Introduction
Venous thromboembolism (VTE) is a disease which
consists of deep vein thrombosis (DVT) and pulmonary
thromboembolism (PTE) (1).
Pulmonary embolism (PE), which can be subdivided into acute
pulmonary embolisms (APE) and chronic thromboembolic pulmonary
hypertension (CTEPH), is a global health problem with high
incidence, mortality and misdiagnosis rates (2). The risk factors for VTE include
environmental and hereditary influences; however, environmental
factors are more commonly linked to the disease (3). Nine editions of guidelines for the
prevention, diagnosis and treatment of VTE in surgical patients
have been published by The American College of Chest Physicians
since 1995 (4,5); however, the incidence rate of
symptomatic VTE has not been reduced (6). One possible explanation for this may
be that the pathogenesis of VTE has not yet been fully
elucidated.
Smeeth et al (7) reported an association between acute
infections and the increased risk of VTE. Another study reported
that VTEs were identified in multiple organ samples, including the
lungs, spleen, pancreas, kidneys and adrenal glands of patients
that succumbed to severe acute respiratory syndrome (8). In addition, a previous study
demonstrated that messenger (m)RNA expression of genes associated
with natural killer (NK) and T cells were significantly
downregulated in patients with symptomatic PE (9), and cellular immune function was
decreased in patients with acute PE and CTEPH (10,11).
These studies indicated that the occurrence and progress of
symptomatic PE were closely associated with the innate and adaptive
immune system.
The innate and adaptive immune system influence and
interact with each other. T and B cells, as key components of the
adaptive immune system, co-operate in order to form the immune
response to non-self antigens (12). A previous study reported that mRNA
expression of CD19 was downregulated in patients with symptomatic
VTE (13). The present study aimed
to investigate the expression of a wide range of B-cell-associated
genes involved in B-cell activation using human complementary
(c)DNA microarray analysis in order to identify differential gene
expression at different stages of B-cell activation between PE
patients and healthy controls.
Materials and methods
Patients
A total of 20 PE patients were enrolled in the
present study from Tongji Hospital of Tongji University (Shanghai,
China) between 2007–2008. PE patients were required to meet any two
of the following three criteria prior to enrollment in the study:
i) a selective pulmonary angiography showing pulmonary artery
obstruction or filling defects; ii) a lung ventilation/perfusion
scan showing single or multiple blood perfusion defects, normal or
abnormal ventilation and a ventilation/perfusion mismatch; and iii)
a clinical diagnosis indicating that there are risk factors for PE
and other cardiovascular diseases, which may have been demonstrated
through clinical performance, an electrocardiogram and chest film,
arterial blood gas analysis indicating hypoxemia and hypocapnia,
D-dimer detection, echocardiography or chest computed tomography. A
total of 20 healthy volunteers were enrolled in the present study
as controls. The groups were divided as follows: PE group, 20
patients (11 males and nine females; mean age, 70±14 and range,
44–89) which included three cases of CTEPH; control group, 20 age-
and gender-matched volunteers (11 males and nine females; mean age,
70±14 and range, 44–91) who passed health examinations for PE, DVT,
arterial thrombosis and congenital coagulation abnormalities. There
was no statistically significant difference in age between the two
groups (P>0.05). The present clinical trial was approved by the
Ethics Committee of Tongji University and written informed consent
was obtained from all PE patients and healthy volunteers.
Gene expression profiling
Agilent G4112A Whole Human Genome Oligo Microarrays
were obtained from Agilent Technologies (Santa Clara, CA, USA).
Microarrays were composed of 44,290 spots, including 41,675 genes
or transcripts, 314 negative control spots, 1,924 positive control
spots and 359 blank spots. The functions of more than 70% of the
genes in the microarray had been previously elucidated. Blood
samples from all participants were subjected to microarray
analysis.
Total RNA isolation
EDTA-anti-coagulated peripheral fasted blood samples
(5 ml) were drawn from PE patients and healthy volunteers
immediately following admittance to the hospital. Mononuclear cells
were obtained from the samples using density gradient
centrifugation at 7,000 × g for 30 min with Ficoll solution
(Agilent Technologies) and the remaining red blood cells were
ablated using erythrocyte lysis buffer (Qiagen, Limburg,
Netherlands). Total RNA was extracted from mononuclear cells using
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) and then purified using a Qiagen RNeasy column (Qiagen)
according to the manufacturer’s instructions. Isolated total RNA
was evaluated and quantified using a Nanodrop ND-1000
spectrophotometer (Nanodrop Technology, Inc., Wilmington, DE,
USA).
Detection of gene expression
Reverse transcription of ~1 μg total RNA was
performed using an Agilent Low RNA Input Linear Amplification kit
(Agilent Technologies) in order to produce double-stranded cDNA.
Following purification, an Agilent Low RNA Input Linear
Amplification kit (Agilent Technologies) was used to perform in
vitro amplification and modified α-D-glucose-1-phosphate
uridylyltransferase [UTP; aaUTP, 5-(3-aminoallyl)-UTP; Agilent
Technologies] was used to replace UTP. The integrated aaUTP
interacted with cyanine-3-N-hydroxysuccinimide ester
(Agilent Technologies) forming fluorescent products, which were
then used for hybridization. The integration rate of fluorescence
was determined using a NanodropND-1000 spectrophotometer. The
hybridization mixture was then prepared using an Agilent
Oligonucleotide Microarray in situ Hybridization Plus kit
(Agilent Technologies). Fluorescent products (~750 ng) were
fragmented at 60°C and hybridization was conducted using Human
Whole-Genome 60-mer oligo-chips (G4112F; Agilent Technologies) at
60°C for 17 h at 600 × g. Following hybridization, the chips were
washed with Agilent Gene Expression Wash-Buffer (Agilent
Technologies) according to manufacturer’s instructions. The
original signals were then obtained using an Agilent scanner
(G2655AA; Agilent Technologies) and Feature Extraction software
(GeenSpring GX; Agilent Technologies). The standardization of
original signals was performed using the robust multiarray
averaging method; standardized signal values were then used for the
screening of differentially expressed genes.
