Introduction
Reverse transcription quantitative polymerase chain
reaction (RT-qPCR) is frequently used in gene expression studies
and is currently considered the gold standard for accurate,
sensitive and rapid measurements of gene expression(1). Relative quantification is an
important and commonly used technique to evaluate RT-qPCR data,
while the expression levels of target genes are compared to those
of a stably expressed endogenous control gene, determined
simultaneously in the same biological sample (2,3).
Therefore, the gene expression levels require normalization using
reference genes in order to obtain reliable data. The
identification of appropriate reference genes is a crucial stage
involved in this approach. It is important for the ideal reference
genes to be universally valid under the experimental conditions
(1–3). In general, cellular maintenance genes
are selected as reference genes to examine the variability between
clinical samples. Several studies have demonstrated that the
expression levels of these reference genes vary in different
tissues or between treatments in the same tissue (4–5), as
well as across cell types (6).
Gallbladder carcinoma is the most common type of
malignant tumor of the biliary system worldwide; this type of tumor
is highly fatal, with an overall 5-year survival rate of <5%
(7). In the majority of cases,
this disease is rapid and silent, resulting in a poor prognosis,
which has not improved over the last few decades. An effective
therapeutic approach requires early diagnosis and timely surgery.
Despite this potential for cure, <10% of patients have tumors
that are resectable at the time of surgery, whilst almost 50% have
lymph node metastasis (8).
Gallbladder carcinoma has been regarded as one of the most
difficult conditions to treat. Previous gene expression studies in
gallbladder carcinoma tissue and normal gallbladder tissue
counterparts have been performed to identify new predictive and
prognostic molecular markers associated with gallbladder carcinoma
(9–11). RT-qPCR is a frequently used
technique to investigate these markers, thus, a review of the
normalization standards used in the quantitative gene expression
studies of gallbladder carcinoma was necessary. In the present
study, the keywords gallbladder carcinoma or gallbladder cancer and
RT-PCR were used in a PubMed search of previous studies. GAPDH is
the most frequently used standard.(12,13),
followed by ACTB (14,15). The search results revealed that no
systematic study has been performed on the selection of suitable
reference genes for investigating target gene profiling in
gallbladder carcinoma.
The present study aimed to identify the most
suitable reference gene or set of genes for target gene profiling
of gallbladder carcinoma. The stability of a panel of 12 common
reference genes in gallbladder carcinoma tissues and paired normal
gallbladder tissues from 16 patients were validated. The 12
candidate genes: ACTB, ALAS1, GAPDH, TBP, HPRT1, RPL29, PBGD, PPIA,
PUM1, GUSB, B2M and 18S rRNA are frequently used as endogenous
controls in the context of, but not restricted to, gallbladder
carcinoma. A number of these genes have been identified as optimal
reference genes in certain other cancer types, including HPRT1 and
ACTB (5,16). To investigate these genes, three
common software packages, geNorm (17), NormFinder (18) and Bestkeeper (19) were used and to determine their
validity, candidate reference genes were used to measure C-myc
levels, which are closely associated with gallbladder carcinoma
(20). The aim was to provide
useful information for the selection of suitable reference genes in
further gene expression studies on gallbladder carcinoma
tissues.
Materials and methods
Gallbladder carcinoma samples
A total of 16 gallbladder carcinoma samples were
obtained between January 2008 and December 2013 with prior consent
from untreated patients who underwent tumor resection surgery.
Paired normal samples were collected from the adjacent non-tumor
gallbladder tissues. All the specimens were obtained from patients
at the China-Japan Union Hospital, Jilin University (Changchun,
China) and snap-frozen in liquid nitrogen immediately following
excision prior to storing at −80°C until further processing. Only
histologically confirmed tumor and non-neoplastic tissue samples
were used for RNA analysis. Tumor stage was determined according to
the International Union Against Cancer American Joint Committee on
Cancer and International Union Against Cancer (21). The clinicopathological
characteristics of the patients are summarized in Table I. The present study was approved by
the Ethics Committee of the China-Japan Union Hospital.
 | Table IClinicopathological characteristic of
patients. |
Table I
Clinicopathological characteristic of
patients.
| Clinicopathological
characteristic | Patients with
gallbladder carcinoma |
|---|
| Age (mean ± standard
deviation) | 50±16.7 |
| Gender |
| Male | 10 |
| Female | 6 |
| Histopathological
type |
| Adenocarcinoma | 16 |
| Squamous cell
carcinomas | 0 |
| TNM stagea |
|
T1aN0M0 | 3 |
|
T1bN0M0 | 6 |
|
T2aN1M0 | 3 |
|
T2bN1M0 | 4 |
RNA extraction and RT
A total of 50–100 mg tissue samples were homogenized
in 1 ml TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA) and purified using an RNeasy Mini kit (Qiagen, Valencia, CA,
USA). DNaseI was used to eliminate genomic DNA contamination. The
concentrations and quality of the isolated RNA were measured using
a Synergy HT enzyme standard instrument (BioTek, Winooski, VT,
USA). The purity of total RNA was determined using the A260/A280
ratio. The standard of including RNA samples was 260/280 between
1.9 and 2.2. The integrity of the RNA samples was determined by
electrophoresis on a 1% agarose gel (Invitrogen Life Technologies).
