Introduction
Low-density lipoprotein cholesterol (LDL-C) has been
identified to have a crucial and causal role in the genesis of
coronary heart disease and atherosclerotic cardiovascular disease
(ASCVD) (1). There is notable
evidence to suggest that higher LDL-C levels are correlated with
greater ASCVD risk, and that lowering cholesterol levels reduces
ASCVD events (2–5). Hepatic LDL receptor (LDLR) is
essential for the uptake of extracellular LDL-C (6). As such, it is a determinant regulator
of the LDL-C metabolism. One of the most optimal strategies to
lower LDL-C is to upregulate and stabilize hepatic LDLR
expression.
The abundance of LDLR is noted on the
transcriptional and post-transcriptional levels. On the
transcriptional level, LDLR is tightly regulated by sterol response
element binding protein 2 (SREBP2) (7,8).
Cellular cholesterol depletion activates the nuclear translocation
of SREBP2, and subsequently SREBP2 activates LDLR and proprotein
convertase subtilisin/kexin type 9 (PCSK9) gene expression
(7–9). PCSK9 plays a pivotal role in the
post-transcriptional regulation of LDLR, in which it binds to the
extracellular domain LDLR and directs the trafficking of it to the
lysosomes for degradation (10).
Previous studies have identified a new significant degrader of
LDLR, named inducible degrader of the LDLR (Idol), which is an E3
ubiquitin ligase that triggers ubiquitination of LDLR on its
cytoplasmic domain and promotes lysosomal degradation (11). Distinct from PCSK9, Idol is
regulated by another sterol-dependent nuclear receptor, liver X
receptor α (LXRα), which is activated in response to cellular
cholesterol excess (12).
Omega-3 fatty acids, including docosahexanoic acid
(DHA, 22:6n−3) and eicosapentaenoic acid (EPA, 20:5n−3), are also
known as marine fatty acids, and are delivered from dietary fish
oil. In vivo and in vitro studies confirmed that DHA
and EPA are potent inhibitors of LXRα (13,14).
In addition, several studies have identified that omega-3 fatty
acids upregulate LDLR abundance (15,16).
However, little is known about the mechanism by which fatty acids
regulate LDLR. We considered that the newly identified LXRα target
gene Idol may participate in this process. In the present study, we
selected DHA as one type of omega-3 fatty acid to search for the
possible mechanism by which these fatty acids exert their
regulatory effects on LDLR. The results revealed that DHA increased
hepatic LDLR abundance through the suppression of Idol expression
rather than through gene expression. Furthermore, this repression
of LXRα activity by DHA and the subsequent inhibition of the
expression of Idol is one of multiple mechanisms.
Materials and methods
Reagents
Palmitic acids (PA), DHA and 25-hydroxycholesterol
were purchased from Sigma-Aldrich (St. Louis, MO, USA). Fatty
acid-free bovine serum albumin (BSA) was obtained from MP
Biomedicals (Santa Ana, CA, USA). Cell culture reagents were
purchased from Gibco (Carlsbad, CA, USA). BCA protein assay was
purchased from Thermo Fisher Scientific (Waltham, MA, USA). PVDF
membranes and the ECL western blot system were provided by Merck
Millipore (Billerica, MA, USA). Anti-LXR alpha antibody (ab176323)
and anti-Idol antibody (ab74562) were obtained from Abcam
(Cambridge, MA, USA); anti-LDLR antibody (10007665) from Cayman
Chemical Co. (Ann Arbor, MI, USA); and anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH) antibody, peroxidase-conjugated
anti-mouse antibody and anti-rabbit antibody from Proteintech Group
(Chicago, IL, USA). The Ultrapure RNA kit was purchased from CWBIO
(Beijing, China). The RevertAid First Strand cDNA synthesis kit
(K1622) was purchased from Thermo Fisher Scientific. iTaq™
Universal SYBR® Green supermix (172-5121) was purchased
from Bio-Rad (Hercules, CA, USA).
Cell cultures and treatments
The human hepatoma HepG2 cell line, obtained from
Laboratory Animal Center (Sun Yat-sen University, Guangzhou,
China), was cultured in low-glucose Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in a
humidified atmosphere of 5% CO2. For experiments, cells
were seeded at a density of 1×106 cells per well and
allowed to adhere for 24 h. Each fatty acid was dissolved in
ethanol and mixed with fatty acid-free BSA at 2.5:1 molar ratios,
working as a fatty acid/BSA complex. 25-hydroxycholesterol was
dissolved in ethanol and used at a concentration of 10 μmol/l.
