Introduction
Glioma is the most common malignant and fatal
primary tumor in the central nervous system (CNS) in adults. Though
progress has been made in the development of treatments to improve
patient outcomes during past decades, prognosis of glioma patients
remains poor, with a median survival time of 12–14 months. The poor
prognosis of patients is mainly attributed to the high tendency of
tumor invasiveness, resistance to therapy and high frequency of
recurrence (1). However, the
molecular mechanisms underlying these molecular events of glioma
cells remain elusive.
Deregulation of microRNAs (miRNAs) has been
implicated in the development and progression of almost all tumor
types (2). miRNAs are
single-stranded, 19–25 bp short, non-coding RNAs, and elicit their
regulatory effects in post-transcriptional regulation of genes by
binding to the 3′untranslated region (3′UTR) of target messenger
RNA (mRNA), mainly leading to translational repression or target
mRNA degradation (3). It has been
estimated that miRNAs regulate up to one-third of human genes at
the post-transcriptional level, suggesting that miRNAs have pivotal
roles in numerous physiological and pathophysiological processes,
including development, differentiation, proliferation, stress
response, metabolism and apoptosis (4–7).
Deregulated miRNAs may function as tumor suppressors or tumor
promoters due to the diversity of miRNAs themselves (8). Emerging evidence has implicated
miR-203 loss in cancer pathology, including cancer cell
proliferation, invasion and drug resistance (9,10).
However, the function of miR-203 in glioma cells remains to be
fully elucidated. The present study investigated miR-203 expression
in aggressive U87MG glioma cells and glioma tissues, and assessed
the effect of ectopic expression of miR-203 on invasion of U87MG
cells and their sensitivity to temozolomide (TMZ) by targeting
E2F3.
Materials and methods
Cell culture and tissue specimens
The human glioma cell lines A172, U87MG and U251MG,
and the human embryonic kidney cell line (HEK-293T) (Cancer
Research Institute of Central South University, Cambridge, MA, USA)
were cultured in Dulbecco’s modified Eagle’s medium supplemented
with 10% fetal bovine serum (HyClone, Logan City, UT, USA) at 37°C
in a humidified atmosphere containing 5% CO2. Fifty-two
paired glioma and non-cancerous brain tissues were obtained from
the Department of Neurosurgery at Xiangya Hospital of Central South
University (Changsha, China) between September 2011 and March 2013.
Ethical approval for human subjects was obtained from the research
and ethics committee of the Xiangya Hospital of Central South
University (Changsha, China) and informed consent was obtained from
each patient. All tumor and normal tissue samples were divided into
two sections and immediately snap-frozen in liquid nitrogen. Half
of each sample was used for miRNA isolation and the remaining
section was used for immunohistochemical (IHC) analysis.
Plasmid transfection
miR-203 expression plasmid pSilencer2.1-U6-miR-203
or empty control vector (Cancer Research Institute of Central South
University) was introduced into U87MG cells with Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) in accordance with the
manufacturer’s instructions in 24-well plates. Following 24 h of
incubation, the cells were selected by incubation with 4 μg/ml
puromycin (Sigma-Aldrich, St. Louis, MO, USA), for two weeks and
the individual stable clones were analyzed by quantitative
polymerase chain reaction (qPCR). U87MG cells were transfected with
pRNAT-U6.1/sh-E2F3 plasmids (Genscript, Nanjing, Jiangsu, China) to
knockdown E2F3 by using Lipofectamine 2000, selected with 800 μg/ml
G418 and validated by western blot analysis. The miR-203 inhibitor
(Qiagen, Hilden, Germany) was transfected in a 10-cm dish using
Lipofectamine 2000. Cells were harvested 48 h later and the
subsequent experiments were performed.
miRNA-specific qPCR
miRNAs from cultured cells and tissues were isolated
and purified using an miRNA isolation system (OMEGA Bio-Tek,
Norcross, GA, USA). cDNA was generated with the miScript II RT Kit
(Qiagen) and the qPCR was performed using the miScript SYBR Green
PCR Kit (Qiagen) following the manufacturer’s instructions. The
miRNA sequence-specific qPCR primers for miR-203 and endogenous
control RNU6 were purchased from Qiagen, and qPCR analysis was
performed using the 7500 Real-Time PCR System (Applied Biosystems,
Foster City, CA, USA). The gene expression threshold cycle (CT)
values of miRNAs were calculated by normalizing to the internal
control RNU6 and relative quantification values were
calculated.