Quantitative polymerase chain reaction
(qPCR)
Three genes were selected at random from the genes
with differential expressions between the two groups, and then
subjected to qPCR. Relative gene expression levels were normalized
to that of GAPDH (2−ΔΔCt). Differences in gene
expression between the two groups were analyzed using a
standardized melting curve and the 2−ΔΔCt method.
Results from qPCR were found to be consistent with microarray
analysis.
Statistical analysis
Statistical tests were performed using SPSS 17.0
software (International Business Machines, Armonk, NY, USA). Values
are presented as the mean ± standard deviation. Independent-Samples
t-tests were used to compare mRNA levels in samples from patients
with symptomatic PE and controls. Prior to t-testing, tests for
equality of variances were performed in order to test the
homogeneity of variance between two independent samples, if
variances were not equal, the results of t-tests would be corrected
accordingly. P<0.05 was considered to indicate a statistically
significant difference between values.
Results
Expression of B-cell receptor-associated
B-cell activation genes
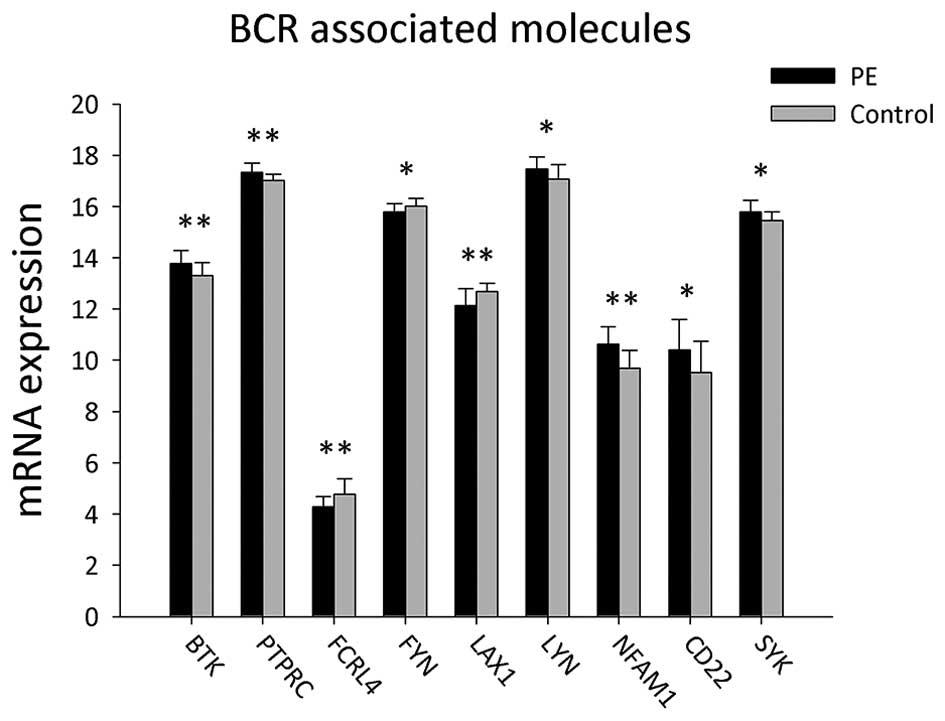
A total of 22 B cell receptor-associated B-cell
activation genes were detected in the peripheral blood mononuclear
cells (PBMCs) of patients with PE and the control group (Table I and Fig. 1). The results showed that gene
expression levels of LYN, CD22, SYK, BTK, PTPRC and NFAM1 were
significantly upregulated in PE patients compared to those of the
control group (P<0.05). However, expression levels of FYN, FCRL4
and LAX1 were significantly lower in PBMCs from PE patients
compared to those in the control group (P<0.05).
 | Table IExpression of genes associated with B
cell receptor-mediated B-cell activation. |
Table I
Expression of genes associated with B
cell receptor-mediated B-cell activation.
