Introduction
Ubiquitously expressed transcript (UXT), located on
the Xp11.23-p11.22 chromosome, is a widely expressed gene in humans
and mice and is upregulated in certain tumors (1). UXT has two isoforms: UXT-V2, the
short form that consists of 157 amino acids and is primarily
expressed in the nucleus; and UXT-V1, which is 169 amino acids in
length and is predominantly expressed in the cytoplasm (2,3). UXT
interacts with the N-terminus of the androgen receptor (AR) and
regulates androgen-responsive genes (3,4); it
has also been described as a suppressor of cell transformation and
a coregulator of nuclear factor-κB (5,6).
High expression of UXT has been demonstrated to result in
mitochondrial aggregation (7), and
one study identified that UXT-V1 protects the cells from
TNF-induced apoptosis (8). Under
conditions of infection and inflammation, UXT has dual opposing
effects on SARM (sterile α and HEAT/armadillo motif
protein)-induced apoptosis. UXT-V2 transfection previously resulted
in cell death and a reduced mitochondrial membrane potential when
cotransfected with SARM (9). By
contrast, UXT-V1 cotransfected with SARM has been demonstrated to
led to a significant reduction in caspase 8 activity (9).
Protein inhibitor of activated signal transducer and
activator of transcription 2 (PIAS2) is a member of the PIAS
protein family (10,11). In mammals, four members of the PIAS
protein family have been identified: PIAS1 (12), PIAS2 (13), PIAS3 (14) and PIAS4 (15). PIAS proteins were previously
thought to be inhibitors of activated STAT only (16), but are now known to interact with
and modulate several other proteins, including AR and p53 (13,17).
In addition, PIAS proteins act as E3 ligases in sumoylation, but
appear to possess functions beyond the modification process of
sumoylation (18,19). The PIAS2 gene encodes two splice
variants, PIASxα/androgen receptor-interacting protein-3 (ARIP3)
and PIASxβ, which have different C termini (13,20).
PIAS2 is highly expressed in the testis and its protein is detected
until stage XII of the seminiferous epithelium in spermatogonia and
pachytene spermatocytes, in addition to Sertoli cells, suggesting a
role for PIAS2 in testicular function (21).
The functional regulators of PIAS2 in
spermatogenesis remain largely unknown, thus, in the current study,
cDNAs encoding PIAS2-binding proteins were screened using the yeast
two-hybrid system. UXT was identified as a novel PIAS2-binding
protein and was suggested to physically interact with PIAS2.
Materials and methods
Reagents and antibodies
Mouse anti-human monoclonal anti-c-Myc Tag antibody,
was purchased from EMD Millipore (cat no. 4A6, 05–724; Billerica,
MA, USA). Rabbit anti-green fluorescent protein (GFP) polyclonal
antibody was purchased from Epitomics (cat no. ab137828;
Burlingame, CA, USA).
Construction of plasmids
The cDNA fragment of the mouse PIAS2 gene (9–401 aa
corresponding to nucleotides 213–1391 of NM_008602) was cloned into
a pGBKT7 vector (cat no. 630489; Clontech Laboratories, Inc.,
Mountainview, CA, USA) containing the GAL4 DNA-binding domain. This
generated the bait plasmid, pGBKT7-PIAS2. This construct was not
observed to produce toxic effects or autonomous transcriptional
activation following transformation into the yeast strain, Y187.
This plasmid was used for the yeast two-hybrid screening.
Polymerase chain reaction (PCR) was used to amplify
the common cDNA fragment of the mouse PIAS2 gene, which was then
cloned into the pCMV-c-Myc (cat no. 631604; Clontech Laboratories,
Inc.) and pDsRed-Express-1 vectors (cat no. 632413; Clontech
Laboratories, Inc.) to generate pCMV-c-Myc-PIAS2 and
pDsRed-Express-1-PIAS2, respectively. Full-length UXT was cloned
into a pEGFP-N1 vector (cat no. 6085-1; Clontech, Laboratories,
Inc.) to generate pEGFP-N1-UXT. pCMV-c-Myc-PIAS2 and pEGFP-N1-UXT
were used in the Co-IP assay, while pDsRed-Express-1-PIAS2 and
pEGFP-N1-UXT were used in the colocalization analysis.