(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)
(MTS) assay
The Cell Titer 96® AQueous One Solution
Cell Proliferation Assay kit (Promega, Madison, WI, USA) was used
to determine the sensitivity of cells to TMZ. In brief, cells were
seeded in 96-well plates at a density of 4×103
cells/well (0.20 ml/well) for 24 h prior to use. The culture medium
was replaced with fresh medium containing various concentrations of
TMZ and incubated for 72 h. Subsequently, MTS (0.02 ml/well) was
added to the culture medium. Following a further 2 h of incubation,
the absorbance at 490 nm of each well was recorded on an ELX800
(Biotek, Winoosky, VT, USA). The growth rate was calculated as the
ratio of the absorbance of the experimental well to that of the
control well. The inhibition rate and the IC50
(concentration of drug resulting in 50% of control value) were also
calculated.
Western blot analysis
Total proteins were extracted from corresponding
cells using the RIPA buffer (Pierce, Rockford, IL, USA) in the
presence of Protease Inhibitor Cocktail (Pierce). The protein
concentration of the lysates was measured using a BCA Protein Assay
Kit (Pierce), loaded and separated by 10% SDS-PAGE and then
transferred to a polyvinylidene fluoride membrane (Millipore,
Billerica, MA, USA). The primary antibodies used for analysis
included mouse anti-E2F3 monoclonal antibody (1:800) and mouse
anti-β-actin monoclonal antibody (1:2,000; Santa Cruz, Dallas, TX,
USA).
Luciferase reporter assay
Two single strands of the wild-type 3′UTR with
miR-203 binding site and two single strands of the mutant type with
seven bases deleted in the miR-203 binding site (as mutant
control), of E2F3 were synthesized with restriction sites for
SpeI and HindIII located at both ends of the
oligonucleotides for further cloning. The single strand DNA
sequences were: Wild-type 3′UTR of E2F3 sense,
5′-CTAGTCAGCAATCTTCCTTAATAGCATTTCAAGCCG
TGCCTTCTCCCGCAGAATGCA-3′ and antisense,
5′-AGCTTGCATTCTGCGGAGAAGGCACGGCTTGAAAT
GCTATTAAGGAAGATTGCTGA-3′; and the mutant type 3′UTR of E2F3
sense, 5′-CTAGTCAGCAATCTTCCTT
AATAG-------AGCCGTGCCTTCTCCGCAGAATGCA-3′ and antisense,
5′-AGCTTGCATTCTGCGGAGAAG
GCACGGCT-------CTATTAAGGAAGATTGCTGA-3′. The corresponding
sense and antisense strands were annealed and subsequently cloned
into pMir-Report plasmids downstream of the firefly luciferase
reporter gene. Cells were seeded in 96-well plates and
co-transfected with pMir-Report luciferase vector, pRL-TK
Renilla luciferase vector and miR-203 expression vector
(Cancer Research Institute of Central South University). Following
48 h of incubation, luciferase activity was determined using a
Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI,
USA) where Renilla luciferase activity was used as an
internal control and the firefly luciferase activity was calculated
as the mean ± standard deviation (SD) after being normalized
against Renilla luciferase activity.
Wound healing assay
Cells were synchronized to ensure a homogeneous and
viable cell monolayer prior to wounding. Equal numbers of cells
were seeded into six-well culture plates. When cell confluence
reached ~90% at 48 h post-transfection, an artificial homogenous
wound was created on the monolayer with a sterile plastic 100-μl
micropipette tip. Following wounding, debris was removed by washing
the cells with serum-free medium. Migration of cells into the wound
was observed after 36 h. Images were captured of cells which
migrated into the wounded area or cells with extended protrusions
from the border of the wound. Related observation was done under
the contrast phase model on microscope from Leica (Wetzlar,
Germany).