The RT reaction was performed using an All.in.One™ First.Strand
cDNA Synthesis kit (GeneCopoeia Inc., Rockville, MD, USA) in a
total volume of 25 μl according to the manufacturer’s
instructions.
RT-qPCR
The primers of 12 putative reference genes were
designed using Primer Premier 5.0 software (Premier Biosoft, Palo
Alto, CA, USA) and were synthesized by Sangon company (Beijing,
China) as shown in Table II. A
Roche LightCycler 480 detection system (Roche Diagnostics, GmbH,
Mannheim, Germany) was used for RT-qPCR. Reactions were performed
using All.in.One™ qPCR Mix (GeneCopoeia, Inc.) according to the
manufacturer’s instructions. All the samples were run in triplicate
on 96-well plates. The PCR volume was 20 μl, containing 2 μl cDNA.
The following cycling conditions were used: 55°C for 5 min; 95°C
for 5 min; 40 cycles of 95°C for 20 sec, 55°C for 20 sec and 72°C
for 4 min. This cycle was followed by melting curve analysis, the
baseline and cycle threshold values (Ct values) were automatically
determined for all the plates using Roche LightCycler 480 software
(Roche Diagnostics, Mannheim, Germany). A standard curve was
constructed for each primer pair to determine the product
specificity.
 | Table IIPrimer sequences, product size and
polymerase chain reaction (PCR) efficiency. |
Table II
Primer sequences, product size and
polymerase chain reaction (PCR) efficiency.
| Gene | Primer sequence | Product size
(bp) | PCR efficiency |
|---|
| 18SrRNA | F:
CGGCTACCACATCCAAGGAA
R: GCTGGAATTACCGCGGCT | 186 | 2.11 |
| GAPDH | F:
GACAGTCAGCCGCATCTTCT
R: TTAAAAGCAGCCCTGGTGAC | 127 | 1.99 |
| B2M | F:
AGCGTACTCCAAAGATTCAGGTT
R: ATGATGCTGCTTACATGTCTCGAT | 206 | 1.97 |
| ACTB | F:
AGAAAATCTGGCACCACACC
R: TAGCACAGCCTGGATAGCAA | 173 | 1.97 |
| ALAS1 | F:
GGCAGCACAGATGAATCAGA
R: CCTCCATCGGTTTTCACACT | 150 | 2.02 |
| GUSB | F:
AGCCAGTTCCTCATCAATGG
R: GGTAGTGGCTGGTACGGAAA | 160 | 1.79 |
| HPRT1 | F:
GACCAGTCAACAGGGGACAT
R: CCTGACCAAGGAAAGCAAAG | 132 | 1.96 |
| PBGD | F:
AGTGTGGTGGGAACCAGC
R: CAGGATGATGGCACTGAACTC | 144 | 2.20 |
| PPIA | F:
AGACAAGGTCCCAAAGAC
R: ACCACCCTGACACATAAA | 118 | 1.96 |
| PUM1 | F:
CAGGCTGCCTACCAACTCAT
R: GTTCCCGAACCATCTCATTC | 211 | 2.01 |
| RPL29 | F:
GGCGTTGTTGACCCTATTTC
R: GTGTGTGGTGTGGTTCTTGG | 120 | 2.00 |
| TBP | F:
TGCACAGGAGCCAAGAGTGAA
R: CACATCACAGCTCCCCACCA | 132 | 2.16 |
| C-myc | F:
GCCACGTCTCCACACATCAG
R: TGGTGCATTTTCGGTTGTTG | 132 | 1.98 |
The Ct values were identified by
quantitative comparison of the amplification of the candidate
genes. The Ct values were calculated to relative
quantities (Q) for data analysis, in view of the PCR efficiencies
of the candidate genes according to the equation:
Q=2−ΔC.
PCR efficiency
A random pool of cDNA from the samples was selected
and used for 2-fold serial dilutions, ranging between 1X and
100,000X. The PCR were run in triplicate, as mentioned previously.
The PCR efficiency was calculated using the slopes of the
calibration curve and by the formula: E = 10−1/slope
(22). All PCR efficiencies are
shown in Table II.