Yeast two-hybrid system
cDNA libraries for the mouse stem cells were
constructed in a pGADT7-Rec vector containing a GAL4 activation
domain using Matchmaker Library Construction and Screening Kits
(cat no. 630445; Clontech Laboratories, Inc.) and then transformed
into the yeast AH109 strain. Yeast two-hybrid screening was
conducted using the Matchmaker Gold Yeast Two-Hybrid System (cat
no. 630489; Clontech Laboratories, Inc.). Positive clones were
selected based on ability to grow on synthetic dropout medium (cat
no. 630412; Clontech Laboratories, Inc.) with an absence of
leucine, tryptophan, histidine and adenine (SD/-LTHA)/X-α-Gal (cat
no. 630407; Clontech Laboratories, Inc.) and for α-galactosidase
activity.
In the Matchmaker Two-Hybrid assay, pGBKT7 expresses
proteins as fusions to the GAL4 DNA-BD, while pGADT7-Rec expresses
proteins as fusions to the GAL4 AD. A bait gene is expressed as a
fusion to the GAL4 DNA-binding domain (DNA-BD), while another gene
or cDNA is expressed as a fusion to the GAL4 activation domain
(AD). When bait (recombined pGBKT7) and library fusion proteins
interact in a yeast reporter strain such as AH109, the DNA-BD and
AD are brought into proximity and activate transcription of the
reporter genes (ADE2, HIS3, lacZ, and MEL1). It is known that P53
can interact with Simian virus 40 T large antigen (SV40), thus,
these were used as a positive control. When cotransformed
pGBKT7-p53 and pGADT7-SV40 into AH109, they can interact and
activate transcription of and thus induce expression of the
reporter genes (ADE2, HIS3, lacZ, and MEL1). Thus, the transformed
AH109 strain can grow on an SD/-LTHA plate. X-α-Gal is a
chromogenic substrate for α-galactosidase. In the AH109 strain,
this enzyme is encoded by the MEL1 gene, which is regulated by
several GAL genes. Secretion of this enzyme in response to GAL4
activation leads to hydrolysis of X-α-Gal in the medium, resulting
in yeast colonies developing a blue color.
Plasmid DNA of prey clones was isolated using
QIAprep Spin Miniprep kit according to the manufacturer’s
instructions (cat no. 27104; Qiagen, Hilden, Germany) and
transformed into E. coli DH5α (cat no. D9057; Takara Bio
Inc. Otsu, Japan). Subsequently, prey clones were recovered in the
lysogeny broth medium (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) containing ampicillin. and cDNA inserts were amplified by PCR
using the Advantage 2 PCR kit (cat no. 639207; Clontech
Laboratories, Inc.). The primer sequences used for PCR were as
follows: Forward: 5′-TTCCACCCAAGCAGTGGTATCAACGCAGAGTGG-3′ and
Reverse: 5′-GTATCGATGCCCACCCTCTAGAGGCCGAGGCGGCCGACA-3′. PCR was
performed at 94°C for 3 min; 30 cycles of 94°C for 30 sec and 68°C
for 3 min; 68°C for 3 min; and then maintained at 15°C. The PCR
products were sequenced and then analyzed using the basic local
alignment search tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). Interaction
between the bait and identified prey clones was verified by
cotransforming the purified prey plasmid with the bait pGBKT7-PIAS2
construct into the yeast AH109 strain, followed by selection on
SD/-LTHA medium. Cotransformation of pGBKT7-p53 with pGADT7-SV40
was used as a positive control. A negative control was also
produced by the cotransformation of pGBKT7-UXT with the pGADT7
vector into the AH109 yeast cells.
Mammalian cell culture and transient
transfection assay
A human cervical carcinoma (HeLa) and SV40
T-antigen-expressing human embryonic kidney (HEK 293T) cell lines
were cultured and maintained in Dulbecco’s modified Eagle’s medium
(DMEM; Invitrogen, Life Technologies, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS; GE Healthcare Life
Sciences, Logan, UT, USA) and 50 U/ml each of penicillin and
streptomycin (Invitrogen Life Technologies), at 37°C in a
humidified atmosphere with 5% CO2. For the transient
transfections, HEK 293T and HeLa cells were seeded into 6-well
plates at 70~90% confluence, washed once with DMEM and transfected
with 2 μg pCMV-c-Myc-PIAS2 and 2 μg pEGFP-N1-UXT (or 2 μg
pDsRed-Express-1-PIAS2 and 2 μg pEGFP-N1-UXT) using 12 μl
Lipofectamine 2000 reagent (Invitrogen Life Technologies) in a
total of 1 ml serum-free DMEM per 25-mm well, in accordance with
the manufacturer’s instructions. Approximately 3 h following
transfection, the transfection mixture was replaced with 2 ml
DMEM-10% FBS, rested for 3–5 h and then replaced again with fresh
DMEM-10% FBS.