Following the replacement of serum-free medium at 24 h, fatty
acid/BSA complexes were added to the culture dishes at a
concentration of 100 μmol/l fatty acid and 0.25% BSA, with or
without 25-hydroxycholesterol. Control cells were treated with BSA
vehicle with or without 25-hydroxycholesterol. For dose-response
experiments, 25-hydroxycholesterol and increasing concentrations
(50, 100 and 200 μmol/l) of DHA were added to the culture media.
After 24 h, cells were either harvested for protein extraction or
RNA isolation.
Western blot analysis
Following the treatments, cells were washed with
phosphate-buffered saline, lysed in cell lysis buffer and incubated
at 4°C for 20 min, then centrifuged at 10,000 × g for 10 min at
4°C. Protein concentration was measured using the BCA method.
Twenty micrograms of total proteins from each extract were
separated by 8% or 10% SDS-polyacrylamide gels and transferred onto
PVDF membranes in a cooling system at 100 V for 2 h. Membranes were
blocked by 5% non-fat dried milk in Tris-buffered saline with 0.1%
Tween (TBS-T) for 1 h at room temperature. Membranes were then
incubated with anti-GAPDH (diluted 1:20,000), anti-LDLR (diluted
1:200), anti-LXRα (diluted 1:1,000) or anti-Idol (diluted 1:1,000)
overnight at 4°C, washed three times with TBS-T and incubated with
peroxidase-conjugated secondary antibodies (anti-mouse or
anti-rabbit, respectively, diluted 1:20,000) for 1 h at room
temperature. Specific bands were then detected by the ECL western
blot system. Antibodies against GAPDH were used as the normalizing
control.
RNA isolation, cDNA synthesis and
quantitative polymerase chain reaction (qPCR)
Total RNA was isolated with the Ultrapure RNA kit
and cDNA synthesized with the RevertAid First Strand cDNA synthesis
kit from 1 μg total RNA. Primers for mRNA detection were designed
and synthesized by Sangon Biotech (Shanghai, China). qPCR was
carried out on a CFX96 real-time machine (Bio-Rad) using the SYBR
Green polymerase by the ΔΔCt method. Values were normalized to
GAPDH levels.
Statistical analysis
Duplicates were used in all experiments and
experiments were repeated at least three times. Significant
differences between the control and treatment groups were assessed
by one-way ANOVA with a Bonferroni post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
DHA increases the amount of LDLR protein
in a dose-dependent manner
To examine the effect of DHA on the expression of
LDLR in vitro, the abundance of LDLR in HepG2 cells was
detected by western blot analysis following exposure to the
respective fatty acid for 24 h. Compared with HepG2 cells cultured
with DMEM, LDLR protein decreased by 77% (P<0.05) when treated
with 25-hydroxycholesterol (10 μmol/l; Fig. 1A). In the presence of
25-hydroxycholesterol, HepG2 cells were treated with BSA vehicle,
PA and DHA, respectively. BSA plus 25-hydroxycholesterol had no
significant effect on LDLR protein compared with
25-hydroxycholesterol alone. With co-treatment of
25-hydroxycholesterol, DHA, but not PA, upregulated LDLR protein
levels 1.4-fold compared with control cells treated with BSA
vehicle (i.e., DHA significantly attenuated the suppressive effects
of 25-hydroxycholesterol on LDLR protein abundance; Fig. 1A). However, there was no
significant difference in LDLR expression between the cells treated
with DHA and with DMEM only (data not shown).