Transwell cell invasion assay
Cells were detached and resuspended in serum-free
medium. Cells (1×105 cells/well) were then plated into
Matrigel®-coated invasion chambers (Becton Dickinson,
Franklin Lakes, NJ, USA) and allowed to invade for 24 h. The
remaining cells in the chambers were removed by cotton swabs and
the invading cells on the lower surface of the chambers were fixed
with 70% ethanol and then stained with hematoxylin (Sigma-Aldrich).
The number of invading cells was calculated by counting three
different fields under a phase contrast microscope (Leica Wetzlar,
Germany).
IHC analysis
Formalin-fixed, paraffin-embedded tissue specimens
were cut into 4-μm sections. The specimens were deparaffinized in
xylene (Sigma-Aldrich) and rehydrated using a series of graded
alcohols following being dried at 62°C for two hours. The tissue
slides were then treated with 3% hydrogen peroxide (Sigma-Aldrich)
in methanol for 15 min. To exhaust endogenous peroxidase activity,
the antigens were retrieved in 0.01 M sodium citrate buffer (pH
6.0) (Sigma-Aldrich) using a microwave oven. Following one hour of
preincubation in 10% goat serum, the specimens were incubated with
primary antibody (1:50; Santa Cruz) at 4°C overnight. The tissue
slides were treated with a non-biotin horseradish peroxidase
detection system according to the manufacturer’s instructions
(DAKO, Carpinteria, CA, USA). Two pathologists who specialize in
liver cancer evaluated the results of IHC.
Statistical analysis
Each experiment was repeated at least three times.
Statistical analysis was performed using SPSS 16.0 (International
Business Machines, Armonk, NY, USA). Student’s t-test was used to
analyze the statistical difference. Results were presented as the
mean ± SD. P<0.05 was considered to indicate a statistically
significant difference between values.
Results
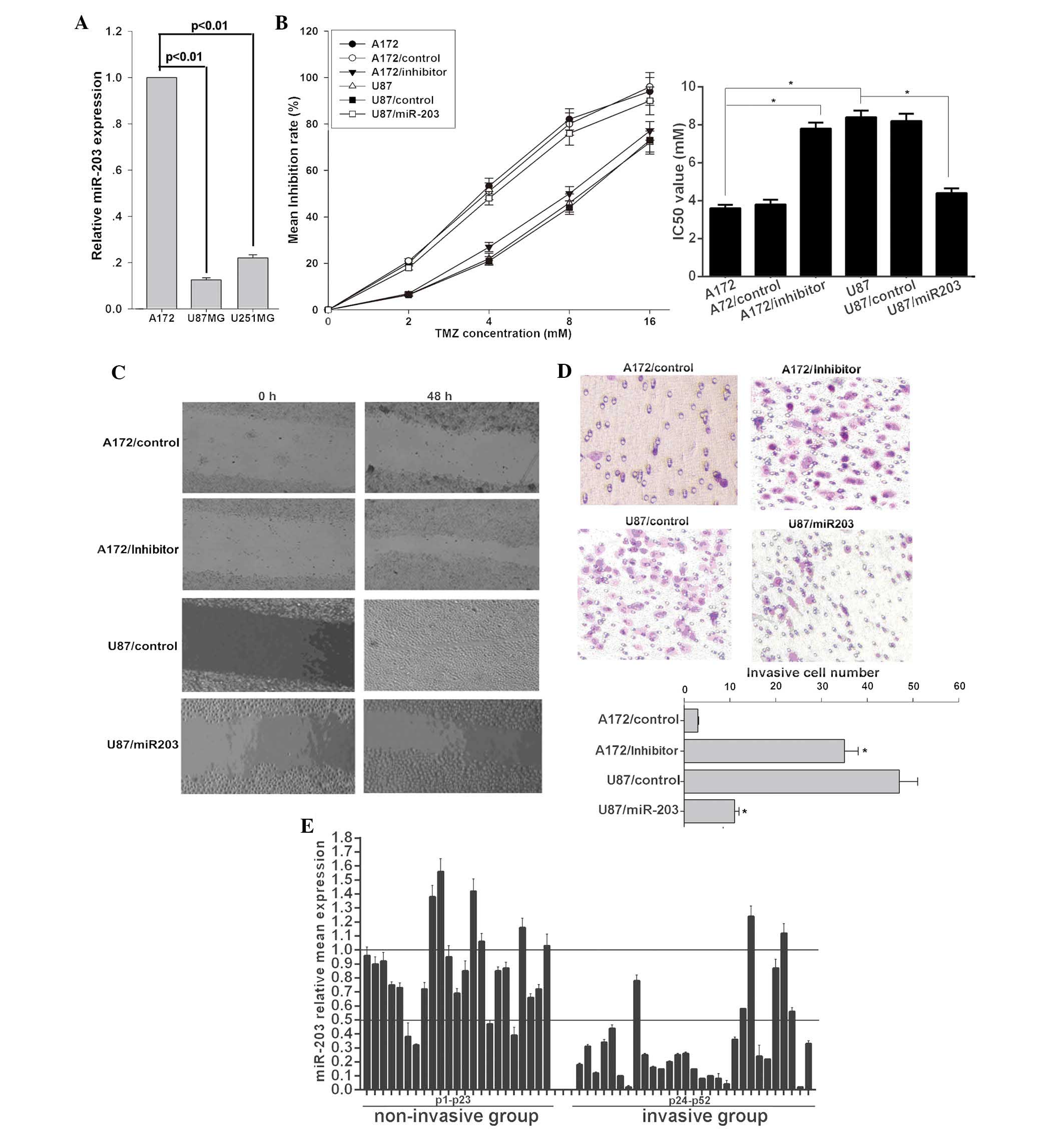
miR-203 sensitizes glioma cells to TMZ
and inhibits their invasion
To determine the role of miR-203 in glioma, the
miR-203 expression pattern in selected glioma cell lines with