Co-IP
The HEK 293T cells were transiently transfected with
pCMV-c-Myc-PIAS2 and pEGFP-N1-UXT constructs, then were maintained
as described above for 48 h. The cells were then washed with
ice-cold phosphate-buffered saline (PBS; cat no. ZLI-9061; Origene,
Beijing, China) and harvested in 500 ml lysis buffer [150 mM NaCl;
50 mM Tris, pH 8.0; 1% Nonidet P-40; 0.5% deoxycholate; and a
protease inhibitor mixture (Roche Diagnostics GmbH, Mannheim,
Germany)]. Subsequent to 60-min incubation in lysis buffer on ice,
lysates were clarified by centrifugation for 20 min at 16,873 × g
at 4°C. Anti-Myc or anti-GFP antibody (8 μg) was added to the
supernatant and incubated for 120 min at 4°C prior to the addition
of 50 μl protein G-agarose (cat no. 11719386001; Hoffmann-La Roche
Ltd., Basel, Switzerland) and incubation at 4°C overnight. Samples
were then centrifuged for 1 min in a microcentrifuge at 16,873 × g
(5424; Eppendorf, Hamburg, Germany) and washed with 1 ml lysis
buffer, 1 ml washing buffer (500 mM NaCl; 50 mM Tris, pH 7.5; 0.1%
Nonidet P-40; 0.05% deoxycholate) and 1 ml washing buffer (10 mM
Tris, pH 8.0; 0.1% Nonidet P-40; 0.05% deoxycholate).
Western blotting
Initial lysates and immunoprecipitated proteins were
analyzed using SDS-PAGE. SDS buffer and 12% polyacrylamide gels
were prepared with reagents purchased from Invitrogen Life
Technologies. Electrophoresis was carried out using the
Mini-PROTEAN Tetra Cell-2 gel system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Immunoblotting was carried out using anti-GFP
or anti-Myc antibody. Proteins were transferred to nitrocellulose
membranes (GE Healthcare Life Sciences, Chalfont, UK) by
electrophoresis and were probed with the appropriate antibodies.
Anti-GFP or anti-Myc antibody was visualized with horseradish
peroxidase (HRP)-conjugated anti-rabbit (cat no. A9169;
Sigma-Aldrich, St. Louis, MO, USA) or HRP conjugated goat anti
mouse IgG antibodies (cat no. A9044; Sigma-Aldrich) using the
Pierce ECL Plus western blotting substrate (Thermo Fisher
Scientific, Inc.).
Fluoresence microscopy
To detect PIAS2 and UXT, HEK 293T and HeLa cells
plated on round coverslips were transiently transfected with the
pDsRed-Express-1-PIAS2 and pEGFP-N1-UXT constructs using
Lipofectamine 2000. Cells were washed twice with PBS 48 h later and
visualized using a scanning laser confocal microscope (Nikon A1;
Nikon Corporation, Tokyo, Japan).
Results
UXT interacts with PIAS2
To further understand the function of PIAS2, the
common cDNA fragment of the mouse PIAS2 gene was used as an
interaction trap for interactive substrates expressed by a mouse
stem cell cDNA library with the Matchmaker Library Construction and
Screening kit. Several positive clones that demonstrated strong
growth on the SD/-LTHA medium were isolated, sequenced and aligned
using the NCBI BLAST. Among these positive clones, two clones that
encoded the same region of UXT, spanning a region of 147 amino
acids (UXT residues 11–157), were observed to interact with the
PIAS2 protein. The interaction specificity between PIAS2 and UXT
was confirmed by the direct yeast two-hybrid assay. Strong growth
on SD/-LTHA medium was observed in yeast cells cotransformed with
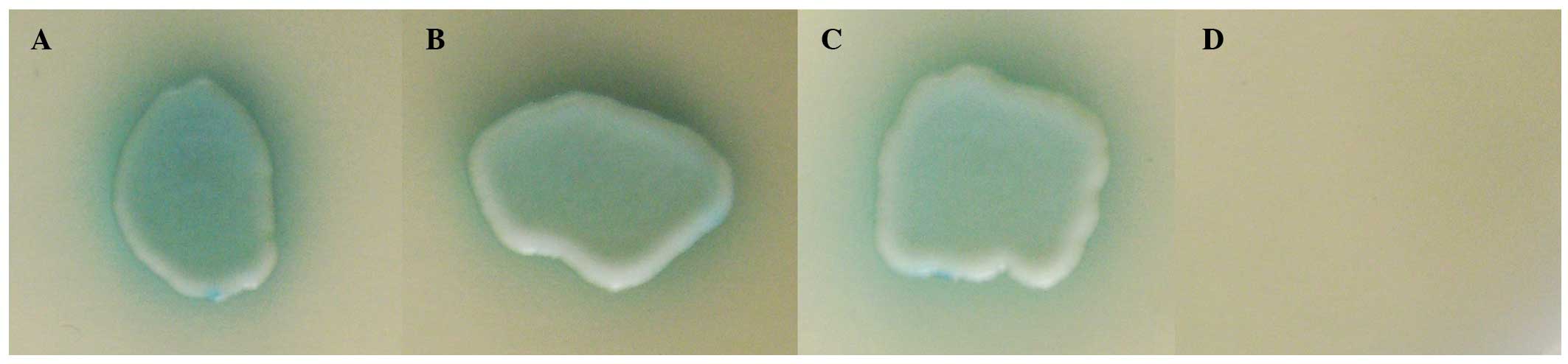
pGBKT7-PIAS2 and pGADT7-UXT (Fig. 1A
and B), indicating an interaction between UXT and PIAS2 in
yeast. Several positive clones that demonstrated strong growth on
the SD/-LTHA medium were isolated, sequenced and aligned using the
NCBI BLAST. Among these positive clones, two clones that encoded
the same region of UXT, spanning a region of 147 amino acids (UXT
residues 11–157), were observed to interact with the PIAS2 protein.
