furthermore,
BRSK2 and AKRIB10 were included in the selected submodules. BRSK2
has been identified as a member of the AMP-activated protein kinase
related kinases (48); however,
its function in metastatic OS was previously unclear. The results
of the present study showed that the molecular function of BRSK2
was associated with nucleotide binding and protein amino acid
phosphorylation. Furthermore, research regarding AKRIB10 is
limited; however, the present study demonstrated that it was mainly
enriched within the cytoplasm, and was involved in the bisphenol A
degradation and bile acid biosynthesis pathways. These results may
provide novel information regarding the potential mechanisms of
metastatic OS.
In conclusion, 965 characteristic DEGs were
identified in metastatic, as compared with non-metastatic, OS
samples by microarray analysis. Functional annotation and pathway
enrichment analyses of the DEGs were also performed, and a total of
182 co-expressed gene pairs were identified in the metastatic OS
samples. A regulatory network of co-expressed genes and DEGs was
constructed, and the submodules were shown to contain BRSK2,
AKRIB10 and other genes. Certain hub nodes identified in the
present study, such as MMP1, SMO, EWSR1 and FEZ1, may have the
potential to become targets for the diagnosis and treatment of
metastatic OS. In addition, BRSK2 and AKRIB10 may have important
functions in metastatic OS. The present study provided novel
information that may be beneficial for further research regarding
the therapy of metastatic OS. However, these results require
further confirmation.
Acknowledgements
The present study was supported by the Liaoning
Province Science and Technology Plan Project (grant no. 2012408002)
and the National Natural Science Foundation (grant no.
81271538).
Abbreviations:
|
OS
|
osteosarcoma
|
|
DEGs
|
differentially expressed genes
|
|
CGH
|
comparative genomic hybridization
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
|
GO
|
Gene Ontology
|
|
MMP 1
|
matrix metalloproteinase 1
|
|
SMO
|
smoothened
|
|
EWSR1
|
ewing sarcoma breakpoint region 1
|
|
SLMAP
|
sarcolemmal membrane-associated
protein
|
|
FEZ1
|
fasciculation and elongation protein
ζ-1
|
|
BRSK 2
|
brain selective kinase 2
|
|
AKRIB 10
|
aldo-keto reductase family 1 member B
10
|
References
|
1
|
Bielack SS, Kempf-Bielack B, Heise U,
Schwenzer D and Winkler K: Combined modality treatment for
osteosarcoma occurring as a second malignant disease. Cooperative
German Austrian-Swiss Osteosarcoma Study Group. J Clin Oncol.
17:1164. 1999.
|
|
2
|
Bielack SS, Kempf-Bielack B, Delling G, et
al: Prognostic factors in high-grade osteosarcoma of the
extremities or trunk: an analysis of 1,702 patients treated on
neoadjuvant cooperative osteosarcoma study group protocols. J Clin
Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: where do we go from here? Pediatric Drugs.
10:315–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kager L, Zoubek A, Pötschger U, et al:
Cooperative German-Austrian-Swiss Osteosarcoma Study Group: Primary
metastatic osteosarcoma: presentation and outcome of patients
treated on neoadjuvant Cooperative Osteosarcoma Study Group
protocols. J Clin Oncol. 21:2011–2018. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meyer WH, Pratt CB, Poquette CA, et al:
Carboplatin/ifosfamide window therapy for osteosarcoma: results of
the St Jude Children’s Research Hospital OS-91 trial. J Clin Oncol.
19:171–182. 2001.PubMed/NCBI
|
|
7
|
Ferguson WS, Harris MB, Goorin AM, et al:
Presurgical window of carboplatin and surgery and multidrug
chemotherapy for the treatment of newly diagnosed metastatic or
unresectable osteosarcoma: pediatric oncology group trial. J
Pediatr Hematol Oncol. 23:340–348. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Link MP, Goorin AM, Miser AW, et al: The
effect of adjuvant chemotherapy on relapse-free survival in
patients with osteosarcoma of the extremity. N Engl J Med.