varying aggressive potential was examined. miR-203 was markedly
decreased in U87MG and U251MG cell lines with highly invasive
potential compared to that in the less invasive A172 cells
(Fig. 1A). In order to verify the
correlation between the biological characteristics of glioma cells
and miR-203 expression, a series of assays were employed. The
present study demonstrated that U87MG cells were more resistant to
TMZ (Fig. 1B) and more invasive
(Fig. 1C and D) than A172 cells.
Ectopic miR-203 expression significantly sensitized U87MG cells to
TMZ and inhibited U87MG cell migration and invasion (Fig. 1C and D). A wound healing assay
demonstrated that miR-203-transduced U87MG cells exhibited markedly
slower migration compared with vector-control cells (Fig. 1C). Through the cell invasion assay,
the effect of miR-203 on cell invasion, a key determinant of
malignant progression and metastasis, was assessed. As demonstrated
in Fig. 1D, miR-203 led to
significantly decreased invasion (miR-203 group, 11±1 cells per
field; control group, 47±4 cells per field) of U87MG cells.
Inversely, the miR-203 inhibitor attenuated the sensitivity of A172
cells to TMZ (Fig. 1B) and
promoted migration (Fig. 1C) and
invasion (miR-203 inhibitor group, 35±3 cells per field; control
group, 3±0.2 cells per field) (Fig.
1D). To investigate the association between miR-203 and glioma
invasion, a total of 52 clinical glioma tissue samples and paired
non-cancerous brain samples were collected and divided into
non-invasive (23 samples) and invasive groups (29 samples). The
present study demonstrated that miR-203 was significantly
downregulated in the invasive group with ~79.31% of the cases
displaying >2-fold downregulation. However, in the non-invasive
group only ~13.04% of the cases displayed >2-fold downregulation
(P<0.01) (Fig. 1E). These
results revealed a functional role for miR-203 in cell invasion and
chemosensitivity in glioma cells, thus suggesting a mechanism by
which downregulation of miR-203 may contribute to tumor progression
and chemotherapy resistance in glioma.