| Symbol | PE (X±SD) | Control (X±SD) | P |
|---|
| BLK | 12.50±0.80 | 12.55±0.69 | 0.823 |
| BLNK | 10.76±0.66 | 10.74±0.57 | 0.909 |
| BTK | 13.77±0.52 | 13.30±0.51 | 0.007a |
| CD5 | 13.68±0.73 | 13.58±0.65 | 0.653 |
| CD19 | 13.93±0.95 | 14.02±0.69 | 0.736 |
| CR2 | 10.34±0.68 | 10.18±0.61 | 0.430 |
| PTPRC | 17.33±0.37 | 17.01±0.25 | 0.003a |
| CD79A | 14.76±0.79 | 14.70±0.70 | 0.809 |
| CD79B | 13.20±0.68 | 13.19±0.49 | 0.967 |
| CD81 | 15.17±0.53 | 15.20±0.44 | 0.851 |
| FCRL1 | 11.28±0.80 | 11.03±0.85 | 0.357 |
| FCRL2 | 10.83±0.79 | 10.64±0.62 | 0.409 |
| FCRL3 | 11.18±0.74 | 11.40±0.69 | 0.344 |
| FCRL4 | 4.29±0.38 | 4.77±0.61 | 0.004a |
| FCRL5 | 11.07±0.93 | 11.34±0.87 | 0.346 |
| FYN | 15.79±0.32 | 16.01±0.31 | 0.033a |
| LAX1 | 12.14±0.66 | 12.68±0.33 | 0.002a |
| LYN | 17.47±0.46 | 17.06±0.57 | 0.016a |
| NFAM1 | 10.63±0.68 | 9.69±0.69 | 0.000a |
| PIR | 6.32±0.74 | 6.07±0.64 | 0.257 |
| CD22 | 10.41±1.17 | 9.51±1.23 | 0.023a |
| SYK | 15.78±0.46 | 15.44±0.34 | 0.011a |
Expression of genes associated with T
cell-dependent B-cell activation
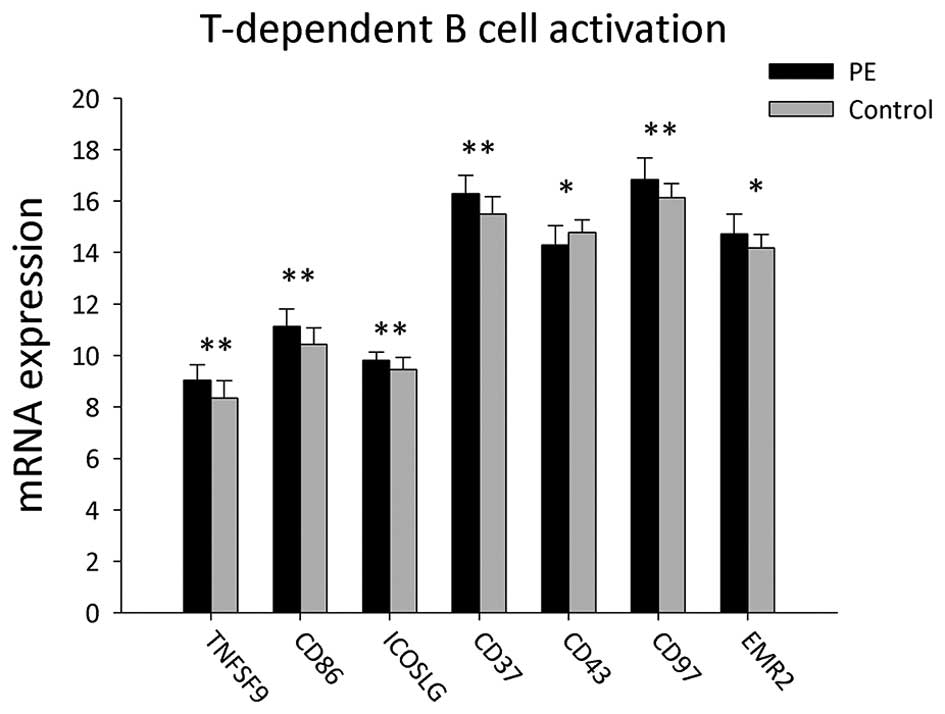
A total of 22 genes associated with T cell-dependent
B-cell activation were detected in the PBMCs of PE patients and
healthy volunteers (Table II and
Fig. 2). In PE patients, the mRNA
expression of EMR2, TNFSF9, CD86, ICOSLG, CD37 and CD97 were
significantly increased compared to those of the healthy volunteers
(P<0.05), whereas SPN (CD43) mRNA was significantly decreased
(P<0.05).
 | Table IIExpression of genes associated with T
cell-dependent B-cell activation. |
Table II
Expression of genes associated with T
cell-dependent B-cell activation.
