Introduction
Breast cancer is the most prevalent type of
malignant cancer in females worldwide, of which invasive ductal
breast carcinoma (IDBC), also known as infiltrating ductal
carcinoma, accounts for 70–80% of invasive breast cancer cases
(1). Previous studies have
indicated that IDBC has multiple stages of development, initiating
from premalignant hyperplastic breast lesions, which progress to
ductal breast cancer in situ (DCIS) and then to IDBC
(2–4). It was reported that an interim stage,
DCIS with microinvasion, may also have an important role in the
progression from DCIS to metastatic IDBC (5). This linear multi-step model of human
breast cancer progression serves as a starting point for current
understanding of breast cancer pathogenesis; however, numerous
studies have contradicted this model (4,6).
Therefore the complex pathogenesis of IDBC remains to be
elucidated.
mRNA conveys the genetic information in DNA into the
translation of amino acids. Many studies have reported that the
expression of mRNAs was altered in breast cancer tissues (7–9);
therefore, mRNA expression may be used to predict the prognosis of
patients with IDBC (10). A
previous study into the transcriptomic landscape of breast cancer
using in-depth mRNA sequencing revealed numerous novel and
annotated transcripts in breast cancer tissue; this therefore
reflected the limited current understating of mRNAs in the
pathogenesis of the disease (8).
Long non-coding RNAs (lncRNA) are non-coding RNAs
consisting of >200 nucleotides. lncRNAs were previously
considered to be ‘junk DNA’; however, studies have demonstrated
that lncRNAs participated in the regulation of protein
transcription and epigenetic modification, and were reported to be
involved in a variety of developmental processes as well as several
diseases (11–13). Only a small number of lncRNAs have
been studied extensively; therefore, the function of numerous
lncRNAs remains to be elucidated (14). In addition, the identification of
novel lncRNAs and exploration of their underlying regulatory
mechanisms in the initiation and progression of diseases is
essential for a deeper understanding of disease pathogenesis.
Previous studies have also demonstrated the altered expression of
lncRNAs in breast cancer, including Hox transcript antisense
intergenic RNA (HOTAIR) and growth arrest-specific 5 (15–17).
However, the biological functions of the majority of lncRNAs in
association with IDBC remain to be elucidated. In the present
study, a pilot study was conducted to explore novel mRNAs and
lncRNAs that exhibit aberrantly altered expression in patients with
IDBC, which may therefore potentially be involved in the
pathogenesis of IDBC.
Materials and methods
Participants
In June, 2013, three female patients aged ≥60 years
and diagnosed with IDBC underwent a modified radical mastectomy
without chemotherapy at the Department of Breast and Thyroid
Surgery of the Third Xiangya Hospital (Changsha, China). The
criteria for IDBC diagnosis was as follows: Pathological
examination which revealed a tumor with a diameter >2cm and
<5cm in the presence of lymph node metastasis. Pathological
examinations were performed by experienced clinicians in clinical
pathology at the Third Xiangya Hospital. All three participants
were diagnosed with stage III IDBC according to the
Bloom-Richardson grading system (18). Informed consent was obtained from
each participant, and the study was conducted in adherence to the
tenets of the Declaration of Helsinki. This study was approved by
the ethics committee of the Third Xiangya Hospital of Central South
University.
Resection of breast specimens
Fresh breast cancer specimens and normal breast
tissues were obtained from the participants during a modified
radical mastectomy, and each surgery was performed by the same
experienced surgeon. Normal breast tissue samples were resected
from breast glands >5cm distance from the tumor tissue.
Following surgery, the breast tissue samples were diagnosed by
pathological clinicians, then preserved in liquid nitrogen within
30 min and stored at −80°C.
RNA microarray and hybridization
Total RNA was extracted using the mirVana RNA
Isolation kit (Life Technologies, Grand Island, NY, USA). RNA
quality control, labeling and hybridization were performed by
Shanghai Biochip Co., Ltd. (Shanghai, China) according to the
manufacturer’s instructions of the Agilent microRNA Microarray
System 2.4 (Agilent Technologies, Santa Clara, CA, USA). Arrays
were then scanned using an Agilent Microarray Scanner (G2505C;
Agilent Technologies) and the fluorescence intensities of the
labeled samples were normalized according to the median of the
total signals on the arrays. Images were captured using Scanner
Control Software 7.0 (Agilent Technologies) and signal intensities
were analyzed using ArrayVision 6.0 software (Imaging Research, St.
Catharines, ON, Canada).
Target gene analysis
TargetScan Human release 6.0 online software
(http://www.targetscan.org/vert_60/)
was used to predict microRNA targets as previously described
(19,20). The Database for Annotation,
Visualization and Integrate Discovery v6.7 (http://david.abcc.ncifcrf.gov/) was then used to
annotate the biological functions of predicted targets as
previously described (21,22).
Statistical analysis
All experiments were performed in triplicate.
Screening for differentially expressed mRNA or lncRNA was performed
using paired t-test (criterion 1), and significance was indicated
by a threshold of ≥2-fold change in expression and a corresponding
P-value of ≤0.05. False discovery rate (FDR) analysis was used to
adjust for multiple testing (criterion 2) and Q≤0.05 was considered
to indicate a significant change in expression between groups.