Statistical analysis
All the samples were divided into three groups:
Gallbladder carcinoma, normal matching gallbladder and total sample
groups. In order to better evaluate the stability of the reference
genes, three frequently used software programs (geNorm, http://medgen.ugent.be/~jvdesomp/genorm/http://medgen.ugent.be/~jvdesomp/genorm/;
NormFinder, http://www.mdl.dk/publica-tionsnormfinder.htm; and
BestKeeper, http://www.gene-quantification.de/bestkeeper.html)
were selected. GeNorm is designed to establish reference genes for
RT-qPCR and can be used to analyze and determine the M-value, which
refers to the stability of the reference gene expression (17). The default value suggested by
geNorm is M=1.5. The higher the M-value, the less stable and the
lower the M value, the more stable. If M is >1.5, it is not
suitable for use as a reliable reference gene. GeNorm software can
also be used to analyze the pairwise variation value of the
normalization factor (V), which has a default value of 0.15. The
value of Vn/Vn+1 can be used to determine
whether adding a new reference gene affects the normalization
factor. If the value of Vn/Vn+1 is >0.15,
it is necessary to use the n+1 reference genes as internal
controls. If it is <0.15, then it is not necessary to use new
reference genes. NormFinder software is a tool designed to identify
the optimal reference gene among a set of candidates and it has a
similar operation principle to geNorm (4). This programme analyzes expression
data, ranks the set of candidate normalization genes according to
their expression stability and considers the gene with the minimum
expression data as the most stable gene (19). This software can also be used to
compare the stability of inter- and intra-group reference genes.
BestKeeper evaluates candidate reference gene stability based on
the standard deviation (SD) and correlation coefficient (r). An
SD>1, is unsuitable for use as a stable and reliable reference
gene. The remaining genes were ranked according to their r value,
the higher the r value, the more the stable and reliable the
gene.
Target gene relative expression
analysis
The C-myc proto-oncogene is involved in the process
of malignant tumor formation (23). The present study measured C-myc as
a target gene with the primer sequence shown in Table II. The relative expression levels