| Symbol | PE (X±SD) | Control (X±SD) | P |
|---|
| TNFSF9 | 9.04±0.60 | 8.35±0.68 | 0.002a |
| CD80 | 5.56±0.76 | 5.68±0.78 | 0.616 |
| CD86 | 11.14±0.67 | 10.43±0.64 | 0.001a |
| ICOSLG | 9.81±0.33 | 9.45±0.47 | 0.008a |
| CD276 | 4.92±0.94 | 5.11±1.15 | 0.562 |
| BTLA | 10.03±0.69 | 10.07±0.81 | 0.865 |
| CD28 | 11.46±0.85 | 11.70±0.57 | 0.301 |
| CD37 | 16.28±0.73 | 15.49±0.69 | 0.001a |
| CD40 | 10.10±0.60 | 9.73±0.58 | 0.053 |
| SPN (CD43) | 14.30±0.75 | 14.79±0.49 | 0.020a |
| CD72 | 11.14±0.79 | 11.35±0.59 | 0.349 |
| CD84 | 12.27±0.57 | 11.91±0.67 | 0.069 |
| CD97 | 16.83±0.85 | 16.14±0.54 | 0.004a |
| LY9 | 12.52±0.61 | 12.65±0.50 | 0.451 |
| CTLA4 | 7.51±1.00 | 7.59±0.62 | 0.772 |
| CD226 | 12.14±0.71 | 12.04±0.39 | 0.598 |
| EMR2 | 14.73±0.77 | 14.18±0.53 | 0.011a |
| MS4A1 | 13.01±0.97 | 13.11±0.73 | 0.708 |
| TNFSF4 | 11.82±0.67 | 11.56±0.51 | 0.171 |
| PDCD1 | 11.50±0.56 | 11.86±0.58 | 0.052 |
| SLAMF1 | 11.29±0.53 | 11.53±0.55 | 0.158 |
Expression of genes associated with T
cell-independent B-cell activation
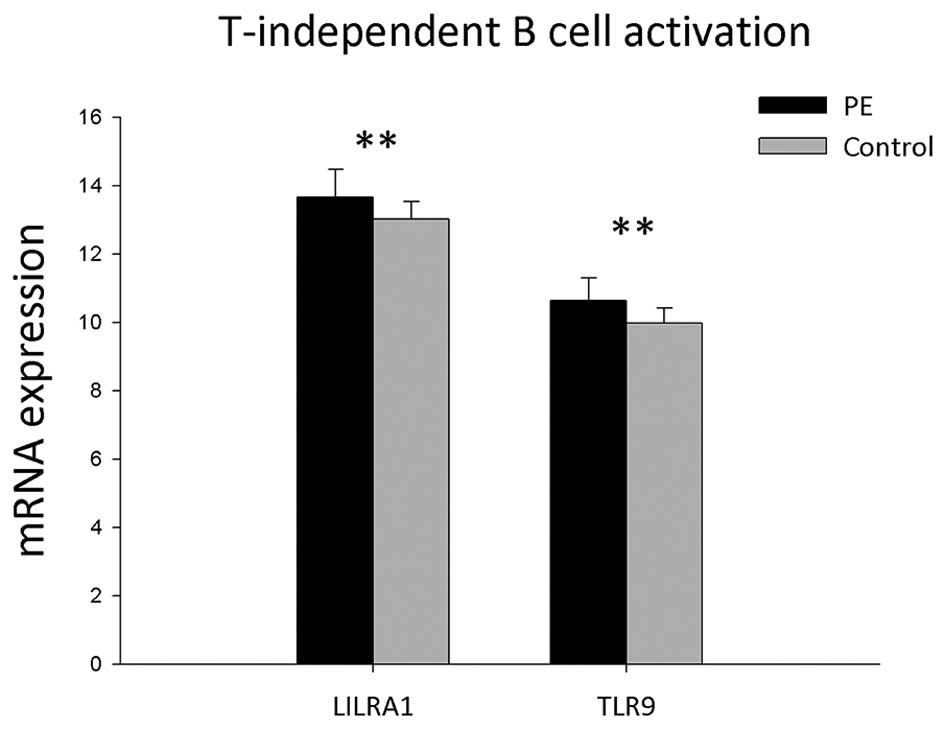
The results showed that mRNA expressions of seven
genes associated with T cell-independent B-cell activation in PBMCs
were detected in the PBMCs of the PE and control groups (Table III and Fig. 3). mRNA expression levels of LILRA1
and TLR9 were found to be significantly upregulated in PE patients
compared with those of the control group (P<0.05).
 | Table IIIExpression of genes associated with T
cell-independent B-cell activation. |
Table III
Expression of genes associated with T
cell-independent B-cell activation.
| Symbol | PE (X±SD) | Control (X±SD) | P |
|---|
| SLAMF8 | 10.83±0.91 | 11.20±0.58 | 0.131 |
| UBD | 6.23±0.82 | 6.51±0.50 | 0.196 |
| FCGR2B | 13.18±0.54 | 12.92±0.62 | 0.161 |
| FCGR3A | 18.76±0.38 | 18.72±0.39 | 0.702 |
| LILRA1 | 13.67±0.81 | 13.03±0.52 | 0.005a |
| CD180 | 11.70±0.57 | 11.42±0.57 | 0.128 |
| TLR9 | 10.64±0.66 | 9.97±0.46 | 0.001a |
Expression of B-cell activation
regulatory genes
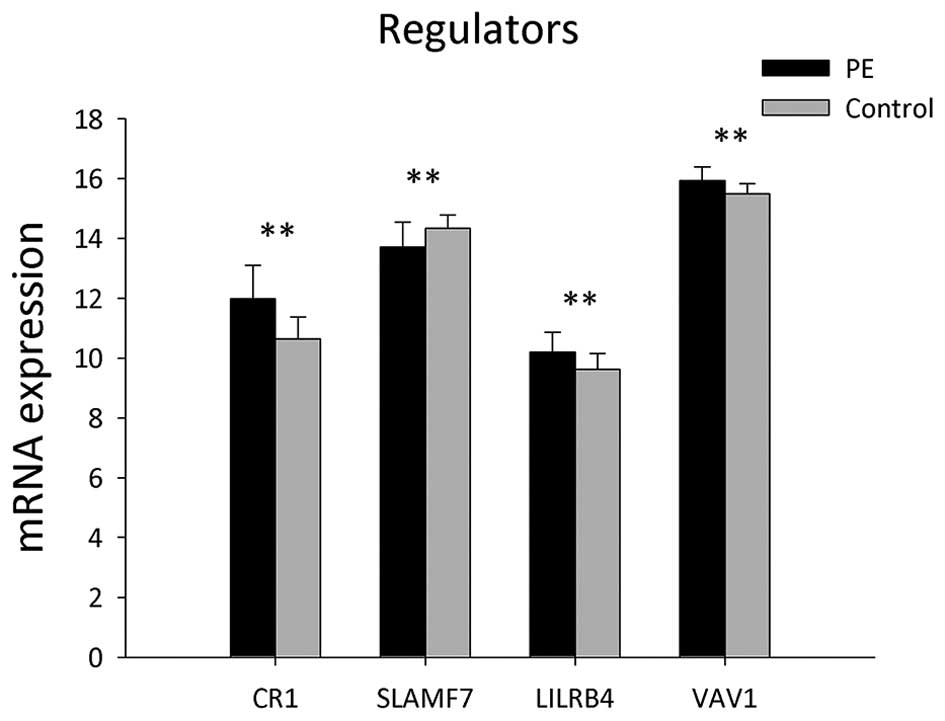
Ten genes involved in the regulation of B-cell
activation were identified in the PBMCs of the PE and control
groups (Table IV and Fig. 4). The expression levels of genes,
including CR1, LILRB4 and VAV1, were significantly increased in PE
patients compared to those of the heathy controls (P<0.05),
whereas expression levels of SLAMF7 were significantly decreased in
PE patients (P<0.05).