Sure independence screening procedure based on the
distance correlation (DC-SIS), was then performed in order to
compare the findings (criterion 3) (23). DC-SIS is a novel statistical method
for screening important characteristics for ultra-high dimensional
data; in addition, DC-SIS does not make any model assumption (e.g.
linear model) for the response (e.g. breast cancer or not) and the
predictors (e.g. expression of mRNAs or lncRNAs), which therefore
makes model misspecification highly unlikely. Sure independence
ensures that all important variables may be selected with
sufficient sample size, which enables DC-SIS to be a more flexible
and reliable for screening important predictors compared to
conventional statistical methods such as the t-test. Due to the
limited sample size in the present study, a model of size
6[n/log(n)], where n is the sample size and [n/log(n)] is the
integer part of n/log(n), was selected in order to reduce the
possibility of missing important probes. Statistical analyses were
performed using R software (www.R-project.org) and SAS 9.3 (SAS Institute Inc.,
Cary, NC, USA).
Results
Interrogated probes
A total of 44,244 probes, which consisted of 22,078
mRNA and 22,166 lncRNA probes (49.9 and 50.1%, respectively), were
interrogated and included in the final analyses.
Aberrant expression of mRNAs and lncRNAs
in IDBC tissue
Paired t-tests located 3,510 probes with
statistically significant expression levels changes of ≥2-fold
(P≤0.05) in IDBC tissue compared with those of normal breast
tissue. A total of 2,090 mRNAs (9.5% of interrogated mRNA probes)
demonstrated significant changes, of which 722 (34.5%) showed
elevated expression; in addition, 1,420 lncRNAs (6.4% of
interrogated lncRNA probes) showed significantly altered expression
in IDBC tissue, of which 304 (21.4%) exhibited elevated expression
(Table I).
 | Table IDifferential expression of mRNAs and
lncRNAs in invasive ductal breast carcinoma samples compared to
that of normal breast tissue from the same patients. |
Table I
Differential expression of mRNAs and
lncRNAs in invasive ductal breast carcinoma samples compared to
that of normal breast tissue from the same patients.
| mRNA | lncRNA |
|---|
|
|
|
|---|
| Statistical
criteria | Increased | Decreased | Total | Increased | Decreased | Total |
|---|
| Paired t-test (n,
%)a | 722 (34.5) | 1368 (65.5) | 2090 | 304 (21.4) | 1116 (78.6) | 1420 |
| FDR | 79 (40.5) | 116 (59.5) | 195 | 37 (76.6) | 121 (76.6) | 158 |
| DC-SIS | 3 (25.0) | 9 (75.0) | 12 | 0 (0) | 6 (100) | 6 |
FDR analysis revealed that a total of 353 probes
demonstrated significant changes in expression levels in IDBC
tissue compared with those of normal breast tissue. Of note, 195
mRNAs (0.9%) showed significant changes, of which 79 (40.5%)
exhibited elevated expression; in addition, 158 lncRNAs (0.7%)
showed significantly altered expression, of which 37 (23.4%)
demonstrated increased expression levels (Table I).
DC-SIS feature screening identified 18 probes which
demonstrated significantly altered expression in IDBC tissue
compared with that of normal breast tissue. Of these 18 identified
probes, 12 were mRNAs (0.05%), three (25%) of which exhibited
elevated expression, and six were lncRNAs (0.03%), all of which
showed significantly downregulated expression (Table I). These 18 probes were therefore
demonstrated to have altered expression levels in IDBC tissues by
all the three criteria. Table II
provides detailed information regarding these 18 selected
probes.
 | Table IIDetailed information for the 18
probes identified using DC-SIS. |
Table II
Detailed information for the 18
probes identified using DC-SIS.