miR-203 directly targets E2F3 in glioma
cells
To analyze the molecular mechanisms of miR-203
involvement in glioma cells, its target gene was sought. The public
database TargetScan (http://www.targetscan.org) was used to predict
potential targets of miR-203. E2F3, with critically conserved
binding sites, was selected for further characterization (Fig. 2A). To validate the potential
targeting of E2F3 by miR-203, protein expression of E2F3 in glioma
cells that had been subjected to miR-203 expression interference
was examined. The present study demonstrated that E2F3 protein
expression in U87MG and U251MG cells was markedly greater than that
in A172 cells (Fig. 2B). E2F3 in
U87MG cells was significantly decreased following ectopic miR-203
expression. However, miR-203 inhibitor upregulated E2F3 protein
level in A172 cells. To assess whether E2F3 is a direct target of
miR-203, a luciferase reporter vector containing the putative E2F3
3′UTR target site for miR-203 downstream of the luciferase gene
(pMir-E2F3-Wt) and a mutant version containing a deletion of 7 bp
in the seed region was constructed (pMir-E2F3-Mut). As demonstrated
in Fig. 2C, the luciferase
activity of pMir-E2F3-Wt in U87MG was ~44.78% higher than that of
the A172 cells, while luciferase activity of pMir-E2F3-Mut was
unaffected, suggesting that endogenous miR-203 suppressed gene
expression with the seed region in the 3′UTR of miRNA. Moreover,
the luciferase reporter assay performed on HEK293T cells revealed
that miR-203 reduced the luciferase activity of the vector with the
wild-type E2F3 3′UTR by ~52.27%. However, the mutant version
abrogated the repressive ability of miR-203 (Fig. 2D). These results demonstrated the
specificity of miR-203 targeting of E2F3.
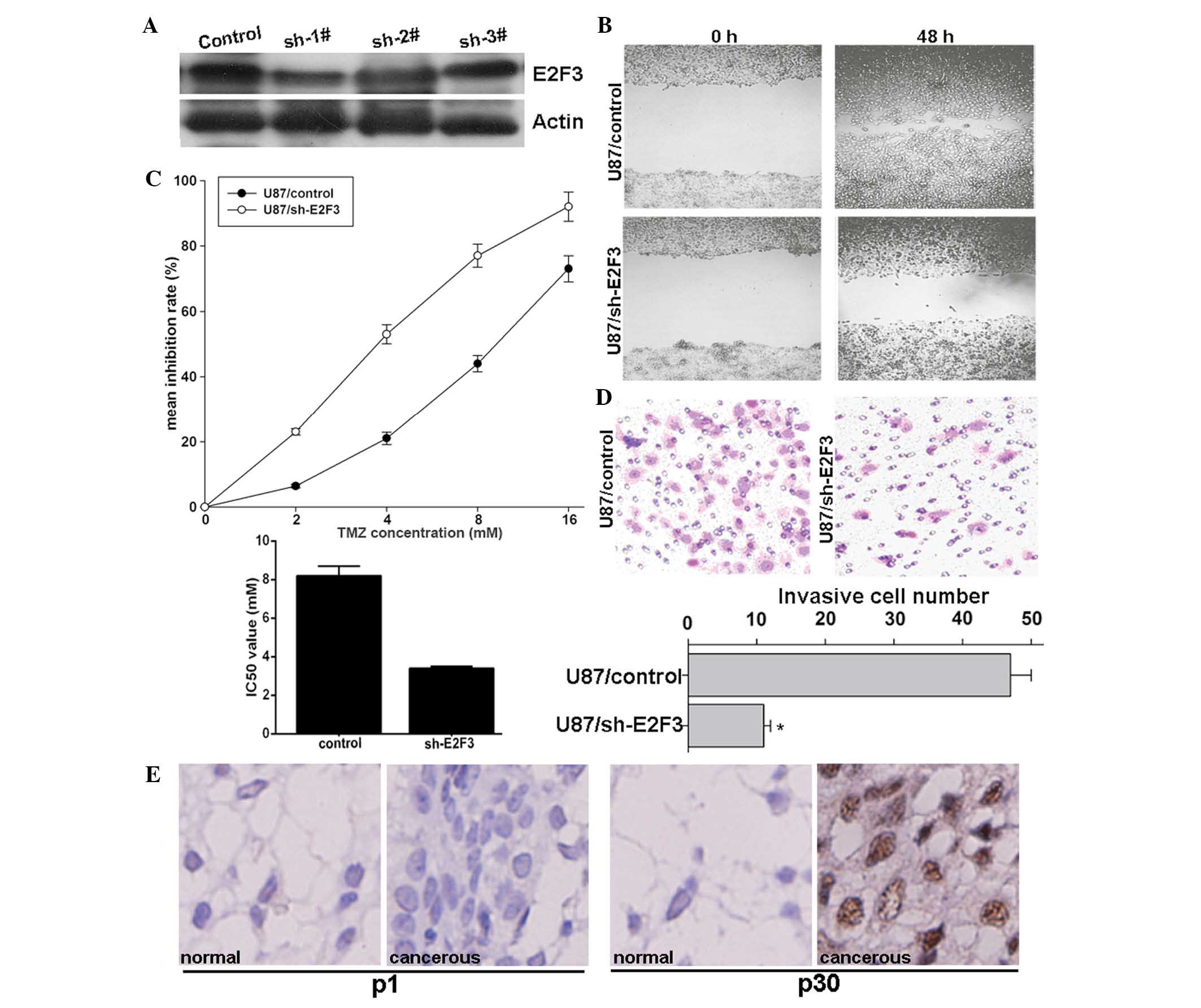
E2F3 knockdown enhances the effect of TMZ
and inhibits invasion of glioma cells
To investigate whether miR-203 enhances
chemotherapeutic sensitivity and inhibits invasion through
inhibition of E2F3 expression, E2F3 was knocked down using
pRNAT-U6.1/sh-E2F3 plasmid expressing a short hairpin RNA (shRNA)
targeting E2F3. Compared with the control, sh-1# markedly decreased
E2F3 protein expression (Fig. 3A),
and therefore, was selected for further study. The functional