 | Table IVGene expression of B-cell
activation-associated regulators. |
Table IV
Gene expression of B-cell
activation-associated regulators.
| Symbol | PE (X±SD) | Control (X±SD) | P |
|---|
| CR1 (CD35) | 11.98±1.11 | 10.63±0.74 | 0.000a |
| SLAMF7 | 13.71±0.83 | 14.34±0.44 | 0.005a |
| IL4I1 | 10.97±0.80 | 11.01±0.65 | 0.867 |
| LILRB1 | 12.94±0.70 | 12.61±0.94 | 0.207 |
| LILRB4 | 10.19±0.66 | 9.62±0.52 | 0.004a |
| LILRB2 | 17.42±0.50 | 17.21±0.38 | 0.135 |
| LILRB3 | 18.05±0.47 | 17.82±0.42 | 0.112 |
| LILRA3 | 12.88±1.60 | 12.22±1.30 | 0.165 |
| LAIR1 | 13.53±0.52 | 13.22±0.50 | 0.061 |
| VAV1 | 15.93±0.45 | 15.49±0.33 | 0.001a |
| TLR9 | 10.64±0.66 | 9.97±0.46 | 0.001a |
Expression levels of B-cell
activation-associated cytokine mRNAs
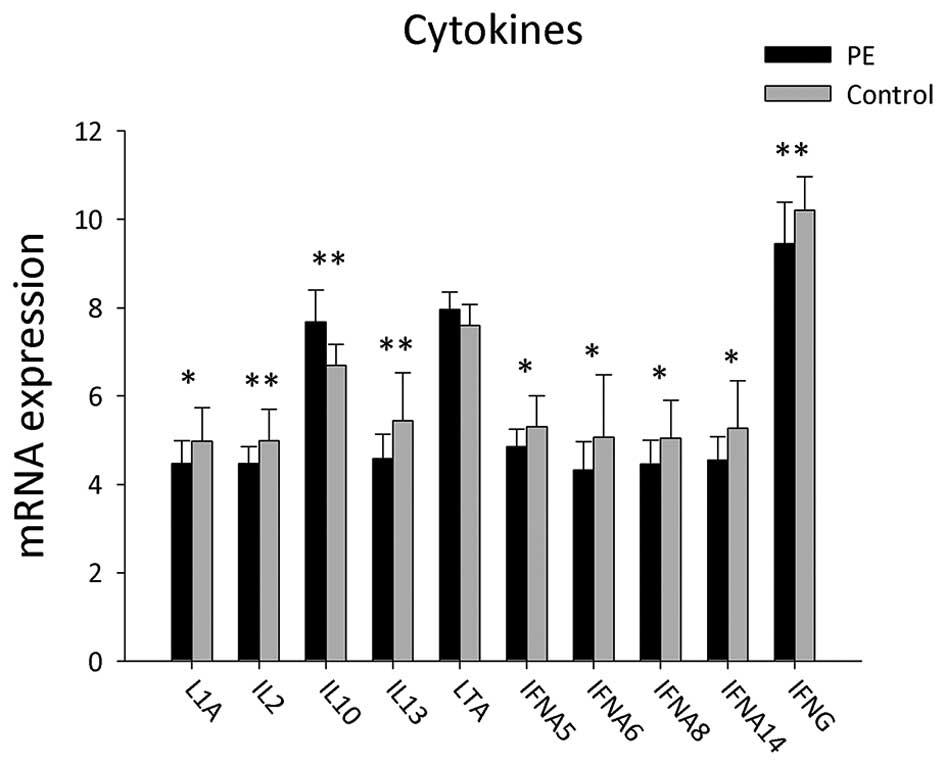
A total of 22 genes of cytokines involved in B-cell
activation were detected in the PBMCs of PE patients and healthy
controls (Table V and Fig. 5). mRNA expression levels of
cytokines, including LTA and IL10, were significantly upregulated
in PE patients compared to those in the control group (P<0.05),
whereas the expressions levels of IL1A, IFNA5, IFNA6, IFNA8,
IFNA14, IL2, IL13 and IFNG were significantly decreased in the
PBMCs of PE patients (P<0.05).
 | Table VGene expression of B-cell
activation-associated cytokines. |
Table V
Gene expression of B-cell
activation-associated cytokines.