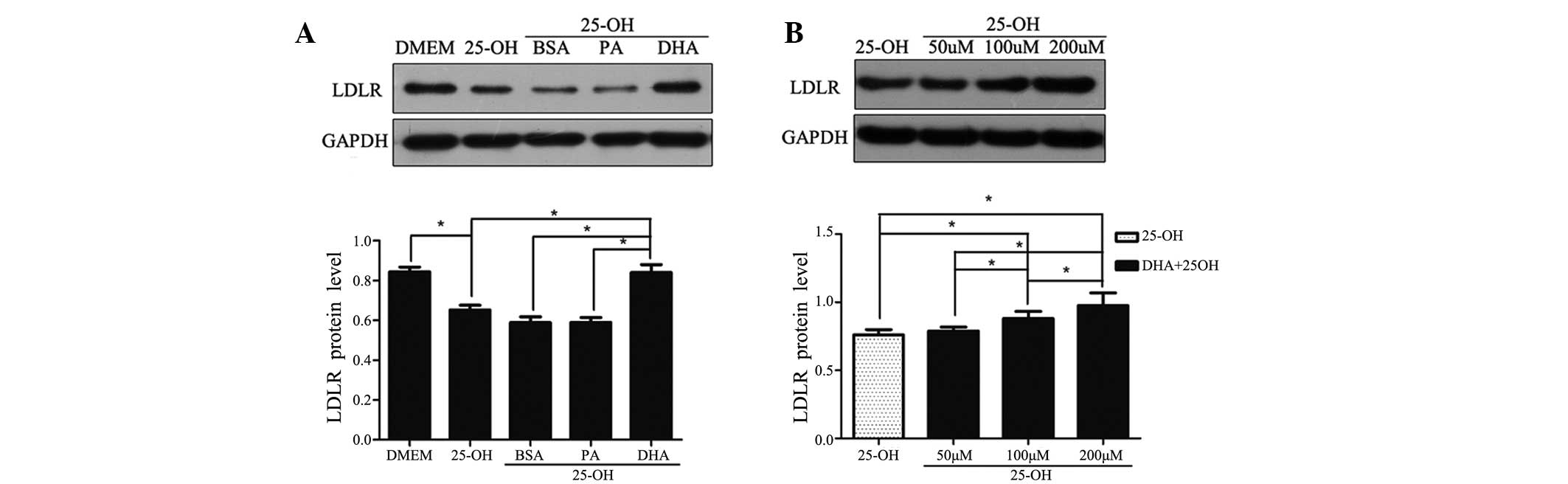 | Figure 1Regulatory effects of LDLR protein on
fatty acid treatment. Following overnight culture in a serum-free
medium, HepG2 cells were exposed to 25-hydroxycholesterol (10
μmol/l) with or without fatty acid/BSA complexes (100 μmol/l) for
24 h (A). HepG2 cells were exposed to 25-hydroxycholesterol (10
μmol/l) with DHA at various concentrations (50, 100 and 200 μmol/l)
(B). At the end of the treatment, total cell lysates were isolated
for western blot analysis for LDLR and GAPDH. The data shown are
the mean (± SEM) of at least three separate experiments
(*P<0.05). LDLR, low-density lipoprotein receptor;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; DMEM, Dulbecco’s
modified Eagle’s medium; 25-OH, 25-hydroxycholesterol; BSA, bovine
serum albumin; PA, palmitic acid; DHA, docosahexanoic acid. |
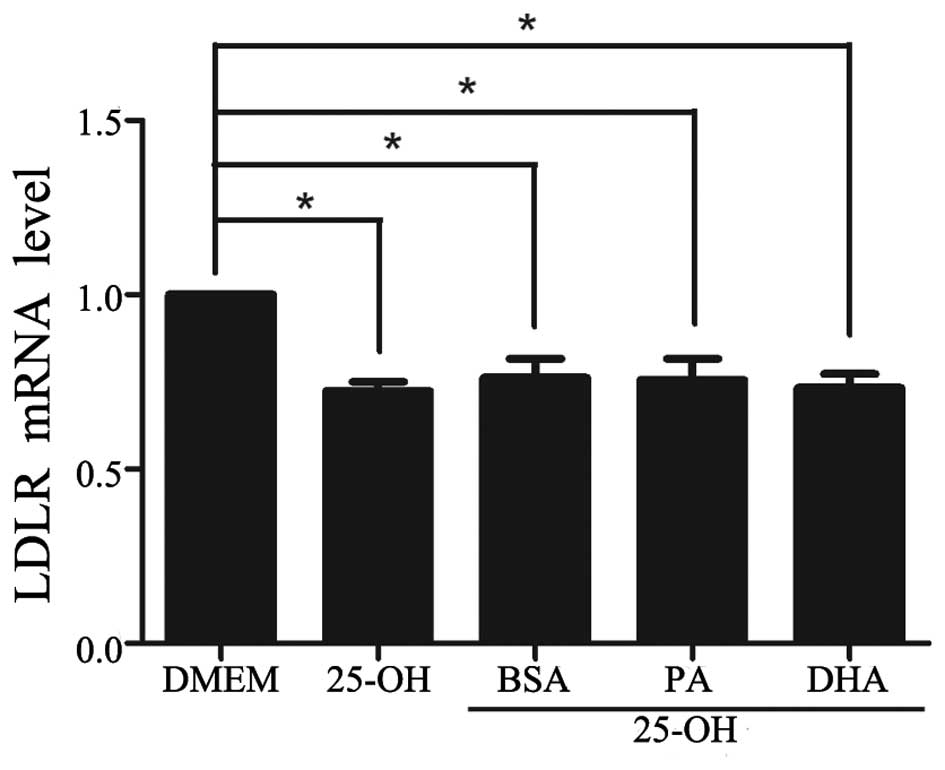
Experiments were performed to determine LDLR
expression in response to various concentrations of DHA with
25-hydroxycholesterol co-treatment. At a concentration of 50
μmol/l, the amount of LDLR protein did not differ from that
observed in control cells treated with BSA vehicle. A
dose-dependent increase in LDLR protein abundance was only observed
in DHA-treated cells above concentrations of 100 μmol/l, confirming
its inductive effect on LDLR expression (Fig. 1B).