of the target gene C-myc were calculated in the 16 paired samples
according to the 2−ΔΔC method (24), with different candidate reference
genes used as standards.
Results
RNA quality
To avoid erroneous results, only high-quality RNA
samples were included in this study. The concentration, purity and
integrity of the total RNA sample were determined. The mean
A260/280 ratio of the RNA samples was 2.01±0.045 (25) and the integrity of RNA samples was
characterized by the 28S/18S ratio (>1.5) on 1% agarose
gels.
The primers sequences, corresponding length of the
amplified products and PCR amplification efficiency is shown in
Table I. There are two methods to
verify the specificity of the primers, the RT-qPCR amplification
products were detected by 1% agarose gel electrophoresis. The gel
imaging system indicated that the size of the amplified fragment
was consistent with the expected size, with a clear band and
without primer dimers and nonspecific bands. In addition, the
melting curve of each gene fragment amplified by qPCR revealed that
all curves exhibited a single signal peak. For the candidate
reference gene and target gene, the amplification efficiency range
of the standard curve was 1.79–2.20 and all correlation
coefficients were >0.98.
Gene expression levels
The expression level of the candidate reference
genes was determined by the Ct value, which is inversely
proportional to the expression level of the gene. Higher Ct values
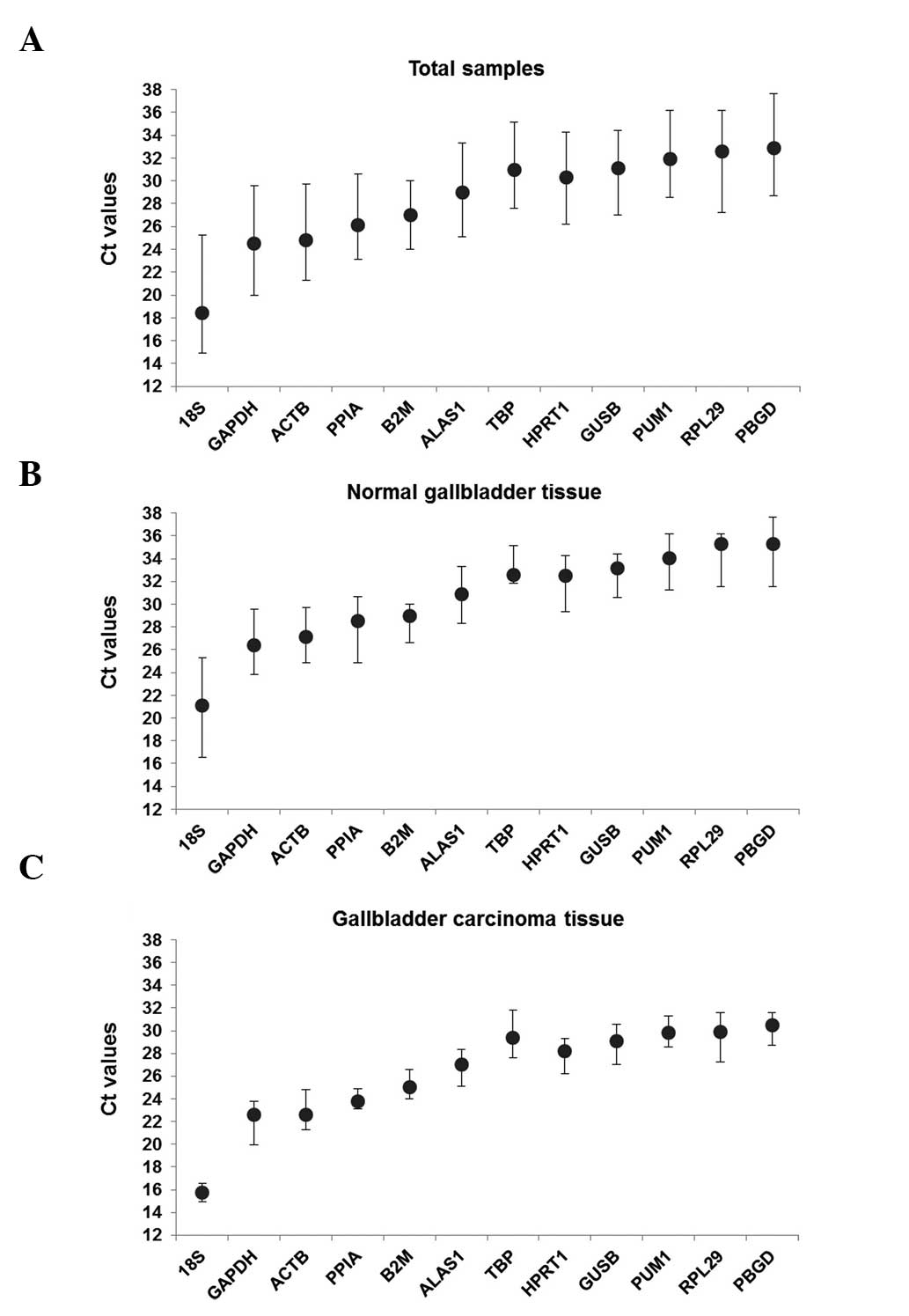
indicated smaller the expression quantities. As shown in Fig. 2, the Ct value of all the samples
ranged between 14.92 and 37.64. In all groups, 18SrRNA had the
smallest Ct values of 18.41±3.49, 21.09±4.52 and 15.72±0.85 and
PBGD had the greatest CT values of 32.88±4.19, 35.29±3.73 and
30.47±1.47. There was a significant difference in the expression
levels of the candidate reference genes between the gallbladder
carcinoma tissues and its paired normal gallbladder tissues.
Overall, the change in the Ct value of each group of candidate
genes indicates that the expression level changed under different
experimental conditions.
Stability analysis of the candidate
reference gene
Theoretically, 12 reference genes constitute an
appropriate internal for controlling genes. The measure used by the