Fig. 1A and B exhibit two
different cDNA clones screened from the cDNA library. Growth was
also observed in the positive control (Fig. 1C). However, the yeast strain
cotransformed with pGADT7-UXT and pGBKT7 did not grow on SD/-LTHA
medium and exhibited negative α-galactosidase activity (Fig. 1D).
UXT interacts with PIAS2 in mammalian
cells
In order to confirm the results of the two-hybrid
screen using an independent biochemical method, it was determined
whether PIAS2 and UXT proteins bind tightly enough to each other to
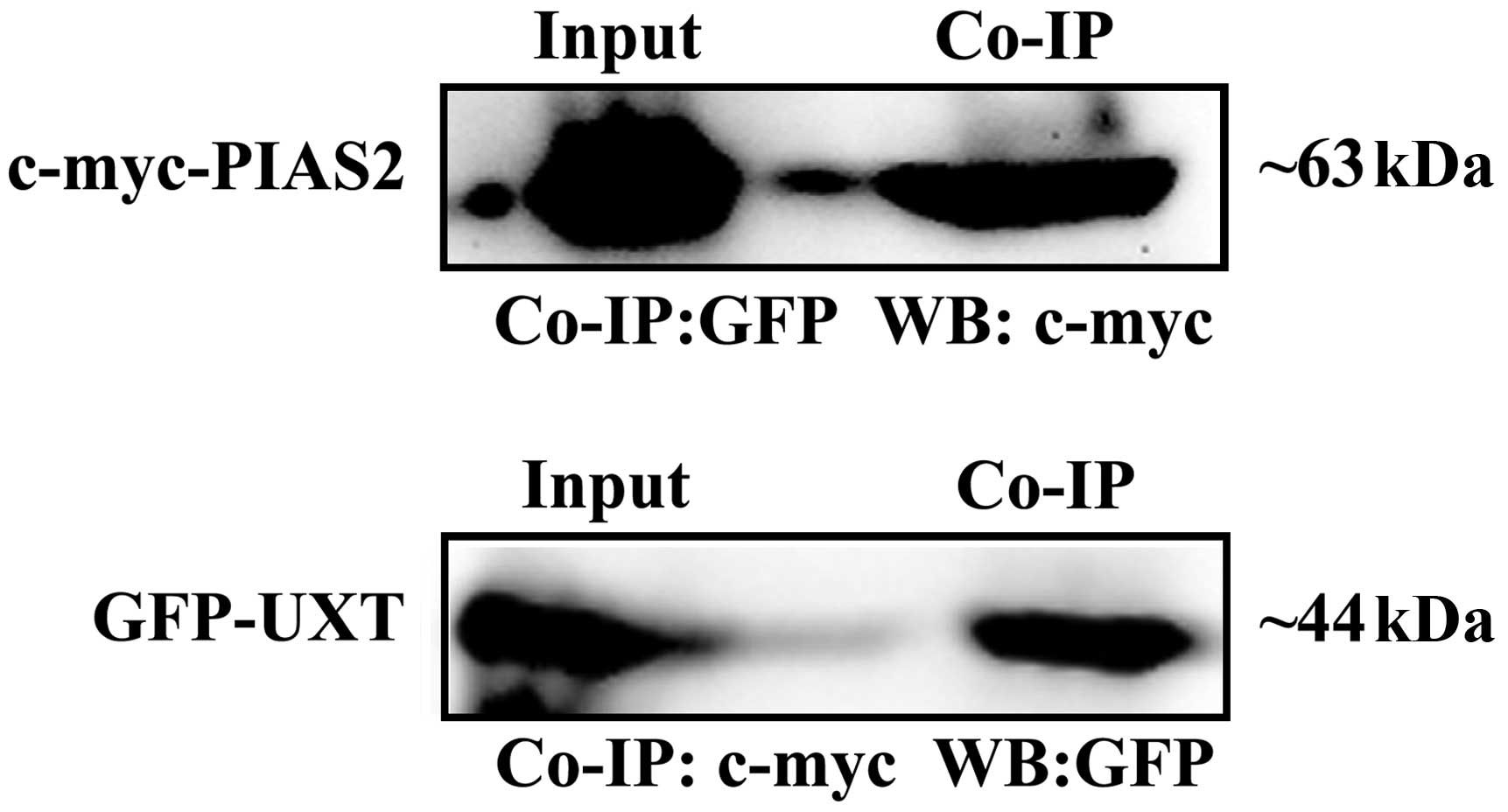
be co-immunoprecipitated. The expression vectors for c-myc-tagged
PIAS2 and GFP-tagged UXT were cotransfected into HEK 293T cells.