effect of E2F3 knockdown on glioma cells was then assessed. The
preset study revealed that E2F3 knockdown significantly inhibited
U87MG-cell migration as demonstrated by the wound-healing assay
(Fig. 3B), enhanced sensitivity of
U87MG cells to TMZ as demonstrated by the MTS assay (Fig. 3C) and inhibited invasion as
demonstrated by the Transwell assay (Fig. 3D), similarly to the effect of
ectopic miR-203 overexpression. Importantly, in the selected 52
clinical glioma tissues, high E2F3 protein expression was detected
in the invasive group, compared to that of the non-invasive group.
Representative data were obtained from patient one with high
miR-203 expression and patient 30 with low miR-203 expression,
respectively (Fig. 3E). These
results suggested that E2F3 may be involved in the mechanism of
miR-203 in invasion inhibition in glioma cells.
Discussion
Although the roles of miRNAs in cancer have been
widely studied via analysis of specific genes targeted and their
biological functions as well as their significance in cancer
development, progression and therapeutic response, their
contribution to cancer remains to be fully elucidated (8). For example, knowledge concerning the
role of miR-203 in cancer, particularly in human glioma, remains
incomplete. Recently, miR-203 was found to be frequently
downregulated in cervical cancer tumors and cell lines due to
methylation of the miR-203 promoter. miR-203 was also capable of
suppressing cervical cancer cell proliferation, tumor growth, and
angiogenesis by directly targeting vascular endothelial growth
factor A (11). Zhang et al
(12) observed that
hypermethylated miR-203 was involved in Nickel-induced
tumorigenesis and that miR-203 may suppress tumorigenesis, at least
in part, by targeting Abelson murine leukemia viral oncongene
homolog 1. Another study revealed that miR-203 was significantly
downregulated in esophageal cancer and overexpression of miR-203 in
esophageal cancer cells markedly increased cell apoptosis and
inhibited cell proliferation, migration and invasion as well as
tumor growth (13). The present
study, for the first time, to the best of our knowledge,
demonstrated that miR-203 expression was markedly lower in the more
aggressive U87MG glioma cells than that in A172 glioma cells.