geNorm program uses to calculate the stability of gene expression
is the M-value, among which the lowest M-value indicates the most
stable expression. Based on ranking of M-values, the most unstable
genes are gradually removed and the two most stable genes are
determined simultaneously. The M-value of the 12 candidate
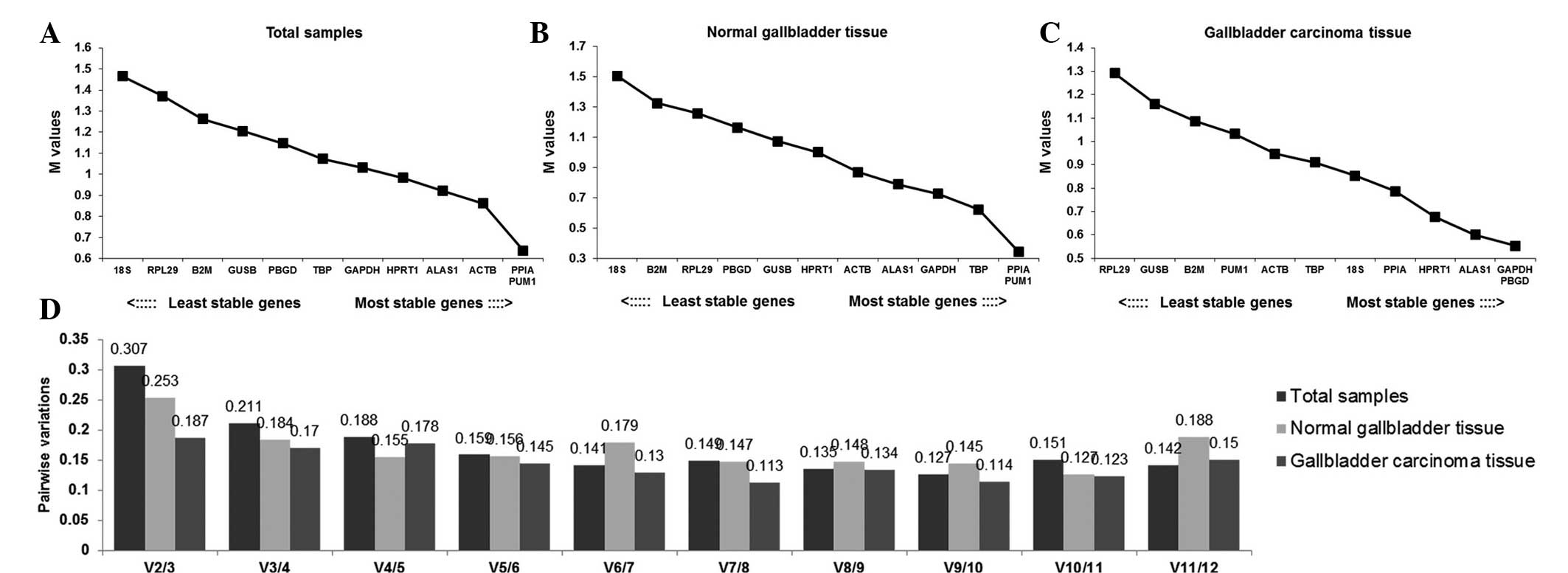
reference genes in each group are shown in Fig. 3. In the total sample group and the
normal gallbladder group, 18SrRNA had the biggest M value,
suggesting that it is the most unstable candidate gene in the two
groups. In these groups, PPIA and PUM1 (M=0.6) were determined to
be the most stable genes. In the gallbladder carcinoma tissue
group, GAPDH and PBGD were the most stable reference genes and
RPL29 was the most unstable. The default threshold V-value is 0.15,
however, 0.15 is not an absolute cut-off value, but an ideal value,
which is dependent on the expression of the genes and the diversity
of the samples assessed (26). A
combination of six reference genes in the total sample group was
optimal (V6/7=0.141), while a combination of five genes was optimal
in the gallbladder carcinoma group (V5/6=0.145). In the normal
gallbladder group, a combination of seven reference genes was
optimal (V7/8=0.147).
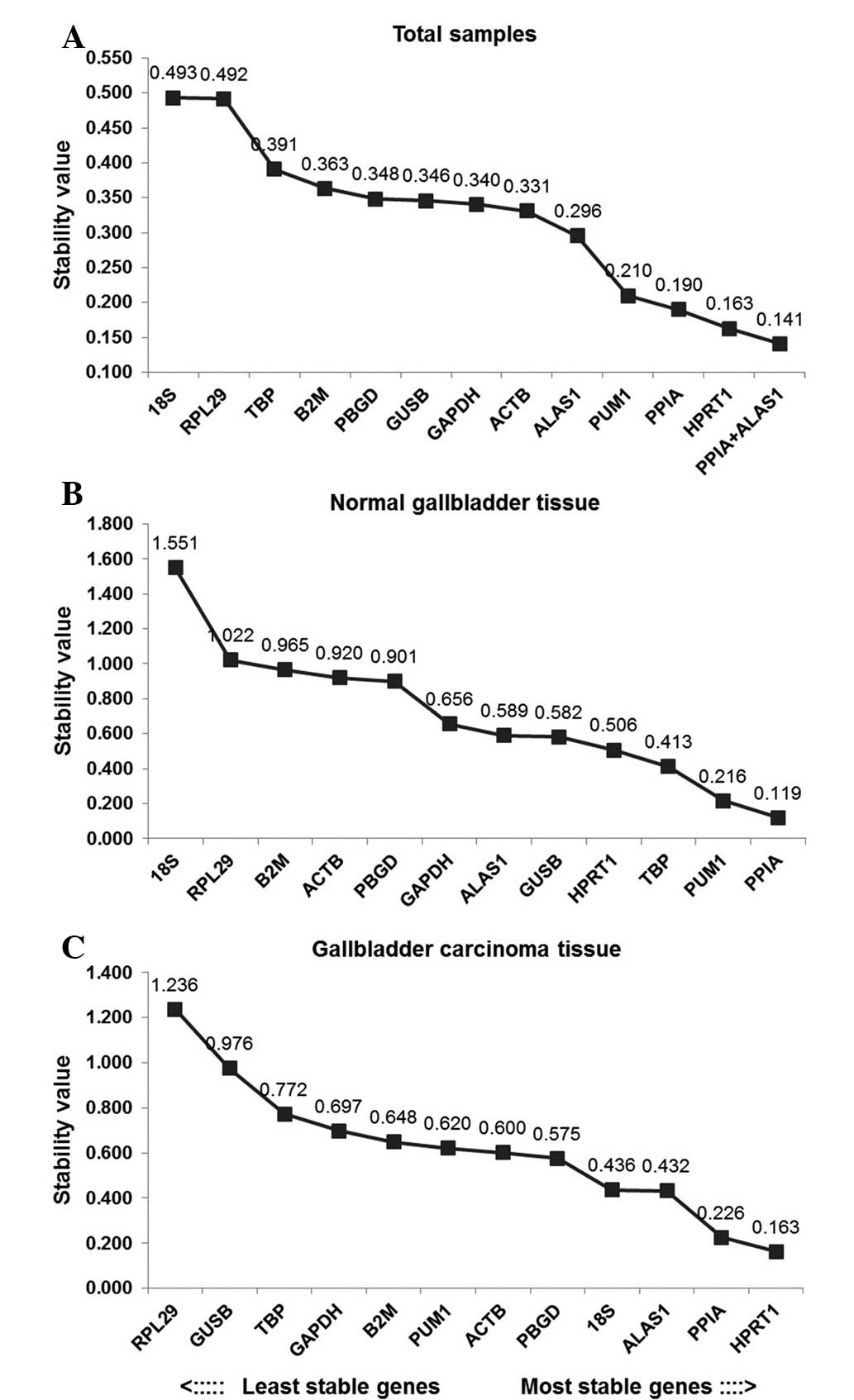
In order to better evaluate the stability of the 12
reference genes, the present study also used the Normfinder
program. As shown in Fig. 4 ALAS1
+ PPIA was the most stable reference gene combination in the total
sample group. HPRT1 was the most stably expressed gene in this
group, followed by PPIA. The least stably expressed gene in the
total sample group was 18SrRNA. In the gallbladder carcinoma group,
HPRT1 was the most stably expressed gene, followed by PPIA. RPL29
was the least stably expressed gene in the gallbladder carcinoma
group. In the paired normal gallbladder group, PPIA was the most
stable reference gene, followed by PUM1, whilst 18SrRNA was the
least stable gene (Fig. 4).
The BestKeeper program can also be used to compare
the stability of internal reference genes. Since the BestKeeper
program can only analyze 10 internal reference genes (19), the two most unstable internal
reference genes indicated by the geNorm analyses were removed in
each group. The BestKeeper analysis demonstrated that the SD values