The cell extract was prepared and the proteins in the extract were
immunoprecipitated with anti-c-myc and anti-GFP, respectively, 24 h
subsequent to transfection. The precipitates were then
immunoblotted against the anti-GFP or anti-c-myc antibodies. The
anti-c-myc antibody precipitated c-myc-PIAS2, whereas GFP-UXT was
detected in the immunoprecipitate with the anti-GFP antibody
(Fig. 2). PIAS2 protein was also
observed to be immunoprecipitated from the transfected HEK 293T
cells, using a rabbit anti-GFP antibody prior to detection of PIAS2
by the anti-c-myc antibody. As demonstrated in Fig. 2, UXT and PIAS2 fusion proteins were
observed in the Co-IP and total lysate (Input). These results
indicate that UXT may interact with PIAS2 in mammalian cells.
UXT colocalized with PIAS2 in HEK 293T
and HeLa cells
The interaction between the PIAS2 and UXT proteins
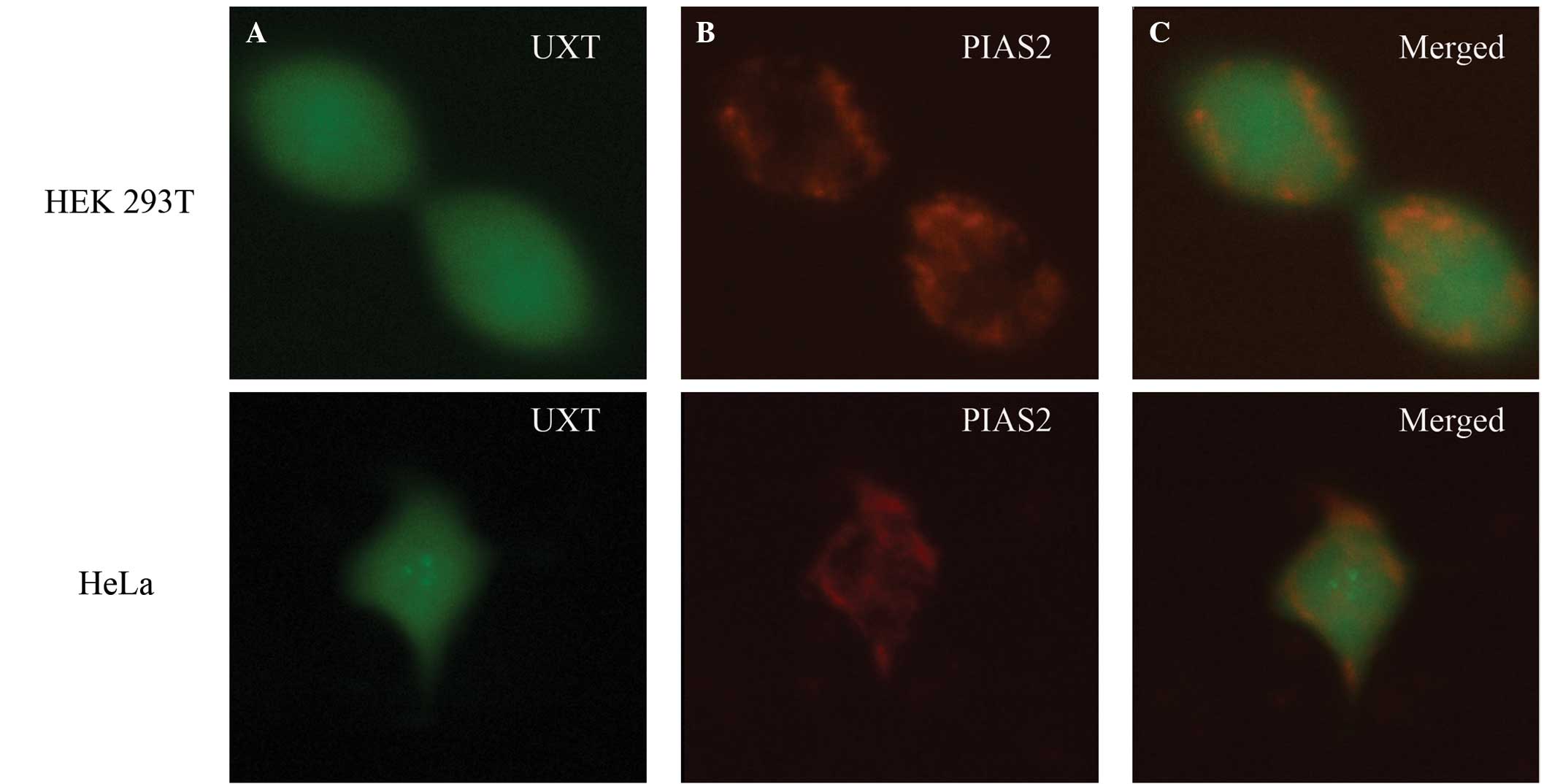
was further determined by colocalization analysis. The
pDsRed-Express-1-PIAS2 and pEGFP-N1-UXT plasmids were cotransfected
into HEK 293T and HeLa cells, respectively, then the cells were
detected using a scanning laser confocal microscope two days
subsequent to transfection. As demonstrated in Fig. 3, GFP-UXT protein was distributed in
the cytoplasm and nucleus of HEK 293T and HeLa cells (Fig. 3A), as was DsRed-PIAS2 protein
(Fig. 3B). Fig. 3C indicates that PIAS2 and UXT were
partially colocalized in these cells, implicating a potential
interaction between UXT and PIAS2 in HEK 293T and HeLa cells.
Discussion
The aim of the current study was to elucidate the
protein interactions of PIAS2. There are two isoforms of human
PIAS2, including PIASxα and PIASxβ, whereas there are five isoforms
of the mouse PIAS2 gene (isoforms 1–5), which have different N- and
C-termini as a result of alternative splicing. Thus, the common
encoding region of mouse PIAS2 as a bait to screen a mouse cDNA
library. UXT protein, which coregulates various transcription
factors, including AR, was observed to specifically interact with
PIAS2 in vitro. In addition, PIAS2 and UXT were observed to
colocalize in the nucleus and cytoplasm, suggesting that they have