| Symbol | PE (X±SD) | Control (X±SD) | P |
|---|
| L1A | 4.47±0.52 | 4.98±0.75 | 0.017a |
| L1B | 14.79±0.86 | 14.56±0.69 | 0.366 |
| IL2 | 4.47±0.38 | 4.99±0.71 | 0.006a |
| IL4 | 6.39±1.32 | 6.92±1.51 | 0.247 |
| IL5 | 5.10±0.96 | 4.86±0.71 | 0.372 |
| IL6 | 6.66±0.71 | 6.55±0.47 | 0.575 |
| IL10 | 7.68±0.72 | 6.69±0.48 | 0.000a |
| IL12A | 7.03±0.74 | 7.36±0.53 | 0.109 |
| IL12B | 5.46±1.48 | 5.39±0.94 | 0.847 |
| IL13 | 4.58±0.55 | 5.44±1.09 | 0.003a |
| TNF | 10.10±0.66 | 9.59±0.95 | 0.056 |
| LTA | 7.96±0.39 | 7.60±0.47 | 0.013a |
| IFNA2 | 4.50±0.60 | 4.87±0.71 | 0.081 |
| IFNA4 | 7.96±0.54 | 7.80±0.47 | 0.350 |
| IFNA5 | 4.85±0.40 | 5.31±0.70 | 0.014a |
| IFNA6 | 4.32±0.65 | 5.07±1.41 | 0.036a |
| IFNA8 | 4.46±0.54 | 5.04±0.86 | 0.014a |
| IFNA10 | 4.84±0.89 | 4.98±0.73 | 0.585 |
| IFNA14 | 4.55±0.53 | 5.27±1.07 | 0.011a |
| IFNA21 | 4.89±1.60 | 5.25±0.90 | 0.388 |
| IFNB1 | 5.05±0.94 | 5.31±1.17 | 0.443 |
| IFNG | 9.45±0.94 | 10.21±0.76 | 0.009a |
Discussion
A previous study demonstrated that mRNA expression
levels of genes associated with NK and T cells were downregulated
in PE patients (9); in addition,
NK-cell and T-cell function as well as mRNA expression of CD19 were
reported to be markedly decreased in VTE patients (10,11,13).
The present study identified the differential gene expression at
different stages of B-cell activation in patients with symptomatic
PE. B-cell activation requires two signals, of which the first is
dependent on antigen binding to the B-cell antigen receptor. If the
antigen is thymus-dependent, the second signal is initiated
following interactions between T cell and B cell; however,
thymus-independent antigens activate the second signal autonomously
(14).
The results of the present study showed that the
mRNA expression of genes associated with B-cell receptor-mediated
B-cell activation, including LYN, CD22, SYK, BTK, PTPRC and NFAM1,
were significantly upregulated in PE patients compared with the
control group. LYN mediates positive as well as negative roles in
B-cell activation (15) and CD22
is an inhibitory co-receptor, which negatively regulates B-cell
receptor signaling (16). B-cell
activation is initiated by SYK-dependent and -independent signaling
cascades (17). In addition, BTK
is involved in the regulation of actin dynamics required for
antigen processing and presentation in B cells (18). Furthermore, PTPRC-deficient B cells
were reported to exhibit impaired proliferation in vitro
(19) and the overexpression of
NFAM1 resulted in severe impairment of early B-cell development
(20).
However, in the present study, the mRNA expression
levels of FYN, FCRL4 and LAX1 genes associated with B-cell
receptor-mediated B-cell activation were significantly
downregulated in PE patients compared with those of the control
group. A previous study using FYN-deficient mice demonstrated that
FYN had a key role in B-cell activation (21). FCRL4 was found to inhibit B cell
receptor signaling in response to chronic antigenic stimulation
(22) and LAX was reported to be a
negative regulator of B-cell activation, as LAX-deficient B cells
were found to be hyper-responsive to B-cell receptor stimulation
(23).
These studies indicated that the differential
expression of genes associated with B-cell receptor-mediated B-cell
activation in PE patients and the control group in the present
study may have positively or negatively regulated B-cell
activation.
In the present study, the mRNA expression levels of
EMR2, TNFSF9, CD86, ICOSLG, CD37 and CD97 genes associated with T
cell-dependent B-cell activation were significantly upregulated in
PE patients compared with those of the control group. CD97, EMR2
and CD86 were previously reported to have important roles in the
interaction of T cells with B cells (24,25)
and the interaction of TNFSF9 and its receptor were shown to
positively regulate the humoral immune response (26). The ICOSLG gene encodes the ligand
for the inducible co-stimulator receptor, which was reported to
have an essential role in T cell-dependent B-cell responses
(27). In addition, one study
demonstrated that interactions between T cells and B cells were
significantly impaired in CD37-deficient mice (28).
The present study also demonstrated that expression
of the T cell-dependent B-cell activation gene SPN was
significantly downregulated in PE patients compared with that of
the control group; SPN downregulation has previously been reported
to enhance interactions between T cells and B cells (29). These studies therefore indicated
that in the present study, the differential gene expression of
genes involved in T cell-dependent B-cell activation may enhance
interactions between T cells and B cells in patients with
symptomatic PE compared to those in the healthy controls.
In the present study, seven genes associated with T
cell-independent B-cell activation were detected, of which LILRA1
and TLR9 were found to be significantly upregulated in the PBMCs of
PE patients compared with those of the control group. A previous
study detected LILRA1 mRNA in B cells (30), which was reported to activate cells
through associating with the Fc receptor gamma chain (31); in addition, TLR9 agonists were
reported to be involved in the activation of human B cells
(32). This therefore indicated
that T cell-independent B-cell activation was enhanced in PE
patients in the present study.