DHA has no significant effect on the
regulation of LDLR mRNA
To study whether the increase in LDLR protein levels
by DHA was due to the upregulation of LDLR gene expression, the
amount of LDLR mRNA was quantified by qPCR. Compared with HepG2
cells cultured with DMEM, LDLR mRNA decreased by 72% (P<0.05)
when treated with 25-hydroxycholesterol (10 μmol/l; Fig. 2). With co-treatment of
25-hydroxycholesterol, neither DHA nor PA had a significant effect
on LDLR mRNA compared with control cells treated with BSA vehicle
(Fig. 2). Despite a DHA-induced
moderate increase in LDLR protein abundance, DHA did not
significantly antagonize the inhibition of 25-hydroxycholesterol on
LDLR mRNA, even at the highest dose applied.
DHA exerts a downregulatory effect on the
expression of LXRα
The expression of LXRα was detected on the mRNA and
protein levels. Compared with HepG2 cells cultured with DMEM, LXRα
protein was increased 1.4-fold (P<0.05) when treated with
25-hydroxycholesterol (10 μmol/l; Fig.
3A). With co-treatment of 25-hydroxycholesterol, DHA, but not
PA, significantly downregulated the protein level of LXRα by 50%
compared with control cells treated with BSA vehicle. Parallel
alteration was observed with the LXRα mRNA levels (Fig. 3B).
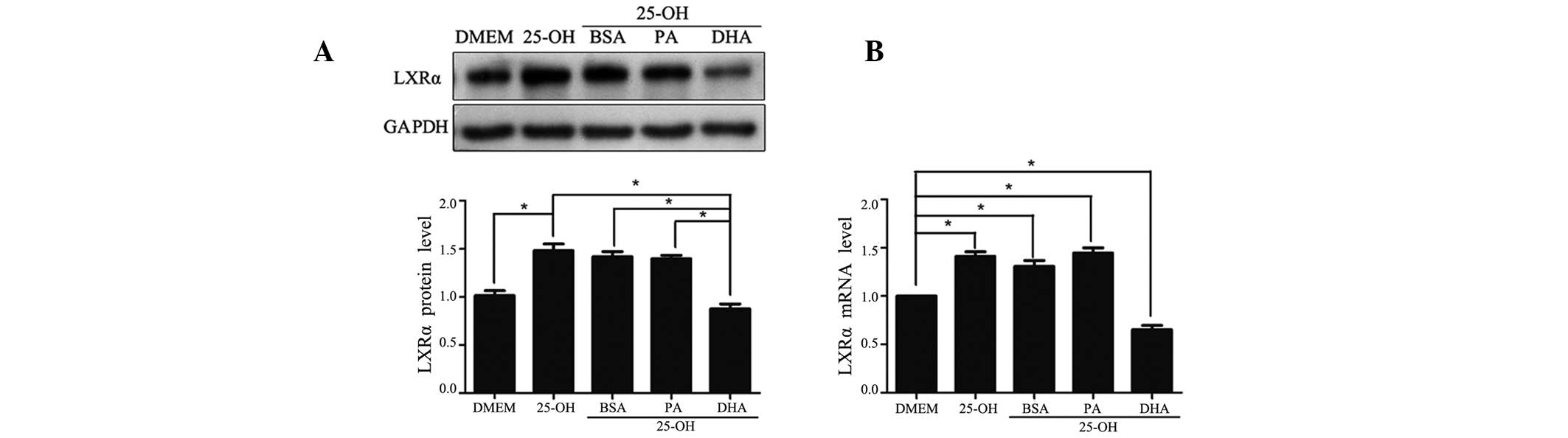 | Figure 3Regulatory effects of LXRα expression
on fatty acid treatment. Following overnight culture in a
serum-free medium, HepG2 cells were exposed to
25-hydroxycholesterol (10 μmol/l) with or without fatty acid/BSA
complexes (100 μmol/l) for 24 h. At the end of the treatment, total
cell lysates were isolated for western blot analysis for LXRα and
GAPDH (A), and total RNA was isolated for quantitative polymerase
chain reaction analysis of target genes (B). The data shown are the
mean (± SEM) of three separate experiments (*P<0.05).