in the total sample group were all >1, however this was not
considered to indicate that the 12 candidate internal reference
genes were all unstable, since analysis using a single software
program is not conclusive. In order of the SD value, the most
stable internal reference gene in the total sample group was TBP.
In the matching normal gallbladder group, the SD values of GUSB,
RPL29 and HPRT1 were all <1. In terms of r-value, HPRT1 was the
most stable internal reference gene. In the gallbladder carcinoma
group, only the SD values of TBP and GAPDH were >1 and the
r-values of the remaining candidate genes indicated that HPRT1 was
the most stable internal reference gene.
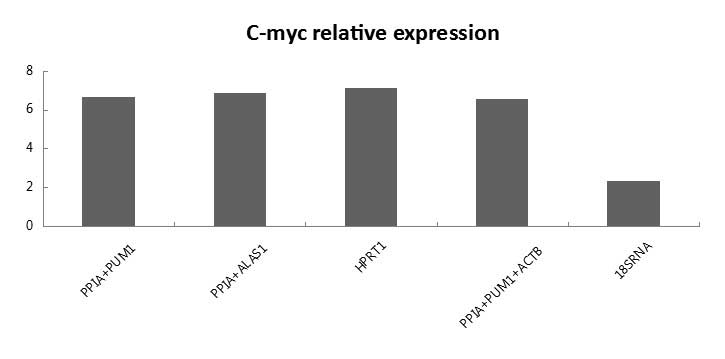
Relative expression of C-myc
The quantification of target gene expression was
affected by selecting different reference genes. As shown in
Fig. 6, when the recommended HPRT1
gene and the gene combinations ALAS1 + PPIA, PPIA + PUM1 and PPIA +
PUM1 + ACTB, were set as references, no significant difference was
observed in the gene expression of C-myc. However, when 18SrRNA was
used as a reference gene for normalization, the relative expression
of C-myc in malignant gallbladder tissue was markedly different,
compared with using the previously mentioned reference gene as a
standard.
Discussion
In the detection of target gene expression, a gene
with a steady expression level is required to normalize the data,
these are internal reference genes (2,3).
Previous studies have indicated that the majority of these commonly
used internal control genes have flaws. Their expression level
varies significantly depending on various experimental conditions,
including different cell types and tissues, different stages of
cell proliferation and organ development and in vitro
culture (4,5,15).
To the best of our knowledge, the present study is first to compare
the stability of commonly used internal reference genes in
gallbladder carcinoma tissue and their benign counterparts. As of
studies investigating gallbladder carcinoma gene profiling develop,
confirming stable and reliable internal control genes is required.
In the present study, the reference genes commonly used in studies
of gene expression in gallbladder carcinoma were used as were those
frequently used in studies examining molecular markers in other
cancer tissues.