Increased gene expression of B-cell activation
regulators, including CR1, LILRB4 and VAV1, was observed in PE
patients compared to that of the control group in the present
study. CR1 inhibits B-cell receptor-mediated B-cell activation
(33) and LILRB4 negatively
regulates the activation of antigen-presenting cells (34); in addition, VAV1-deficient mice
were reported to have activation defects in B cells (35). In the present study, mRNA
expression of the B-cell activation regulator SLAMF7 was found to
be significantly downregulated in PE patients compared to that of
the control group. A previous study showed that SLAMF7 increased
the proliferation of B cells and autocrine cytokine expression on B
cells (36). These previous
studies therefore indicated that in the present study, B-cell
activation was inhibited in patients with symptomatic PE.
The results of the present study also demonstrated
differential gene expression of B-cell activation-associated
cytokines. mRNA expression levels of LTA and IL10 were
significantly increased in the PBMCs of PE patients compared to
those of the healthy controls, whereas expression levels of L1A,
IFNA5, IFNA6, IFNA8, IFNA14, IL2, IL13 and IFNG were found to be
significantly decreased in PE patients.
In addition to producing antibodies against
antigens, B cells also produce cytokines. IL13 is expressed in
activated T cells and has been shown to promote B-cell
proliferation as well as the synthesis of Immunoglobulin E
(37); furthermore, IFNA/B was
reported to induce partial B-cell activation (38). However, IFNG inhibited B-cell
differentiation (39). This
therefore indicated that the downregulation of cytokine genes
impaired B-cell function in PE patients enrolled in the present
study.
In conclusion, the present study demonstrated that
patients with symptomatic PE exhibited downregulation of genes
associated with the B-cell receptor and differential expression of
B-cell activation-regulator genes, indicating that B-cell function
was inhibited. In addition, the differential expression of genes
associated to T cell-dependent B-cell activation suggested that
interactions between T cells and B cells were enhanced in PE
patients, whereas the upregulation of genes associated with T
cell-independent B-cell activation indicated an increase in T
cell-indepenent B-cell activation. Furthermore, the downregulation
of B-cell activation-associated cytokines in PE patients indicated
that cytokine production by B cells was significantly reduced.
These results therefore demonstrated reduced or disorganized B-cell
function in patients with symptomatic PE, which is consistent with
the cytological results of a previous study (13).
Acknowledgements
The present study was supported by a grant from the
‘12th five year’ National Science and Technology Supporting Program
(no. 2011BAI11B16).
References
|
1
|
Geerts WH, Heit JA, Claggett GP, et al:
Prevention of venous thromboembolism. Chest. 119(1 Suppl):
132S–175S. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heit JA: The epidemiology of venous
thromboembolism in the community. Arterioscler Thromb Vasc Biol.
28:370–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martinelli I: Risk factors in venous
thromboembolism. Thromb Haemost. 86:395–403. 2001.PubMed/NCBI
|
|
4
|
Clagett GP, Anderson FA Jr, Heit J, Levine
MN and Wheeler HB: Prevention of venous thromboembolism. Chest.
108(4 Suppl): 312S–334S. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kahn SR, Lim W, Dunn AS, et al: American
College of Chest Physicians: Prevention of VTE in nonsurgical
patients: Antithrombotic therapy and prevention of thrombosis, 9th
ed: American College of Chest Physicians evidence-based clinical
practice guidelines. Chest. 141(2 Suppl): e195S–e226S. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shackford SR, Rogers FB, Terrien CM,
Bouchard P, Ratliff J and Zubis R: A 10-year analysis of venous
thromboembolism on the surgical service: the effect of practice
guidelines for prophylaxis. Surgery. 144:3–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smeeth L, Cook C, Thomas S, Hall AJ,
Hubbard R and Vallance P: Risk of deep vein thrombosis and
pulmonary embolism after acute infection in a community setting.
Lancet. 367:1075–1079. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiang-Hua Y, Le-Min W, Ai-Bin L, et al:
Severe acute respiratory syndrome and venous thromboembolism in
multiple organs. Am J Respir Crit Care Med. 182:436–437. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong Z, Liang AB, Wang LM, et al: The
expression and significance of immunity associated genes mRNA in
patients with pulmonary embolism. Chin J Intern Med. 48:666–669.
2009.(In Chinese).
|
|
10
|
Wang L, Song H, Gong Z, Duan Q and Liang
A: Acute pulmonary embolism and dysfunction of CD3+ CD8+ T cell
immunity. Am J Respir Crit Care Med. 184:13152011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haoming S, Lemin W, Zhu G, et al: T
cell-mediated immune deficiency or compromise in patients with
CTEPH. Am J Respir Crit Care Med. 183:417–418. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O’Leary JG, Goodarzi M, Drayton DL and von
Andrian UH: T cell- and B cell-independent adaptive immunity
mediated by natural killer cells. Nat Immunol. 7:507–516. 2006.
View Article : Google Scholar
|
|
13
|
Duan Q, Gong Z, Song H, et al: Symptomatic
venous thromboembolism is a disease related to infection and immune
dysfunction. Int J Med Sci. 9:453–461. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Janeway CA Jr, Travers P, Walport M and
Shlomchik MJ: Immunobiology: The Immune System in Health and
Disease. 5th edition. Garland Science; New York: 2001
|
|
15
|
Gauld SB and Cambier JC: Src-family
kinases in B-cell development and signaling. Oncogene.
23:8001–8006. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nitschke L and Tsubata T: Molecular
interactions regulate BCR signal inhibition by CD22 and CD72.