314:1600–1606. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hegde P, Qi R, Abernathy K, et al: A
concise guide to cDNA microarray analysis. Biotechniques.
29:548–550. 2000.PubMed/NCBI
|
|
10
|
DeRisi J, Penland L, Brown PO, et al: Use
of a cDNA microarray to analyse gene expression patterns in human
cancer. Nat Genet. 14:457–460. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perou CM, Jeffrey SS, Van De Rijn M, et
al: Distinctive gene expression patterns in human mammary
epithelial cells and breast cancers. Proc Nat Acad Sci USA.
96:9212–9217. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trougakos IP, Chondrogianni N, Amarantos
I, et al: Genome-wide transcriptome profile of the human
osteosarcoma SA OS and U-2 OS cell lines. Cancer Genet Cytogenet.
196:109–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo Y, Deng Z and Chen J: Pivotal
regulatory network and genes in osteosarcoma. 9:569–575. 2013.
|
|
14
|
Wolf M, El-Rifai We, Tarkkanen M, et al:
Novel findings in gene expression detected in human osteosarcoma by
cDNA microarray. Cancer genetics and cytogenetics. 123:128–132.
2000. View Article : Google Scholar
|
|
15
|
Kubista B, Klinglmueller F, Bilban M, et
al: Microarray analysis identifies distinct gene expression
profiles associated with histological subtype in human
osteosarcoma. Int Orthop. 35:401–411. 2011. View Article : Google Scholar :
|
|
16
|
Squire JA, Pei J, Marrano P, et al:
High-resolution mapping of amplifications and deletions in
pediatric osteosarcoma by use of CGH analysis of cDNA microarrays.
Genes, Chromosomes Cancer. 38:215–225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Endo-Munoz L, Cumming A, Rickwood D, et
al: Loss of osteoclasts contributes to development of osteosarcoma
pulmonary metastases. Cancer Res. 70:7063–7072. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bolstad BM; preprocessCore. A collection
of pre-processing functions. R package version:1.0. 2013
|
|
19
|
Bolstad BM, Irizarry RA, Astrand M and
Spreed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kennedy CL, Najdovska M, Tye H, et al:
Differential role of MyD88 and Mal/TIRAP in TLR2-mediated gastric
tumourigenesis. Oncogene. 2013.PubMed/NCBI
|
|
21
|
Zhang S and Cao J: A close examination of
double filtering with fold change and T test in microarray
analysis. BMC Bioinformatics. 10:4022009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smyth GK; Limma. Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. 5. Springer; New York, NY: pp. 397–420.
2005, View Article : Google Scholar
|
|
23
|
Ashburner M, Ball CA, Blake JA, et al:
Gene ontology: tool for the unification of biology. the gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
25
|
Usadel B, Obayashi T, Mutwil M, et al:
Co-expression tools for plant biology: opportunities for hypothesis
generation and caveats. Plant Cell & Environ. 32:1633–1651.
2009. View Article : Google Scholar
|
|
26
|
Barretina J, Caponigro G, Stransky N, et
al: The cancer cell line encyclopedia enables predictive modelling
of anticancer drug sensitivity. Nature. 483:603–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Keshava Prasad TS, Goel R, Kandasamy K, et
al: Human protein reference database-2009 update. Nucleic Acids
Res. 37:D767–D772. 2009. View Article : Google Scholar
|
|
28
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bidkhori G, Narimani Z, Ashtiani SH,
Moeini A, Nowzari-Dalini A and Masoudi-Nejad A: Reconstruction of
an integrated genome-scale co-expression network reveals key
modules involved in lung adenocarcinoma. PLoS One. 8:e675522013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ruggero D and Pandolfi PP: Does the
ribosome translate cancer? Nat Rev Cancer. 3:179–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dickson BJ: Molecular mechanisms of axon
guidance. Science. 298:1959–1964. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ilyin SE, González-Gómez I, Romanovicht A,
Gayle D, Gilles FH and Plata-Salamán CR: Autoregulation of the
interleukin-1 system and cytokine-cytokine interactions in primary
human astrocytoma cells. Brain Res Bull. 51:29–34. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo M and Guan JL: Focal adhesion kinase:
A prominent determinant in breast cancer initiation, progression
and metastasis. Cancer Lett. 289:127–139. 2010. View Article : Google Scholar :
|
|
34
|
Housa D, Housova J, Vernerova Z and
Haluzik M: Adipocytokines and cancer. Physiol Res. 55:2332006.