the ability to synchronously interact with each other in mammalian
cells.
The AR, a member of the steroid receptor
superfamily, is an X-linked nuclear receptor (NR) that is regarded
as critical in sexual differentiation, gonadal maturation,
maintenance of secondary male characteristics and the development
of prostate cancer (3,4). Upon binding to androgen, the AR is
released from heat-shock proteins, forms homodimers and is
translocated into the nucleus. The AR is a single polypeptide with
four functional domains, including the NH2-terminal transactivation
domain; the DNA-binding domain (DBD); the hinge region; and the
ligand-binding domain (LBD) (22).
The transcriptional activation functions (AFs) of AR represent
surfaces that are able to interact with transcription factors and
additional coactivators. Coactivators that interact with the AR
N-terminal AF-1 and the C-terminal AF-2 region, which leads to the
enhancement of AR-dependent gene transcription, have been
identified (23,24). Regions of the AR N-terminus that
are important for transcriptional activation have been identified,
notably AF-1a (residues 154–167) and AF-1b (residues 295–459),
which are necessary for full transcriptional activation mediated by
the receptor (3).
UXT was previously identified as an AR N-terminal
coactivator and is established to interact predominantly with the
AR153–336 (containing AF-1a and a part of AF-1b), which is
localized to the nucleus and increases AR transcriptional activity
when overexpressed in cultured mammalian cells (3). Endogenous UXT interacts with AR in
nuclear extracts from lymph node carcinoma of the prostate (LNCaP)
cells in a ligand-independent manner (3). Multiple studies have demonstrated
that native UXT was a part of multiprotein complex which includes
AR functioning in transcriptional regulation (3,4).
native UXT is part of a multiprotein complex that includes proteins
functioning in transcriptional regulation (3,4).
Certain components of the UXT complex have been identified by mass
spectrometry analysis, and UXT has been observed to associate with
proteins, including RBP5, TIP48, TIP49 and other unidentified
proteins (25).