Trends Immunol. 25:543–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yokozeki T, Adler K, Lankar D and Bonnerot
C: B cell receptor-mediated Syk-independent activation of
phosphatidylinositol 3-kinase, Ras, and mitogen-activated protein
kinase pathways. J Immunol. 171:1328–1335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sharma S, Orlowski G and Song W: Btk
regulates B cell receptor-mediated antigen processing and
presentation by controlling actin cytoskeleton dynamics in B cells.
J Immunol. 182:329–339. 2009. View Article : Google Scholar
|
|
19
|
Huntington ND, Xu Y, Puthalakath H, et al:
CD45 links the B cell receptor with cell survival and is required
for the persistence of germinal centers. Nat Immunol. 7:190–198.
2006. View
Article : Google Scholar
|
|
20
|
Ohtsuka M, Arase H, Takeuchi A, et al:
NFAM1, an immunoreceptor tyrosine-based activation motif-bearing
molecule that regulates B cell development and signaling. Proc Natl
Acad Sci USA. 101:8126–8131. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Horikawa K, Nishizumi H, Umemori H, Aizawa
S, Takatsu K and Yamamoto T: Distinctive roles of Fyn and Lyn in
IgD- and IgM-mediated signaling. Int Immunol. 11:1441–1449. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sohn HW, Krueger PD, Davis RS and Pierce
SK: FcRL4 acts as an adaptive to innate molecular switch dampening
BCR signaling and enhancing TLR signaling. Blood. 118:6332–6341.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu M, Granillo O, Wen R, et al: Negative
regulation of lymphocyte activation by the adaptor protein LAX. J
Immunol. 174:5612–5619. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kwakkenbos MJ, Pouwels W, Matmati M, et
al: Expression of the largest CD97 and EMR2 isoforms on leukocytes
facilitates a specific interaction with chondroitin sulfate on B
cells. J Leukoc Biol. 77:112–119. 2005.
|
|
25
|
McLellan AD, Starling GC, Williams LA,
Hock BD and Hart DN: Activation of human peripheral blood dendritic
cells induces the CD86 co-stimulatory molecule. Eur J Immunol.
25:2064–2068. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu G, Flies DB, Tamada K, Sun Y,
Rodriguez M, Fu YX and Chen L: Progressive depletion of peripheral
B lymphocytes in 4-1BB (CD137) ligand/I-Ealpha-transgenic mice. J
Immunol. 167:2671–2676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mak TW, Shahinian A, Yoshinaga SK, et al:
Costimulation through the inducible costimulator ligand is
essential for both T helper and B cell functions in T
cell-dependent B cell responses. Nat Immunol. 4:765–772. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Knobeloch KP, Wright MD, Ochsenbein AF, et
al: Targeted inactivation of the tetraspanin CD37 impairs
T-cell-dependent B-cell response under suboptimal costimulatory
conditions. Mol Cell Biol. 20:5363–5369. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ostberg JR, Dragone LL, Driskell T, et al:
Disregulated expression of CD43 (leukosialin, sialophorin) in the B
cell lineage leads to immunodeficiency. J Immunol. 157:4876–4884.
1996.PubMed/NCBI
|
|
30
|
Tedla N, Gibson K, McNeil HP, Cosman D,
Borges L and Arm JP: The co-expression of activating and inhibitory
leukocyte immunoglobulin-like receptors in rheumatoid synovium. Am
J Pathol. 160:425–431. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Colonna M, Nakajima H, Navarro F and
López-Botet M: A novel family of Ig-like receptors for HLA class I
molecules that modulate function of lymphoid and myeloid cells. J
Leukoc Biol. 66:375–381. 1999.PubMed/NCBI
|
|
32
|
Hanten JA, Vasilakos JP, Riter CL, et al:
Comparison of human B-cell activation by TLR7 and TLR9 agonists.
BMC Immunol. 9:392008. View Article : Google Scholar
|
|
33
|
Józsi M, Prechl J, Bajtay Z and Erdei A:
Complement receptor type 1 (CD35) mediates inhibitory signals in
human B lymphocytes. J Immunol. 168:2782–2788. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cella M, Döhring C, Samaridis J, et al: A
novel inhibitory receptor (ILT3) expressed on monocytes,
macrophages, and dendritic cells involved in antigen processing. J
Exp Med. 185:1743–1751. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fujikawa K, Miletic AV, Alt FW, et al:
Vav1/2/3-null mice define an essential role for Vav family proteins
in lymphocyte development and activation but a differential
requirement in MAPK signaling in T and B cells. J Exp Med.
198:1595–1608. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JK, Mathew SO, Vaidya SV, Kumaresan PR
and Mathew PA: CS1 (CRACC, CD319) induces proliferation and
autocrine cytokine expression on human B lymphocytes. J Immunol.
179:4672–4678. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Defrance T, Carayon P, Billian G, et al:
Interleukin 13 is a B cell stimulating factor. J Exp Med.
179:135–143. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Braun D, Caramalho I and Demengeot J:
IFN-alpha/beta enhances BCR-dependent B cell responses. Int
Immunol. 14:411–419. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Abed NS, Chace JH, Fleming AL and Cowdery
JS: Interferon-gamma regulation of B lymphocyte differentiation:
activation of B cells is a prerequisite for IFN-gamma-mediated
inhibition of B cell differentiation. Cell Immunol. 153:356–366.
1994. View Article : Google Scholar : PubMed/NCBI
|