|
|
35
|
Lehmann BD, Bauer JA, Chen X, et al:
Identification of human triple-negative breast cancer subtypes and
preclinical models for selection of targeted therapies. J Clin
Invest. 121:2750–2767. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY
and Li DG: Changes in microRNAs associated with hepatic stellate
cell activation status identify signaling pathways. FEBS Journal.
276:5163–5176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Poola I, DeWitty RL, Marshalleck JJ,
Bhatnagar R, Abraham J and Leffall LD: Identification of MMP-1 as a
putative breast cancer predictive marker by global gene expression
analysis. Nat Med. 11:481–483. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Husmann K, Arlt MJ, Muff R, et al: Matrix
metalloproteinase 1 promotes tumor formation and lung metastasis in
an intratibial injection osteosarcoma mouse model. Biochim Biophys
Acta. 1832:347–354. 2013. View Article : Google Scholar
|
|
39
|
Uchibori M, Nishida Y, Nagasaka T, Yamada
Y, Nakanishi K and Ishiguro N: Increased expression of
membrane-type matrix metalloproteinase-1 is correlated with poor
prognosis in patients with osteosarcoma. Int J Oncol. 28:33–42.
2006.
|
|
40
|
Westermarck J and Kähäri VM: Regulation of
matrix metalloproteinase expression in tumor invasion. FASEB J.
13:781–792. 1999.PubMed/NCBI
|
|
41
|
Chen JK, Taipale J, Young KE, Maiti T and
Beachy PA: Small molecule modulation of smoothened activity. Pro
Nat Acad Sci. 99:14071–14076. 2002. View Article : Google Scholar
|
|
42
|
Hirotsu M, Setoguchi T, Sasaki H, et al:
Smoothened as a new therapeutic target for human osteosarcoma. Mol
Cancer. 9:52010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Romeo S and Dei Tos AP: Soft tissue tumors
associated with EWSR1 translocation. Virchows Arc. 456:219–234.
2010. View Article : Google Scholar
|
|
44
|
Debelenko LV, McGregor LM, Shivakumar BR,
Dorfman HD and Raimondi SC: A novel EWSR1-CREB311 fusion transcript
in a case of small cell osteosarcoma. Genes Chromosomes Cancer.
50:1054–1062. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Antonescu CR, Zhang L, Chang NE, et al:
EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A
molecular analysis of sixty-six cases, including soft tissue, bone
and visceral lesions, showing common involvement of the EWSR1 gene.
Genes Chromosomes Cancer. 49:1114–1124. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vecchione A, Ishii H, Baldassarre G, et
al: FEZ1/LZTS1 is down-regulated in high-grade bladder cancer and
its restoration suppresses tumorigenicity in transitional cell
carcinoma cells. Am J Pathol. 160:1345–1352. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tovar C, Rosinski J, Filipovic Z, et al:
Small-molecule MDM2 antagonists reveal aberrant p53 signaling in
cancer: implications for therapy. Proc Natl Acad Sci USA.
103:1888–1893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo Z, Tang W, Yuan J, et al: BRSK2 is
activated by cyclic AMP-dependent protein kinase A through
phosphorylation at THR260. Biochem Biophys Res Commun. 347:867–871.
2006. View Article : Google Scholar : PubMed/NCBI
|