Ectopic expression of miR-203 significantly inhibited the invasion
of U87MG cells and sensitized them to TMZ. Inversely, an miR-203
inhibitor markedly promoted the invasion of A172 cells and
attenuated their sensitivity to TMZ. Moreover, miR-203 expression
was negatively associated with the invasive potential of glioma
cells. In regard to the mechanism of miR-203 in glioma,
bioinformatic analysis combined with a luciferase reporter assay
indicated that miR-203 may directly target E2F3. As expected, E2F3
knockdown demonstrated the same effects as ectopic expression of
miR-203 on U87MG cells.
E2F3 belongs to the E2F transcription factor family,
which is well conserved and widely known for its role in
proliferation and cell-cycle progression (14). Genetic mouse models provided the
first evidence demonstrating that E2F3 largely contributed to
retinoblastoma protein-attributed tumorigenic events, such as the
reduction of retinoblastoma-deficient pituitary adenomas in
E2F3-deficient mice (15), and
additionally inhibited the development of preneoplastic lesions of
the lung (16). Furthermore, the
loss of E2F3, in combination with E2F1 and E2F2, reduced
hyperplasia in intestinal epithelia (17). In human patients, there is mounting
evidence that deregulation of E2F3 protein contributes to tumor
formation. The E2F3 gene, located on chromosome 6p22, was found to
be amplified in a subset of retinoblastoma, breast, and urinary
bladder carcinomas (18–23). In bladder cancer, E2F3
amplification is associated with more malignant and invasive
cancers (20,21). E2F3 overexpression has also been
reported in lung, ovarian and prostate cancer. Importantly, E2F3
levels independently predict clinical outcome in patients with
prostate cancer (24–26). Thus, E2F3 is closely associated
with cancer development and progression. In previous studies, the
overexpression of E2F3 has been mainly attributed to gene
amplification. Recently, studies have focussed greater attention on
the study of the epigenetic mechanisms underlying gene expression
regulation (9). A study by Feng
et al (27) revealed that
miR-200b was capable of targeting E2F3 to reverse the
chemoresistance of docetaxel-resistant human lung adenocarcinoma
cells. In the present study, for the first time, to the best of our
knowledge, it was demonstrated that the role of E2F3 in glioma
cells was associated with the response to chemotherapy and
invasion. The mechanisms responsible for the downregulation of
miR-203 and the role of E2F3 in glioma will be explored in a
forthcoming study.
In conclusion, the present study indicated that
miR-203 was reversely associated with migration and invasion, and
positively associated with chemosensitivity in glioma cells. E2F3
was shown to be a novel target of miR-203 and E2F3 knockdown
exerted a similar effect to that of miR-203 overexpression. These
results indicate that miR-203 may act as a tumor suppressor by
targeting E2F3 in glioma cells and that miR-203/E2F3 may be a novel
candidate for developing rational therapeutic strategies in glioma
treatment.
Acknowledgements
The study was supported by the project of Science
and Technology of Hunan Province, China (no. 2010SK3103).
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hammond SM: MicroRNAs as oncogenes. Curr
Opin Genet Dev. 16:4–9. 2006. View Article : Google Scholar
|
|
3
|
Croce CM and Calin GA: miRNAs, cancer and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar
|
|
7
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z
and Dong JT: Epigenetic silencing of miR-203 upregulates SNAI2 and
contributes to the invasiveness of malignant breast cancer cells.
Genes Cancer. 2:782–791. 2011. View Article : Google Scholar
|
|
10
|
Li Y, Yuan Y, Tao K, Wang X, Xiao Q, Huang
Z, Zhong L, Cao W, Wen J and Feng W: Inhibition of BCR/ABL protein
expression by miR-203 sensitizes for imatinib mesylate. PloS One.