| Probe | Chr | Type | Gene | IDBC | Normal | P-value | Q | Rank |
|---|
|
oebiotech_26202 | 15 | lncRNA | NA | 7.51 | 9.14 |
1.93×10−6 |
1.93×10−2 | 1 |
|
oebiotech_08007 | 17 | lncRNA | NA | 7.89 | 8.08 |
2.57×10−6 |
1.93×10−2 | 2 |
| A_33_P3371999 | 5 | mRNA | TPPP | 2.58 | 5.40 |
4.25×10−6 |
1.93×10−2 | 3 |
| A_23_P144054 | 3 | mRNA | PRKCD | 10.00 | 8.84 |
4.96×10−6 |
1.93×10−2 | 4 |
| A_33_P3320197 | 2 | mRNA | FAM150B | 2.41 | 5.70 |
6.80×10−6 |
2.31×10−2 | 5 |
|
oebiotech_09186 | 21 | lncRNA | NA | 2.41 | 6.18 |
4.94×10−6 |
1.93×10−2 | 6 |
| A_23_P315364 | 4 | mRNA | CXCL2 | 2.59 | 9.56 |
4.47×10−6 |
1.93×10−2 | 7 |
| A_21_P0011386 | 15 | mRNA |
LOC100505679 | 8.19 | 9.68 |
3.64×10−6 |
1.93×10−2 | 8 |
| A_33_P3419691 | 7 | lncRNA | GATS | 7.16 | 7.93 |
9.41×10−6 |
2.84×10−2 | 9 |
| A_33_P3372426 | 21 | mRNA | ADAMTS5 | 2.38 | 5.46 |
1.11×10−5 |
2.97×10−2 | 10 |
|
oebiotech_22954 | 3 | lncRNA | NA | 2.38 | 5.86 |
2.51×10−5 |
3.14×10−2 | 11 |
| A_33_P3300262 | 2 | mRNA | VIT | 2.46 | 5.85 |
2.50×10−5 |
3.14×10−2 | 12 |
|
oebiotech_19472 | 3 | lncRNA | NA | 7.34 | 9.07 |
3.35×10−5 |
3.14×10−2 | 13 |
| A_33_P3290338 | 1 | mRNA | PARP1 | 10.01 | 8.63 |
2.30×10−5 |