LXRα, liver X receptor α; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; DMEM, Dulbecco’s modified Eagle’s medium; 25-OH,
25-hydroxycholesterol; BSA, bovine serum albumin; PA, palmitic
acid; DHA, docosahexanoic acid. |
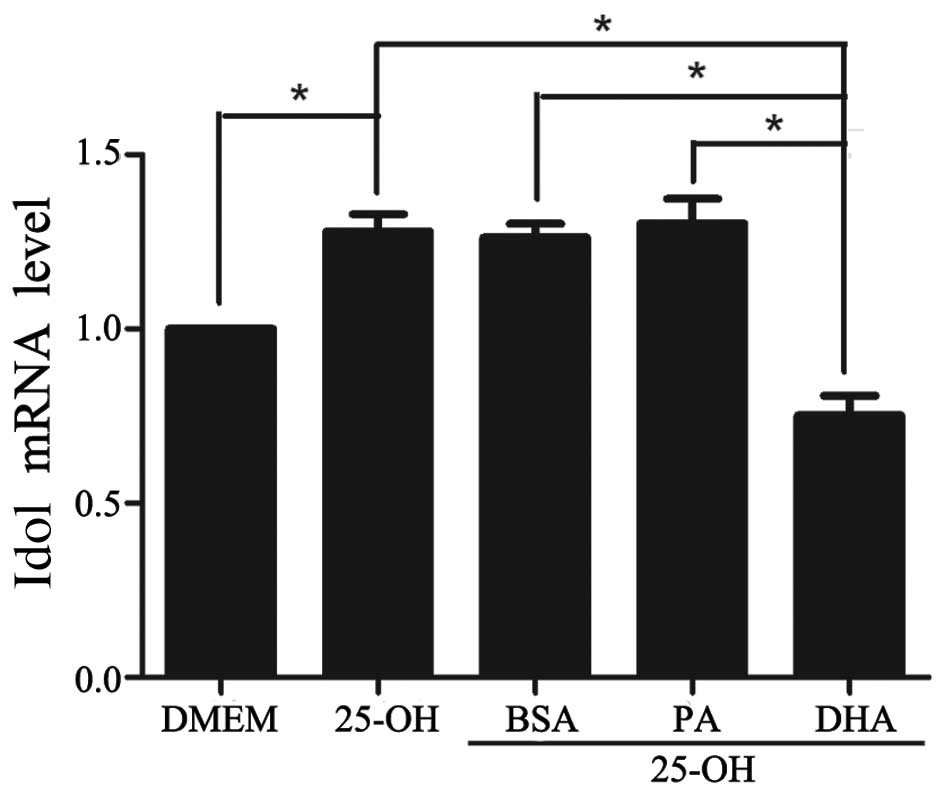
DHA decreases the expression of Idol in a
dose-dependent manner
Since DHA exerted a downregulatory effect on the
expression of LXRα, we evaluated the change in Idol, which is the
downstream protein of the nuclear receptor LXRα. As expected, the
expression of Idol was increased by 25-hydroxycholesterol and
decreased by DHA on the mRNA and protein levels, by 60% and 62%,
respectively, in coordination with the change in LXRα expression
(Figs. 4 and 5). In addition, a dose-dependent decrease
in the amount of Idol protein was observed with the various doses
of DHA (Fig. 4B).