To obtain accurate experimental data and reliable
conclusions, the present study used an experimental process with a
number of characteristics. Malignant and benign specimens from the
same gallbladder were used to minimize differences between
individuals. Due to limitations in the indications for gallbladder
carcinoma surgery, biopsy specimens were not selected by grades and
stages, as according to previous research, the expression of
reference genes is not directly associated with the grades or stage
of a malignant tumor (5,27). The specimens were confirmed by the
Pathology Department of the China-Japan Union Hospital as malignant
and the gallbladder carcinoma samples used were the most common
pathological types of adenocarcinoma. A total of 12 types of common
reference genes were compared in terms of their expression
stability and the geNorm, NormFinder and BestKeeper software
programmes, commonly used to compare stability between reference
genes, were selected for data analysis.
The geNorm program was used for initial analysis.
This software program is based on a pairwise-comparison statistical
model. By calculating the values of M and V, the two most stable
reference genes and the best reference gene combinations were
determined. Following this analysis, the results suggested that in
the total sample group and the paired normal gallbladder group,
PPIA and PUM1 were the most stable reference genes. In the
gallbladder carcinoma group, GAPDH and PBGD were the most stable
reference genes. In addition, by calculating the value of V, the
optimal reference gene combinations of the total sample,
gallbladder and paired normal gallbladder groups consisted of six,
five and seven reference genes, respectively. The boundary value
suggested by geNorm was 0.15, however, rather than a stringent
standard consideration, can provide guidance to determine the
optimal number of reference genes. Regarding the standardized
principle of RT-qPCR, previous studies recommend selecting at least
three internal control genes to perform the relative quantitative
investigation (16). The present
study also recommended that a combination of three reference genes
be most reliable. The recommended combinations for the total sample
group were PPIA + PUM1 + ACTB, the gallbladder carcinoma group were
GAPDH + PBGD + ALAS1 and the paired normal gallbladder group were
PPIA + PUM1 + TBP. The results of the Normfinder software program,
based on the analysis of variance as the statistical model, were
the same. Finally, in order to reduce the one-sidedness of the
computing models of the above-mentioned software programs, the
Bestkeeper program was used for analysis. However, the results of
BestKeeper differed to those from those of geNorm and Normfinder,.
It has been suggested that the Bestkeeper statistical model differs
from these and thus is less effective in ranking reference gene
stability (28). Following
comparison of the results from the three software programs, HPRT1
was the most stably expressed reference gene in the gallbladder
carcinoma group. In the paired normal gallbladder group, the most
stably expressed reference gene was PPIA and in total sample group,
PPIA was the most stably expressed gene.
The gene expression of C-myc differed depending on
the normalization method used, demonstrating the importance of
reference genes to obtain reliable expression data. The C-myc gene
is highly expressed in actively multiplying cells and several tumor
cells. Previous studies have demonstrated that the expression of
C-myc in gallbladder carcinoma tissues is higher than in
gallbladder benign lesion tissues (29). The analysis of the relative
expression level of C-myc in the present study also confirmed this.
The present study used the most stable reference gene HPRT1 and the
reference gene combinations ALAS1 + PPIA, PPIA + PUM1 and PPIA +
PUM1 + ACTB, recommended by the geNorm and Normfinder software, and
also used 18SrRNA, of relatively poor stability as the standard in
relative quantification analysis. The result indicated that the
relative expression levels of C-myc were markedly different,
suggesting the importance of a suitable reference gene for the gene
profiling of gallbladder carcinoma. Similar erroneous
normalizations have been performed in other tissues, including
gastric cancer or in cell lines when inadequate control genes or
normalizing strategies were performed (15,30).
The present study identified the most suitable
reference genes and reference gene combinations for gallbladder
carcinoma tissue and paired normal gallbladder tissue for use in
gene expression profile analysis. A reliable standardized method
has the potential to improve understanding of the biological
mechanisms underlying gallbladder carcinoma in the future. The
relevant clarification of tumor molecular expression markers may
improve the accuracy of diagnosis and estimation of prognostic
factors and provide novel treatments.
Acknowledgements
This study was supported by the Key Foundation of
Jilin Provincial Science and Technology Department (nos.
20130727038YY and 20100942) and the Jilin Provincial Development
and Reform Commission (no. 20101928).
References
|
1
|
Radonić A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004. View Article : Google Scholar
|
|
2
|
Bustin SA, Benes V, Nolan T and Pfaffl MW:
Quantitative realtime RT-PCR - a perspective. J Mol Endocrinol.