3.14×10−2 | 14 |
| A_33_P3360087 | 7 | mRNA | BBS9 | 8.09 | 8.82 |
2.37×10−5 |
3.14×10−2 | 15 |
| A_24_P393958 | 1 | mRNA | DNAJB4 | 6.73 | 8.20 |
1.20×10−5 |
2.97×10−2 | 16 |
| A_24_P189533 | 11 | mRNA | ENDOD1 | 7.73 | 8.52 |
2.30×10−5 |
3.14×10−2 | 17 |
| A_33_P3325914 | 6 | mRNA | TAPBP | 13.16 | 12.19 |
3.90×10−5 |
3.21×10−2 | 18 |
Functional analysis
Gene ontology (GO) analysis was performed in order
to determine the biological functions of genes harboring the 2,090
mRNA probes which were found to be aberrantly expressed in IDBC
tissue compared to that of normal breast tissue. The results of the
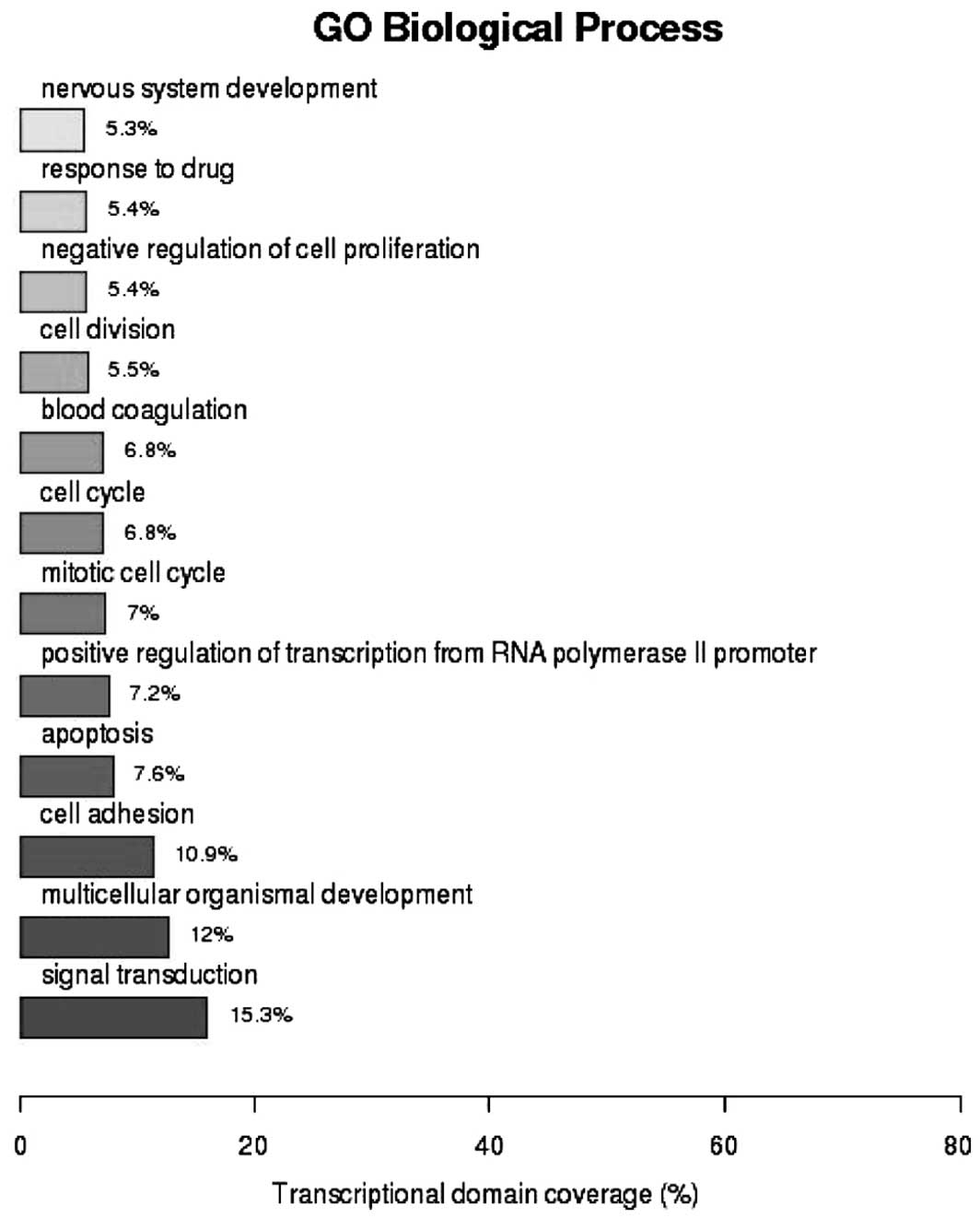
GO analysis of biological functions (Fig. 1) revealed that 15.3% of the genes
were involved in signal transduction [enrichment value
(P)=1.53×10−3], 12% had functions in multi-cellular
organism development (P=6.14×10−10) and 10.9% were
involved in cell adhesion (P=2.01×10−10). As shown in
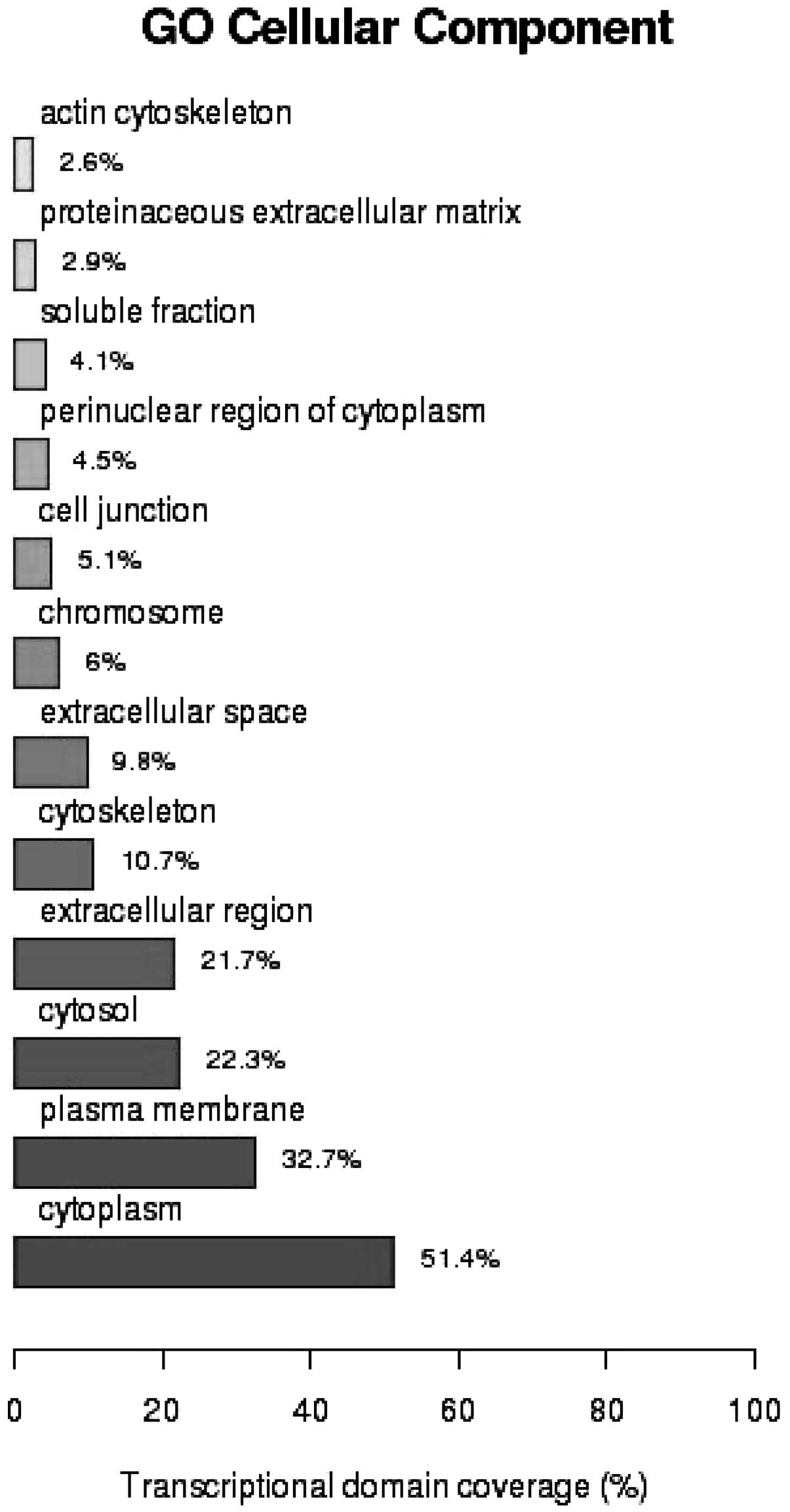
Fig. 2 GO analysis of the cellular
components of the mRNAs showed that 51.4% of the gene products were
located in cytoplasm (P=4.9×10−6), 32.7% in the plasma
membrane (P=4.24×10−2) and 22.3% in the cytosol