ARIP3 (PIASxα) was originally identified as a
testis-specific AR coregulator using yeast two-hybrid analysis with
AR DBD as a bait (11). ARIP3
residues 443–548 have been suggested to be critical for the
AR-ARIP3 interaction to occur, and the majority of the C-terminal
region of ARIP3 containing the AR interaction domain (AR ID,
residues 443–548) is unique to ARIP3 (11). The AR LBD alone was not observed to
associate with the ARIP3 interaction domain (ARIP3 ID) and the
N-terminal half of the AR encompassing AF-1 (residues 5–538) failed
to recognize either ARIP3 ID or full-length ARIP3, indicating that
AR interacts with ARIP3 ID primarily through the DBD (11). To determine whether the ARIP3
mutants maintained their ability to interact with AR, AR and
FLAG-tagged ARIP3 or ARIP3 mutants were ectopically expressed in
COS-1 cells (11). The results
demonstrated that full-length ARIP3 and certain ARIP3 mutants
(Δ1–102, Δ467–547, L23A, L304A, C385S and C388S) displayed
interactions with AR that were not significantly different from
each other, whereas the interaction of ARIP3 Δ347–418 with AR was
markedly weaker (11). In view of
the observation that the region 467–547 is sufficient for the
interaction of ARIP3 with the zinc finger region of AR in yeast, it
is suggested that ARIP3 interacts with AR via multiple domains
(11). ARIP3 interacts with AR
in vitro in addition to in intact mammalian cells, and is
capable of modulating AR-dependent transcriptional activity. The
biphasic responses suggest that ARIP3 belongs to a multisubunit
coactivator complex, and its overexpression has been observed to
lead to the repression of transcription when other limiting
components are titrated out of the complex (11).
In the current study, UXT was demonstrated to
interact with PIAS2. The yeast two-hybrid bait contained residues
9–401 of ARIP3 and the fragment was similar to the ARIP3 mutant
Δ467–547, thus, the ARIP3 bait was able to bind to the AR DBD. UXT
was demonstrated to be able to directly interact with the AR NTB.
Combined with other coregulators, PIAS2, AR and UTX formed a
multiprotein complex and further coregulated the transcriptional
activity of AR. Further investigation into the functional
regulatory network of the coregulators is required.
Acknowledgements
The current study was supported by grants from the
National Natural Science Foundation of China (grant nos. 31071020
and 31371174), the Natural Science Foundation of Jiangsu Province
of China (grant no. BK20131230) and the Key Project on Yangzhou
Social Development (grant no. 2012127).
Abbreviations:
|
UXT
|
ubiquitously expressed transcript
|
|
AR
|
androgen receptor
|
|
STAT
|
signal transducer and activator of
transcription
|
|
PIAS
|
protein inhibitor of activated
STATs
|
|
DMEM
|
Dulbecco’s modified Eagle’s medium
|
|
SD
|
synthetic dropout medium
|
|
SD/-LTHA
|
synthetic complete medium lacking
leucine, tryptophan, histidine and adenine
|
|
GFP
|
green fluorescent protein
|
|
HEK 293T
|
human embryonic kidney cell line
expressing SV40 T-antigen
|
|
HeLa
|
human cervical carcinoma cell line
|
|
SARM
|
sterile α and HEAT/armadillo motif
protein
|
|
Co-IP
|
co-immunoprecipitation
|
References
|
1
|
Schroer A, Schneider S, Ropers H and
Nothwang H: Cloning and characterization of UXT, a novel gene in
human Xp11, which is widely and abundantly expressed in tumor
tissue. Genomics. 56:340–343. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang Y, Liu H, Ge R, Zhou Y, Lou X and
Wang C: UXT-V1 facilitates the formation of MAVS antiviral
signalosome on mitochondria. J Immunol. 188:358–366. 2012.
View Article : Google Scholar
|
|
3
|
Markus SM, Taneja SS, Logan SK, Li W, Ha
S, Hittelman AB, Rogatsky I and Garabedian MJ: Identification and
characterization of ART-27, a novel coactivator for the androgen
receptor N terminus. Mol Biol Cell. 13:670–682. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li W, Cavasotto CN, Cardozo T, Ha S, Dang
T, Taneja SS, Logan SK and Garabedian MJ: Androgen receptor
mutations identified in prostate cancer and androgen insensitivity
syndrome display aberrant ART-27 coactivator function. Mol
Endocrinol. 19:2273–2282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang
B, Shi H and Wang C: UXT is a novel and essential cofactor in the
NF-kappaB transcriptional enhanceosome. J Cell Biol. 178:231–244.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu Z, Li Y, Li X, Ti D, Zhao Y, Si Y, Mei
Q, Zhao P, Fu X and Han W: LRP16 integrates into NF-κB
transcriptional complex and is required for its functional
activation. PLoS One. 6:e181572011. View Article : Google Scholar
|
|
7
|
Moss TN, Vo A, McKeehan WL and Liu L: UXT
(Ubiquitously Expressed Transcript) causes mitochondrial
aggregation. In Vitro Cell Dev Biol Anim; 43:139–146.