34:597–601. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Derveaux S, Vandesompele J and Hellemans
J: How to do successful gene expression analysis using real-time
PCR. Methods. 50:227–230. 2010. View Article : Google Scholar
|
|
4
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of realtime quantitative reverse transcription-PCR
data: a model based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohl F, Jung M, Xu C, Stephan C, Rabien A,
Burkhardt M, Nitsche A, Kristiansen G, Loening SA, Radonić A and
Jung K: Gene expression studies in prostate cancer tissue: which
reference gene should be selected for normalization? J Mol Med
(Berl). 83:1014–1024. 2005. View Article : Google Scholar
|
|
6
|
Mansur NR, Meyer-Stegler K, Wurtz JC and
Sirover MA: Cell cycle regulation of the
glyceraldehydes-3-phosphate dehydrogenase/uracil glycosylase gene
in normal human cell. Nucleic Acids Res. 21:993–998. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai CH and Lau WY: Gallbladder cancer - a
comprehensive review. Surgeon. 6:101–110. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheth S, Bedford A and Chopra S: Primary
gallbladder cancer: recognition of risk factors and the role of
prophylactic cholecystectomy. Am J Gastroenterol. 95:1402–1410.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Legan M: COX-2, P53, GLUT-1 as predictors
of malignancy n the development of gallbladder carcinomas. Bosn J
Basic Med Sci. 10:192–196. 2010.PubMed/NCBI
|
|
11
|
Sun XN, Cao WG, Wang X, et al: Prognostic
impact of vascular endothelial growth factor-A expression in
resected gallbladder carcinoma. Tumour Biol. 32:1183–1190. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sergeant G, Lerut E, Ectors N, et al: The
prognostic relevance of tumor hypoxia markers in resected carcinoma
of the gallbladder. Eur J Surg Oncol. 37:80–86. 2011. View Article : Google Scholar
|
|
13
|
Zhang JT, Sun W, Zhang WZ, Ge CY, Liu ZY,
Zhao ZM, Lu XS and Fan YZ: Norcantharidin inhibits tumor growth and
vasculogenic mimicry of human gallbladder carcinomas by suppression
of the PI3-K/MMPs/Ln-5γ2 signaling pathway. BMC Cancer. 14:1932014.
View Article : Google Scholar
|
|
14
|
Du X, Wu T, Lu J, Zang L, Song N, Yang T,
Zhao H and Wang S: Decreased expression of chromodomain helicase
DNA-binding protein 5 is an unfavorable prognostic marker in
patients with primary gallbladder carcinoma. Clin TranslOncol.
15:198–204. 2013.
|
|
15
|
Huan P, Maosheng T, Zhiqian H, Long C and
Xiaojun Y: TLR4 expression in normal gallbladder, chronic
cholecystitis and gallbladder carcinoma. Hepatogastroenterology.
59:42–46. 2012.PubMed/NCBI
|
|
16
|
Wisnieski F, Calcagno DQ, Leal MF, et al:
Reference genes for quantitative RT-PCR data in gastric tissues and
cell lines. World J Gastroenterol. 19:7121–7128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kubista M, Andrade JM, Bengtsson M,
Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B,
Strömbom L, Ståhlberg A and Zoric N: The realtime polymerase chain
reaction. Mol Aspects Med. 27:95–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goldin RD and Roa JC: Gallbladder cancer:
a morphological and molecular update. Histopathology. 55:218–229.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge S, Byrol BR, Compton CC, Fritz AG,
Greene FL and Troti A: American Joint Committee on Cancer. AJCC
Cancer Staging Manual (M). 7th Edition. Springer; Chicago, IL: pp.
211–217. 2010
|
|
22
|
Ginzinger DG: Gene quantification using
real-time quantitative CR: an emerging technology hits the
mainstream. Exp Hematol. 30:503–512. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brabletz T, Herrmann K, Jung A, et al:
Expression of nuclear betacatenin and c -myc is c orrelated with
tumor size but not with proliferative activity of colorect
aladenomas. Am J Pathol. 156:865–870. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Fleige S, Walf V, Huch S, et al:
Comparison of relative mRNA quantification models and the impact of
RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett.
28:1601–1613. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wan H, Zhao Z, Qian C, et al: Selection of
appropriate reference genes for gene expression studies by
quantitative real-time polymerase chain reaction in cucumber. Anal
Biochem. 399:257–261. 2010. View Article : Google Scholar
|
|
27
|
Li YL, Ye F, Hu Y, Lu WG and Xie X:
Identification of suitable reference genes for gene expression
studies of human serous ovarian cancer by real-time polymerase
chain reaction. Anal Biochem. 394:110–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vinod Kumar, Sharma R, Trivedi P, Vyas C,
Govind K and Khandelwal V: Traditional and novel references towards
systematic normalization of qRT-PCR data in plants. Crop Sci.
511:e1455–e1468. 2011.
|
|
29
|
Yong Cui, Yubing Chen, Gongyu Weng, et al:
Expression and significance of Survivin and C-myc in gallbladder
carcinom. Chinese Journal of Coal Industry Medicine. 10:888–889.
2007.(In Chinese).
|
|
30
|
Liu S, Zhu P, Zhang L, et al: Selection of
reference genes for RT-qPCR analysis in tumor tissues from male
hepatocellular carcinoma patients with hepatitis B infection and
cirrhosis. Cancer Biomark. 13:345–349. 2013.
|