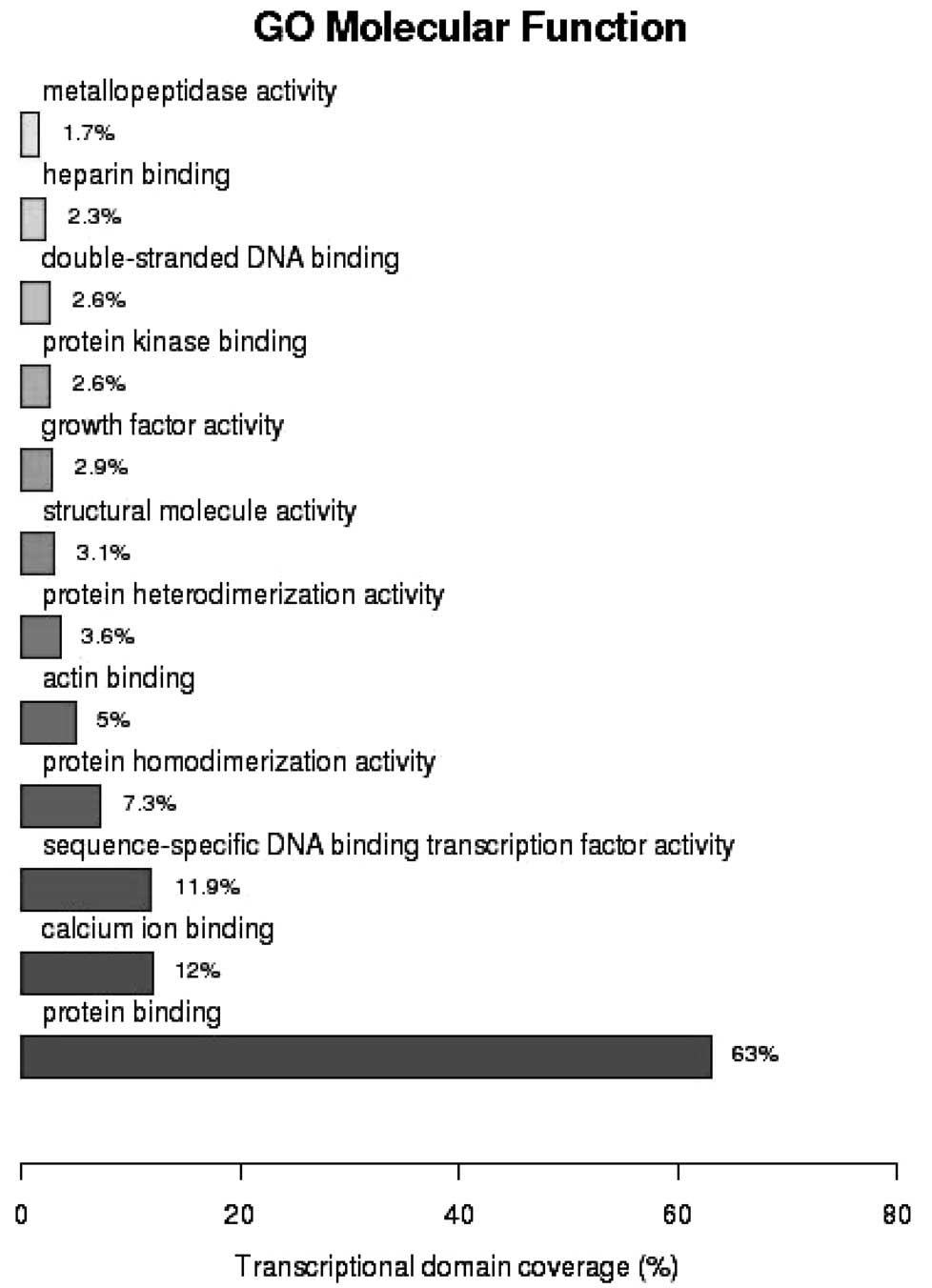
(P=5.18×10−5). GO analysis of molecular function
demonstrated that 63% of the genes were involved in protein binding
(P=5.82×10−5), 12% in calcium ion binding
(P=7.27×10−9) and 11.9% in sequence-specific DNA binding
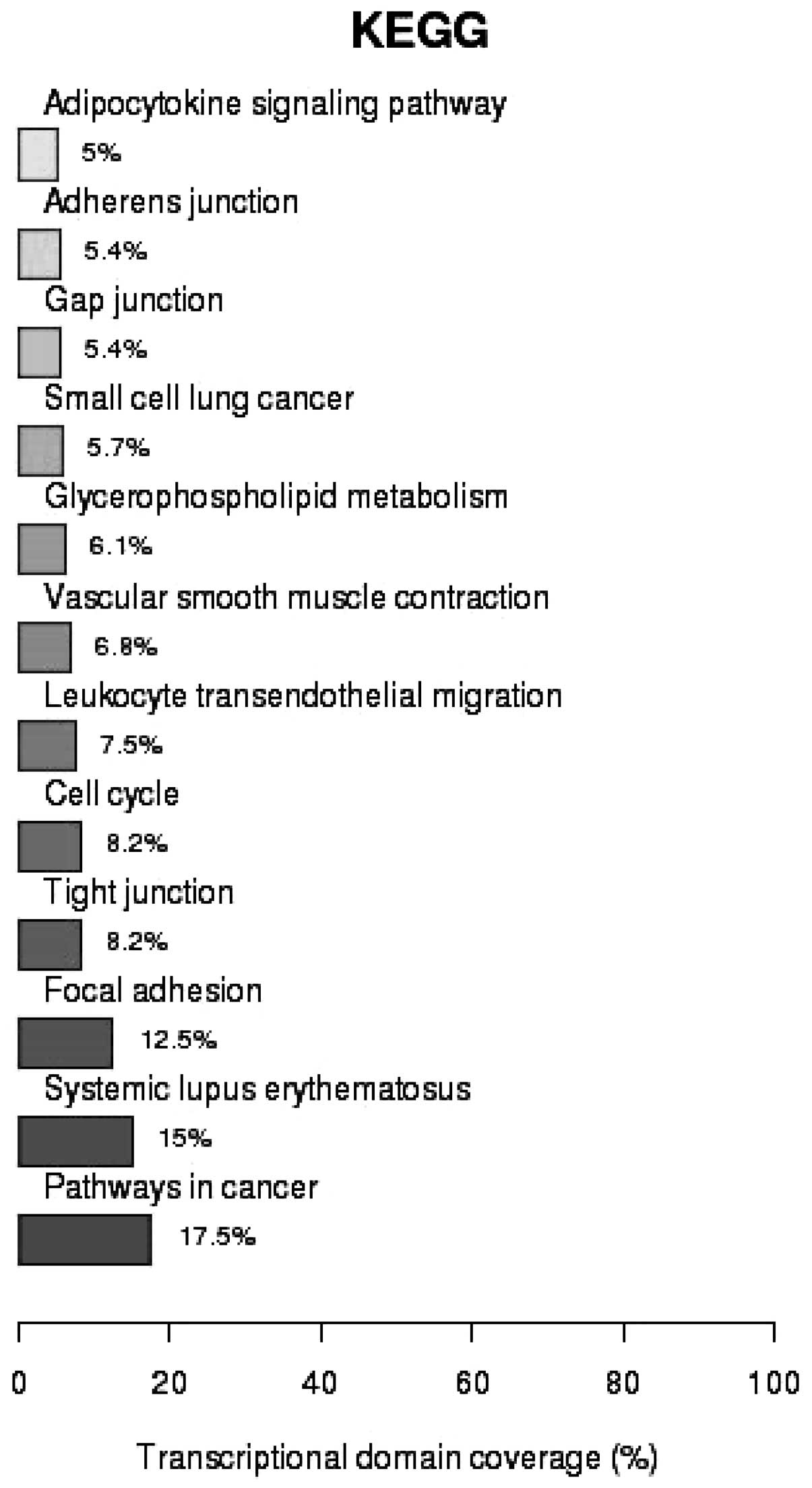
transcription (P=2.16×10−2) (Fig. 3). As shown in Fig. 4, analysis of the Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathways revealed that 17.5% of the
genes identified were associated with cancer pathways
(P=1.37×10−3), 15% were involved in systematic lupus
erythematosus (SLE; P=6.02×10−12) and 12.5% in focal
adhesion (P=4.18×10−4).
Discussion
In the present study, mRNA and lncRNA expression
levels were detected in the normal and cancerous tissues from three
patients with IDBC. Following microarray analysis, numerous
aberrantly expressed mRNAs and lncRNAs were located in IDBC
samples. The majority of genes which harbored the differentially
expressed mRNAs were found to be involved in signal transduction,
protein binding and cancer pathways, and their gene products were
predominantly located in cytoplasm.
One of the identified mRNAs A_23_P315364 is found in
the CXCL2 gene, located on chromosome 4. C-X-C motif ligand
2 (CXCL2) is a chemokine that is highly expressed in metastases
(24). A previous study identified
a paracrine network between tumor and stromal cells comprising of
CXCL1 and 2, which indicated that lung metastasis was associated
with chemotherapy resistance in breast cancer (25). Another mRNA identified in the
present study, A_33_P3290338, is found in the PARP1 gene,
which encodes a nuclear enzyme that has an important role in
regulating DNA repair (26). One
study performed a meta-analysis which showed that PARP1 mRNA
expression was heterogeneous between breast cancer subtypes and was
overexpressed in 58% of breast cancers (9); this was concurrent with the results
of the present study, which found that PARP1 expression was
elevated in patients with IDBC. In addition, mRNA expression of
PARP1 was associated with high medullary histological grade,
tumor size, metastasis-free survival (MFS) and overall survival in
patients with breast cancer, and is an independent prognostic
factor for MFS (9). However,
further studies are required in order to elucidate the exact
biological/molecular functions and pathways of mRNAs identified in
the present study.