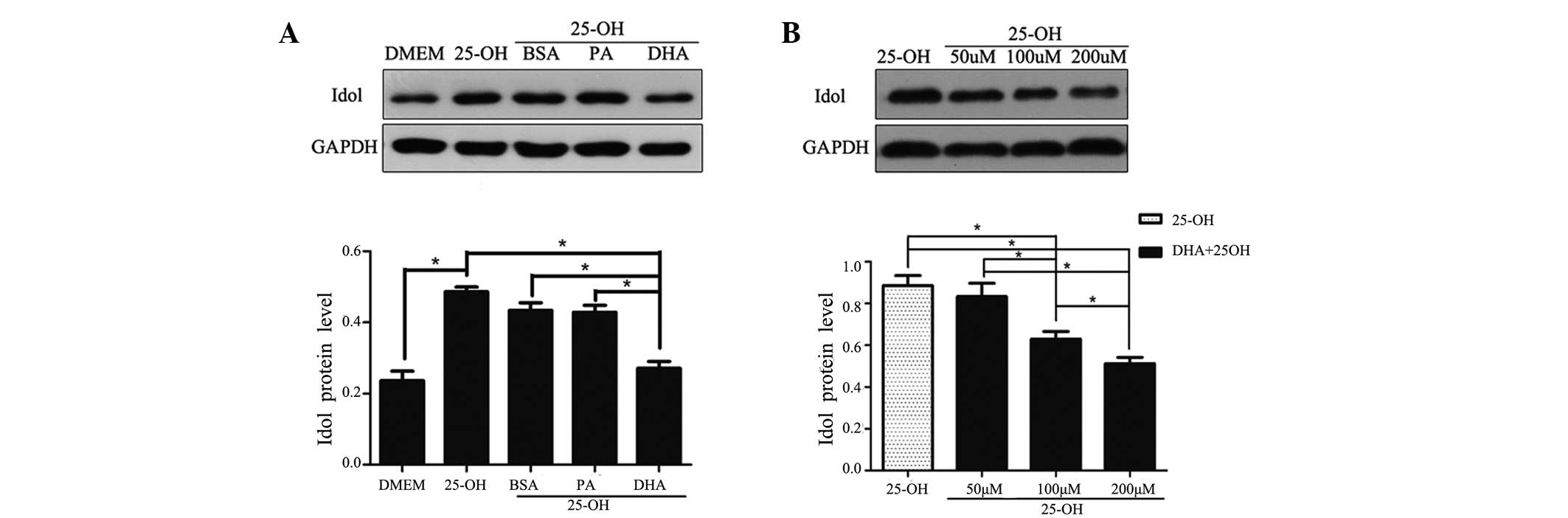 | Figure 4Regulatory effects of Idol protein on
fatty acid treatment. Following overnight culture in a serum-free
medium, HepG2 cells were exposed to 25-hydroxycholesterol (10
μmol/l) with or without fatty acid/BSA complexes (100 μmol/l) for
24 h (A). HepG2 cells were exposed to 25-hydroxycholesterol (10
μmol/l) with DHA at various concentrations (50, 100 and 200 μmol/l)
(B). At the end of the treatment, total cell lysates were isolated
for western blot analysis for Idol and GAPDH. The data shown are
the mean (± SEM) of at least three separate experiments
(*P<0.05). Idol, inducible degrader of the
low-density lipoprotein receptor; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; DMEM, Dulbecco’s modified Eagle’s medium; 25-OH,
25-hydroxycholesterol; BSA, bovine serum albumin; PA, palmitic
acid; DHA, docosahexanoic acid. |
Discussion
Idol has been identified as a novel
post-transcriptional regulator of LDLR abundance, as its full name
implies. Containing a unique C-terminal RING domain, Idol is an E3
ubiquitin ligase that triggers ubiquitination of LDLR and promotes
its internalization and degradation (11,17).
Distinct from LDLR and PCSK9 genes, Idol is directly regulated by
LXRα, which is activated in response to cellular cholesterol excess
(12). Conversely, the expression
of SREBP2 is responsive to cellular cholesterol depletion (7). Therefore, the LXRα-Idol-LDLR pathway
and the SREBP2-PCSK9-LDLR pathway are complementary but independent
pathways in the response to cellular sterol status.