8:e618582013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu X, Er K, Mao C, Yan Q, Xu H, Zhang Y,
Zhu J, Cui F, Zhao W and Shi H: miR-203 suppresses tumor growth and
angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol
Biochem. 2:64–73. 2013. View Article : Google Scholar
|
|
12
|
Zhang J, Zhou Y, Wu YJ, Li MJ, Wang RJ,
Huang SQ, Gao RR, Ma L, Shi HJ and Zhang J: Hyper-methylated
miR-203 dysregulates ABL1 and contributes to the nickel-induced
tumorigenesis. Toxicol Lett. 223:42–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen
X, Gao Z, Xin J, Zhou S, Zhou Z, et al: MiR-203 suppresses tumor
growth and invasion and down-regulates MiR-21 expression through
repressing Ran in esophageal cancer. Cancer Lett. 342:121–129.
2014. View Article : Google Scholar
|
|
14
|
van den Heuvel S and Dyson NJ: Conserved
functions of the pRB and E2F families. Nat Rev Mol Cell Biol.
9:713–724. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ziebold U, Lee EY, Bronson RT and Lees JA:
E2F3 loss has opposing effects on different pRB-deficient tumors,
resulting in suppression of pituitary tumors but metastasis of
medullary thyroid carcinomas. Mol Cell Biol. 23:6542–6552. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parisi T, Yuan TL, Faust AM, Caron AM,
Bronson R and Lees JA: Selective requirements for E2f3 in the
development and tumorigenicity of Rb-deficient chimeric tissues.
Mol Cell Biol. 27:2283–2293. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chong JL, Wenzel PL, Sáenz-Robles MT, Nair
V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, et
al: E2f1–3 switch from activators in progenitor cells to repressors
in differentiating cells. Nature. 462:930–934. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Veltman JA, Fridlyand J, Pejavar S, Olshen
AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson
D, et al: Array-based comparative genomic hybridization for
genome-wide screening of DNA copy number in bladder tumors. Cancer
Res. 63:2872–2880. 2003.PubMed/NCBI
|
|
19
|
Orlic M, Spencer CE, Wang L and Gallie BL:
Expression analysis of 6p22 genomic gain in retinoblastoma. Genes
Chromosomes Cancer. 45:72–82. 2006. View Article : Google Scholar
|
|
20
|
Oeggerli M, Tomovska S, Schraml P,
Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ and
Sauter G: E2F3 amplification and overexpression is associated with
invasive tumor growth and rapid tumor cell proliferation in urinary
bladder cancer. Oncogene. 23:5616–5623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feber A, Clark J, Goodwin G, Dodson AR,
Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, et al:
Amplification and overexpression of E2F3 in human bladder cancer.
Oncogene. 23:1627–1630. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grasemann C, Gratias S, Stephan H, Schüler
A, Schramm A, Klein-Hitpass L, Rieder H, Schneider S, Kappes F,
Eggert A and Lohmann DR: Gains and overexpression identify DEK and
E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene.
24:6441–6449. 2005.PubMed/NCBI
|
|
23
|
Andre F, Job B, Dessen P, Tordai A,
Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al:
Molecular characterization of breast cancer with high-resolution
oligonucleotide comparative genomic hybridization array. Clin
Cancer Res. 15:441–451. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Foster CS, Falconer A, Dodson AR, Norman
AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar
S, et al: Transcription factor E2F3 overexpressed in prostate
cancer independently predicts clinical outcome. Oncogene.
23:5871–5879. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Borczuk A, Gorenstein L, Walter KL, Assaad
AA, Wang L and Powell CA: Non-small-cell lung cancer molecular
signatures recapitulate lung developmental pathways. Am J Pathol.
163:1949–1960. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu KH, Patterson AP, Wang L, Marquez RT,
Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I,
et al: Selection of potential markers for epithelial ovarian cancer
with gene expression arrays and recursive descent partition
analysis. Clin Cancer Res. 10:3291–3300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng B, Wang R, Song HZ and Chen LB:
MicroRNA-200b reverses chemoresistance of docetaxel-resistant human
lung adenocarcinoma cells by targeting E2F3. Cancer. 118:3365–3376.
2012. View Article : Google Scholar
|