In the present study, 18 probes were identified
which were found to have significantly altered expression in IDBC
tissue according to all the three criteria. Six of the identified
probes were lncRNAs, each of which was reported to be downregulated
in the IDBC samples compared with that of the normal breast tissue;
however, further studies into the function of these lncRNA are
required in order to elucidate the mechanism through which they are
involved in the pathogenesis of IDBC, as previous studies are
limited. Of note, in the present study, two lncRNAs were identified
in IDBC tissue which demonstrated a >10-fold decrease in
expression: ENST00000458316 (corresponding probe oebiotech_09186)
is located on chromosome 21 and was reported to be expressed at
higher levels in breast tissue compare various other tissues in the
human body (Illumina Human BodyMap 2.0, ArrayExpress ID,
E-MTAB-513; http://www.ebi.ac.uk/arrayexpress); and NR_072979
(corresponding robe oebiotech_22954) is a transcript variant of
aldehyde dehydrogenase 1 family, member L1 (ALDH1L1). The
gene product of ALDH1L1,10-formyltetrahydrofolate
dehydrogenase (FDH) is a major folate-metabolizing enzyme involved
in the regulation of cell proliferation (27). FDH was reported to be ubiquitously
downregulated in human tumors (27), the mechanism of which was suggested
to proceed via promoter methylation which influenced levels of FDH
(28); however, the exact effect
of this lncRNA on FDH levels and its subsequent influence on cell
proliferations remains to be elucidated.
Previous studies have identified several lncRNAs
involved in the pathogenesis, progression and survival of breast
cancer. HOTAIR, a widely studied lncRNA located on 12q13.13, is
transcribed from the antisense strand of HOXC12 (29) and serves as an interface between
DNA and specific chromatin remodeling. Of note, HOTAIR specifies
the pattern of histone modifications on target genes by providing
binding surfaces for polycomb repressive complex 2 (PRC2) via its
5′ domain as well as providing binding surface for the
lysine-specific demethylase 1A/co-repressor element-1-specific
transcription factor (CoREST)/REST complex via its 3′ domain
(16). A previous study reported
that increased expression levels of HOTAIR in primary breast cancer
tumors was a prognostic factor for metastasis and death (15). In the present study, HOTAIR
expression in IDBC tissue was not found to be significantly
decreased. A previous study also reported that of the 336 tumor
samples analyzed, HOTAIR expression was markedly varied in breast
cancer tissues and 6.5% had undetectable HOTAIR expression; in
addition, no association was found between HOTAIR expression and
the clinical or pathologic characteristics of breast cancer
(30). Furthermore, patients with
higher expression levels of HOTAIR demonstrated a lower risk of
relapse and death than those with lower expression of HOTAIR
(30). These findings are
consistent with the results of the present study; however, further
studies are required to validate the findings.
SLE is an autoimmune rheumatic disease which occurs
primarily in females. Previous studies reported that females with
SLE had a decreased risk of developing breast cancer [odds ratio
(OR)=0.76, P=2.49×10−7] (31) as well as ductal carcinoma (OR=0.95,
P=0.067) (32). However, a study
of ten lupus-associated single nucleotide polymorphism (SNPs) found
less supportive evidence for the association of these SNPs with
breast cancer (33), indicating
that epigenetic factors may have contributed to the decreased risk
of breast cancer in females with SLE. In the present study, KEGG
analysis showed that there was an enrichment of genes involved in
SLE, indicating that epigenetic factors may be involved in
influencing the risk of breast cancer. Of note, the mRNA
ENST00000330452 for PRKCD exhibited a 1.2-fold increase in
expression in IDBC tissues; mutations in PRKCD were
previously reported to result in the reduced expression and
activation of protein kinase C, which in turn may lead to increased
B cell proliferation and susceptibility to SLE (34). The results of the present study
were consistent with the reported decreased risk of breast cancer
in patients with SLE; however, further studies are required in
order to elucidate the mechanism underlying the reduced risk of
breast cancer, which may further current understanding of its
etiology.
The limitations of the present study are due to its
small sample size, which therefore prevented conclusive results
being reached. However, the significant probes identified by the
three criteria represented potential markers and require further
investigation. In addition, all three participants had stage III
IDBC, which therefore prevented the comparison of aberrant RNA
expression among the different stages of IDBC. Furthermore, due to
the cross-sectional nature of the present study, the pattern of
changes throughout the development of IDBC was not analyzed.
In conclusion, microarray analysis was performed in
the present study in order to screen for mRNAs and lncRNAs
exhibiting aberrantly altered expression in patients with IDBC
compared to that in the normal breast tissue of the same patients.
A total of 18 mRNAs and lncRNAs showing significant changes in
expression were identified, of which six were lncRNAs and 12 were
mRNAs. Functional analysis of the identified mRNA probes
demonstrated that they were located in genes involved in various
biological functions, including signal transduction and protein
binding as well as cancer pathways; however, the functions of the
six identified lncRNAs remains to be elucidated. Therefore, further
studies are required in order to determine the functions of the
identified lncRNAs as well as to validate the results of the
present study using larger sample sizes.
Acknowledgements
The present study was supported by a grant from the
Hunan Provincial Natural Science Foundation of China (no.
14JJ7017). The research of Dr Yang was supported by grant no.
R01AG036042 and the Illinois Department of Public Health.
References
|
1
|
Malhotra GK, Zhao X, Band H and Band V:
Histological, molecular and functional subtypes of breast cancers.
Cancer Biol Ther. 10:955–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carter CL, Corle DK, Micozzi MS, Schatzkin
A and Taylor PR: A prospective study of the development of breast
cancer in 16,692 women with benign breast disease. Am J Epidemiol.