As one type of oxysterol, 25-hydroxycholesterol
strongly represses the SREBP2 process and slightly activates the
LXRα pathway (18). Since LDLR is
the downstream protein in the two pathways, the net effect of
25-hydroxycholesterol is the downregulation of LDLR abundance.
Therefore, in the present study, HepG2 cells were initially treated
with 25-hydroxycholesterol to decrease basal levels of the LDLR
protein as previously described (16). In the presence of
25-hydroxycholesterol, DHA significantly increased LDLR protein
level in a dose-dependent manner over 100 μmol/l.
Although numerous in vivo and in vitro
studies have been conducted, the mechanism by which omega-3 fatty
acids exerted their effect on LDLR expression remained unclear.
Previous studies have indicated that multiple mechanisms are
involved in regulating the LDLR gene independently of SREBP1
(16,19). Conversely, a number of studies
demonstrated that DHA inhibited lipogenic gene transcription by
suppressing the expression of SREBP1, possibly at the
post-transcriptional level (20,21).
There was no evidence that LDLR played a role in this; however, DHA
downregulated the hepatic mRNA of SREBP2, and LDLR was observed in
hamsters fed a high cholesterol diet (22).
The present study revealed that DHA had no effect on
LDLR mRNA levels even at the highest dose applied, suggesting that
DHA may affect LDLR via mechanisms other than gene expression. In
addition, a number of studies demonstrated that DHA inhibits the
activity of LXRα (13,14). Furthermore, LXRα controls the
activation of the transcription of Idol (11). Therefore, we speculated that the
unclear mechanism by which DHA increased LDLR abundance is most
likely mediated by suppression of the LXRα-Idol pathway.
When delivered with the appropriate treatment, we
observed that 25-hydroxycholesterol activated LXRα expression since
it is an agonist of LXRα. In line earlier findings, DHA
significantly repressed the expression of LXRα at the mRNA and
protein levels when administered with 25-hydroxycholesterol
(16). As expected, the LXRα
target gene Idol was significantly decreased on DHA treatment.
Consistent with the change in mRNA levels, DHA reduced Idol
protein. Moreover, the reduction of Idol abundance presented in a
dose-dependent manner, corresponding with the alteration of LDLR
protein but in the opposite manner.
All of these results confirmed that DHA suppressed
the LXRα-Idol pathway, and in turn lowered the Idol-induced
degradation of LDLR protein, leading to the upregulation of
LDLR.
However, there was no significant difference in the
LDLR abundance between DHA treatment and BSA vehicle or PA
treatment when 25-hydroxycholesterol was absent. We therefore
speculated that DHA possibly exerted an upregulatory effect of LDLR
abundance under the condition of high cholesterol levels. In our
study, DHA significantly attenuated the suppression effect of
25-hydroxycholesterol on LDLR abundance, as well as the LXRα-Idol
pathway. That is, DHA modified LDLR abundance via suppression of
the LXRα-Idol pathway. The findings of the present study suggest
that in addition to the suppression of the LXRα-Idol pathway, there
are multiple mechanisms participating in the regulation of LDLR by
DHA treatment, and further exploration is required.
In conclusion, we identified that DHA increased
hepatic LDLR abundance in the presence of 25-hydroxycholesterol.
Multiple mechanisms are involved in DHA regulating the LDLR
abundance, and the suppression of LXRα-Idol pathway is one such
mechanism.
References
|
1
|
Stone NJ, Robinson JG, Lichtenstein AH, et
al: 2013 ACC/AHA guideline on the treatment of blood cholesterol to
reduce atherosclerotic cardiovascular risk in adults: a report of
the American College of Cardiology/American Heart Association Task
Force on Practice Guidelines. J Am Coll Cardiol. 63:2889–2934.
2014. View Article : Google Scholar
|
|
2
|
Amarenco P, Labreuche J, Lavallée P and
Touboul PJ: Statins in stroke prevention and carotid
atherosclerosis: systematic review and up-to-date meta-analysis.