2007.PubMed/NCBI
|
|
8
|
Huang Y, Chen L, Zhou Y, Liu H, Yang J,
Liu Z and Wang C: UXT-V1 protects cells against TNF-induced
apoptosis through modulating complex II formation. Mol Biol Cell.
22:1389–1397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sethurathinam S, Singh LP, Panneerselvam
P, Byrne B and Ding JL: UXT plays dual opposing roles on
SARM-induced apoptosis. FEBS Lett. 587:3296–3302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kotaja N, Vihinen M, Palvimo JJ and Jänne
OA: Androgen receptor-interacting protein 3 and other PIAS proteins
cooperate with glucocorticoid receptor-interacting protein 1 in
steroid receptor-dependent signaling. J Biol Chem. 277:17781–17788.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moilanen AM, Karvonen U, Poukka H, Yan W,
Toppari J, Jänne OA and Palvimo JJ: A testis-specific androgen
receptor coregulator that belongs to a novel family of nuclear
proteins. J Biol Chem. 274:3700–3704. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu B, Liao J, Rao X, Kushner SA, Chung
CD, Chang DD and Shuai K: Inhibition of Stat1-mediated gene
activation by PIAS1. Proc Natl Acad Sci USA. 95:10626–10631. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kotaja N, Aittomäki S, Silvennoinen O,
Palvimo JJ and Jänne OA: ARIP3 (androgen receptor-interacting
protein 3) and other PIAS (protein inhibitor of activated STAT)
proteins differ in their ability to modulate steroid
receptor-dependent transcriptional activation. Mol Endocrinol.
14:1986–2000. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chung CD, Liao J, Liu B, Rao X, Jay P,
Berta P and Shuai K: Specific inhibition of Stat3 signal
transduction by PIAS3. Science. 278:1803–1805. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C, Yuan X, Yue L, Fu J, Luo L and
Yin Z: PIASy interacts with p73alpha and regulates cell cycle in
HEK293 cells. Cell Immunol. 263:235–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arora T, Liu B, He H, Kim J, Murphy TL,
Murphy KM, Modlin RL and Shuai K: PIASx is a transcriptional
co-repressor of signal transducer and activator of transcription 4.
J Biol Chem. 278:21327–21330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmidt D and Müller S: Members of the
PIAS family act as SUMO ligases for c-Jun and p53 and repress p53
activity. Proc Natl Acad Sci USA. 99:2872–2877. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishida T and Yasuda H: PIAS1 and
PIASxalpha function as SUMO-E3 ligases toward androgen receptor and
repress androgen receptor-dependent transcription. J Biol Chem.
277:41311–41317. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Santti H, Mikkonen L, Anand A,
Hirvonen-Santti S, et al: Disruption of the murine PIASx gene
results in reduced testis weight. J Mol Endocrinol. 34:645–654.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herold S, Hock A, Herkert B, Berns K,
Mullenders J, Beijersbergen R, Bernards R and Eilers M: Miz1 and
HectH9 regulate the stability of the checkpoint protein, TopBP1.
EMBO J. 27:2851–2861. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan W, Santti H, Jänne OA, Palvimo JJ and
Toppari J: Expression of the E3 SUMO-1 ligases PIASx and PIAS1
during spermatogenesis in the rat. Gene Expr Patterns. 3:301–308.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Chen K, Zhang Q, Cheng H and Zhou
R: Regulation of the transcriptional activation of the androgen
receptor by the UXT-binding protein VHL. Biochem J. 456:55–66.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heinlein CA and Chang C: Androgen receptor
(AR) coregulators: an overview. Endocr Rev. 23:175–200. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He B and Wilson EM: The NH(2)-terminal and
carboxyl-terminal interaction in the human androgen receptor. Mol
Genet Metab. 75:293–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gstaiger M, Luke B, Hess D, Oakeley EJ,
Wirbelauer C, Blondel M, Vigneron M, Peter M and Krek W: Control of
nutrient-sensitive transcription programs by the unconventional
prefoldin URI. Science. 302:1208–1212. 2003. View Article : Google Scholar : PubMed/NCBI
|