128:467–477. 1988.PubMed/NCBI
|
|
3
|
Wiechmann L and Kuerer HM: The molecular
journey from ductal carcinoma in situ to invasive breast cancer.
Cancer. 112:2130–2142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lakhani SR, Chaggar R, Davies S, et al:
Genetic alterations in ‘normal’ luminal and myoepithelial cells of
the breast. J Pathol. 189:496–503. 1999. View Article : Google Scholar
|
|
5
|
Yu KD, Wu LM, Liu GY, et al: Different
distribution of breast cancer subtypes in breast ductal carcinoma
in situ (DCIS), DCIS with microinvasion, and DCIS with invasion
component. Ann Surg Oncol. 18:1342–1348. 2011. View Article : Google Scholar
|
|
6
|
Buerger H, Mommers EC, Littmann R, et al:
Ductal invasive G2 and G3 carcinomas of the breast are the end
stages of at least two different lines of genetic evolution. J
Pathol. 194:165–170. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dziegiel P, Owczarek T, Plazuk E, et al:
Ceramide galactosyltransferase (UGT8) is a molecular marker of
breast cancer malignancy and lung metastases. Br J Cancer.
103:524–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eswaran J, Cyanam D, Mudvari P, et al:
Transcriptomic landscape of breast cancers through mRNA sequencing.
Sci Rep. 2:2642012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gonçalves A, Finetti P, Sabatier R, et al:
Poly(ADP-ribose) polymerase-1 mRNA expression in human breast
cancer: a meta-analysis. Breast Cancer Res Treat. 127:273–281.
2011. View Article : Google Scholar
|
|
10
|
Volinia S and Croce CM: Prognostic
microRNA/mRNA signature from the integrated analysis of patients
with invasive breast cancer. Proc Natl Acad Sci USA. 110:7413–7417.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibb EA, Vucic EA, Enfield KS, et al:
Human cancer long non-coding RNA transcriptomes. PloS One.
6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji P, Diederichs S, Wang W, et al:
MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wan LB and Bartolomei MS: Regulation of
imprinting in clusters: noncoding RNAs versus insulators. Adv
Genet. 61:207–223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai MC, Manor O, Wan Y, et al: Long
noncoding RNA as modular scaffold of histone modification
complexes. Science. 329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
18
|
Bloom HJ and Richardson WW: Histological
grading and prognosis in breast cancer; a study of 1409 cases of
which 359 have been followed for 15 years. Br J Cancer. 11:359–377.
1957. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grimson A, Farh KK, Johnston WK, et al:
MicroRNA targeting specificity in mammals: determinants beyond seed
pairing. Mol Cell. 27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garcia DM, Baek D, Shin C, et al: Weak
seed-pairing stability and high target-site abundance decrease the
proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol.
18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
23
|
Li R, Zhong W and Zhu L: Feature screening
via distance correlation learning. J Am Stat Assoc. 107:1129–1139.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bièche I, Chavey C, Andrieu C, et al: CXC
chemokines located in the 4q21 region are up-regulated in breast
cancer. Endocr Relat Cancer. 14:1039–1052. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Acharyya S, Oskarsson T, Vanharanta S, et
al: A CXCL1 paracrine network links cancer chemoresistance and
metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rouleau M, Patel A, Hendzel MJ, Kaufmann
SH and Poirier GG: PARP inhibition: PARP1 and beyond. Nat Rev
Cancer. 10:293–301. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krupenko SA and Oleinik NV:
10-formyltetrahydrofolate dehydrogenase, one of the major folate
enzymes, is down-regulated in tumor tissues and possesses
suppressor effects on cancer cells. Cell Growth Differ. 13:227–236.
2002.PubMed/NCBI
|
|
28
|
Oleinik NV, Krupenko NI and Krupenko SA:
Epigenetic Silencing of ALDH1L1, a Metabolic Regulator of Cellular
Proliferation, in Cancers. Genes Cancer. 2:130–139. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rinn JL, Kertesz M, Wang JK, et al:
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu L, Zhu G, Zhang C, et al: Association
of large noncoding RNA HOTAIR expression and its downstream
intergenic CpG island methylation with survival in breast cancer.
Breast Cancer Res Treat. 136:875–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bernatsky S, Ramsey-Goldman R, Foulkes WD,
Gordon C and Clarke AE: Breast, ovarian, and endometrial
malignancies in systemic lupus erythematosus: a meta-analysis. Br J
Cancer. 104:1478–1481. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tessier Cloutier B, Clarke AE,
Ramsey-Goldman R, et al: Breast cancer in systemic lupus
erythematosus. Oncology. 85:117–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bernatsky S, Easton DF, Dunning A, et al:
Decreased breast cancer risk in systemic lupus erythematosus: the
search for a genetic basis continues. Lupus. 21:896–899. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Belot A, Kasher PR, Trotter EW, et al:
Protein kinase cdelta deficiency causes mendelian systemic lupus
erythematosus with B cell-defective apoptosis and
hyperproliferation. Arthritis Rheum. 65:2161–2171. 2013. View Article : Google Scholar : PubMed/NCBI
|