Stroke. 35:2902–2909. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
ACCORD Study Group. Ginsberg HN, Elam MB,
Lovato LC, et al: Effects of combination lipid therapy in type 2
diabetes mellitus. N Engl J Med. 362:1563–1574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ingelsson E, Schaefer EJ, Contois JH, et
al: Clinical utility of different lipid measures for prediction of
coronary heart disease in men and women. JAMA. 298:776–785. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pedersen TR, Faergeman O, Kastelein JJ, et
al: High-dose atorvastatin vs usual-dose simvastatin for secondary
prevention after myocardial infarction: the IDEAL study: a
randomized controlled trial. JAMA. 294:2437–2445. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamamoto T, Davis CG, Brown MS, et al: The
human LDL receptor: a cysteine-rich protein with multiple Alu
sequences in its mRNA. Cell. 39:27–38. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hua X, Yokoyama C, Wu J, et al: SREBP-2, a
second basic-helix-loop-helix-leucine zipper protein that
stimulates transcription by binding to a sterol regulatory element.
Proc Natl Acad Sci USA. 90:11603–11607. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawabe Y, Suzuki T, Hayashi M, Hamakubo T,
Sato R and Kodama T: The physiological role of sterol regulatory
element-binding protein-2 in cultured human cells. Biochim Biophys
Acta. 1436:307–318. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeong HJ, Lee HS, Kim KS, Kim YK, Yoon D
and Park SW: Sterol-dependent regulation of proprotein convertase
subtilisin/kexin type 9 expression by sterol-regulatory element
binding protein-2. J Lipid Res. 49:399–409. 2008. View Article : Google Scholar
|
|
10
|
Fisher TS, Lo Surdo P, Pandit S, et al:
Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent
LDL receptor regulation. J Biol Chem. 282:20502–20512. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zelcer N, Hong C, Boyadjian R and Tontonoz
P: LXR regulates cholesterol uptake through Idol-dependent
ubiquitination of the LDL receptor. Science. 325:100–104. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zelcer N and Tontonoz P: Liver X receptors
as integrators of metabolic and inflammatory signaling. J Clin
Invest. 116:607–614. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pawar A, Botolin D, Mangelsdorf DJ and
Jump DB: The role of liver X receptor-alpha in the fatty acid
regulation of hepatic gene expression. J Biol Chem.
278:40736–40743. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vanden Heuvel JP: Cardiovascular
disease-related genes and regulation by diet. Curr Atheroscler Rep.
11:448–455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mustad VA, Ellsworth JL, Cooper AD,
Kris-Etherton PM and Etherton TD: Dietary linoleic acid increases
and palmitic acid decreases hepatic LDL receptor protein and mRNA
abundance in young pigs. J Lipid Res. 37:2310–2323. 1996.PubMed/NCBI
|
|
16
|
Yu-Poth S, Yin D, Kris-Etherton PM, Zhao G
and Etherton TD: Long-chain polyunsaturated fatty acids upregulate
LDL receptor protein expression in fibroblasts and HepG2 cells. J
Nutr. 135:2541–2545. 2005.PubMed/NCBI
|
|
17
|
Sorrentino V, Scheer L, Santos A, Reits E,
Bleijlevens B and Zelcer N: Distinct functional domains contribute
to degradation of the low density lipoprotein receptor (LDLR) by
the E3 ubiquitin ligase inducible Degrader of the LDLR (IDOL). J
Biol Chem. 286:30190–30199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Janowski BA, Willy PJ, Devi TR, Falck JR
and Mangelsdorf DJ: An oxysterol signalling pathway mediated by the
nuclear receptor LXR alpha. Nature. 383:728–731. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Field FJ, Born E and Mathur SN: Fatty acid
flux suppresses fatty acid synthesis in hamster intestine
independently of SREBP-1 expression. J Lipid Res. 44:1199–1208.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Nakamura MT, Cho HP and Clarke SD:
Sterol regulatory element binding protein-1 expression is
suppressed by dietary polyunsaturated fatty acids. A mechanism for
the coordinate suppression of lipogenic genes by polyunsaturated
fats. J Biol Chem. 274:23577–23583. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu J, Teran-Garcia M, Park JH, Nakamura MT
and Clarke SD: Polyunsaturated fatty acids suppress hepatic sterol
regulatory element-binding protein-1 expression by accelerating
transcript decay. J Biol Chem. 276:9800–9807. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen J, Jiang Y, Liang Y, et al: DPA n−3,
DPA n−6 and DHA improve lipoprotein profiles and aortic function in
hamsters fed a high cholesterol diet. Atherosclerosis. 221:397–404.
2012. View Article : Google Scholar : PubMed/NCBI
|