Introduction
Allergic asthma is a complex respiratory disorder,
which is characterized by airway inflammation, bronchial
hyperresponsiveness and reversible airway obstruction (1). In recent decades, an increasing
number of patients have been diagnosed with allergic asthma
(2), and allergic diseases such as
asthma have become a social problem that negatively affects the
quality of life of sufferers. The incidence of allergic diseases,
including asthma, has risen since the mid-20th century, with much
of the increase associated with changes in the environment that
affect the immune system (3). The
exact mechanism for the progression of pediatric allergic diseases
has yet to be elucidated; however, it appears to be a complex
interaction between genetics, environmental exposure and
sensitization (4). Previous
studies have identified small molecular medicines, which may be
used to treat allergic asthma in the future (5); however, adherence rates for asthmatic
patients are problematic, ranging between 30 and 70% (6). Fewer than half of the patients
treated with inhaled asthma medications adhere to their prescribed
regimens (7), and the level of
adherence is similar for children (8). Further molecular and genetic research
is required to elucidate the underlying molecular mechanisms of
allergic asthma.
Microarray DNA hybridization techniques are widely
used in molecular biology research. In a DNA microarray, various
DNA probes are immobilized onto a solid support in groups, forming
an array of microspots. Hybridization to the microarray can then be
performed by applying sample DNA solutions, either in bulk or in a
microfluidic manner. Once the sample DNA has bound to the
immobilized probe DNA through complementary sequence binding,
detection is achieved through the read-out of the tagged markers
attached to the sample target DNA (9). Genome-wide microarray studies of
pooled DNA samples are a valuable tool, which may be used to
identify candidate differentially expressed genes (DEGs) that are
associated with a phenotype in a fast, scalable and economical
manner (10). Previous studies has
used microarray techniques and has reported changes in the
expression of genes associated with viral transcription (RPL3,
RPS10, RPL27, RPS11, RPL27A, RPL37A, EIF5A, EIF5B, and EEF1D) and
lysosome function (ALAS1, ACO1, GPX3, PGD, VKORC1, and DCXR), which
may be associated with the exacerbation of allergic asthma
(5).
Investigating variations in gene expression, which
can be quantitatively measured on a genome-wide scale, is essential
for understanding and interpreting the pathogenic mechanism of
pediatric allergic asthma. The present study used a DNA microarray
method to identify the DEGs between normal and pediatric allergic
asthma samples. The DEGs were then clustered. Functional and
pathway analyses of the potential DEGs were then conducted, and the
pathways were finally annotated based on the Kyoto Encyclopedia of
Genomes and Genes (KEGG). The DEGs present in significant pathways
associated with allergic asthma were further analyzed in order to
explore the pathogenesis of the disease.
Materials and methods
Microarray data
The GSE18965 gene expression profile of pediatric
allergic asthma (11) was
downloaded from the public functional genomics data repository: The
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (11). A total of 16 specimens, including
seven normal samples and nine samples from patients with pediatric
allergic asthma, were available. The gene expression profile was
based on the platform of GPL96 (HG-U133A) Affymetrix Human Genome
U133A Array (Affymetrix, Santa Clara, CA, USA).
Data processing and identification of
DEGs
Based on the annotation platform, 22,283 available
probe IDs were mapped to gene names and 20,952 genes were selected
and their expression profiles were processed (12). The limma package in R software was
used to identify the DEGs between the normal and pediatric allergic
asthma samples (13). The false
discovery rate (FDR) was previously described by Benjamini and
Hochberg (14), and is the
expected proportion of false discoveries, out of the total number
of identified DEGs. Applying a cut-off limit for FDR can help
reduce error from multiplicity, whilst ensuring the identification
of real DEGs. In the present study, cut-off values of log
fold-change (|logFC|)>1.0 and an adjusted P<0.05 were used to
identify DEGs.
Hierarchical clustering of DEGs
Hierarchical clustering is a method used to build a
hierarchy of clusters of DEGs. The process of clustering was based
on the Euclidean distance (15)
between the expression profiles of each of the DEGs filtered from
the samples. The clustering was conducted using pheatmap
(http://cran.r-project.org/web/packages/pheatmap/index.html)
in R (16,17).
Strategies for hierarchical clustering generally
fall into two categories. The ‘bottom up’ approach is where each
DEG begins as a single cluster, and DEGs with similar expression
profiles begin to successively merge as one cluster moves up the
hierarchy. The ‘top down’ approach is where all DEGs begin as one
cluster, and splits are performed recursively as a cluster moves
down the hierarchy. Generally, the merges and splits are determined
in a greedy manner. The results of hierarchical clustering are
usually presented in a dendrogram.
Functional enrichment analysis of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (18,19)
was then used to identify the enriched Gene Ontology (GO)
biological processes that the up- and downregulated DEGs were
associated with (P<0.05). The negative logarithmic P-values of
each enrichment were displayed. The functional enrichments were
presented in a bar chart, with the P-values displayed in a line
chart, using Plotrix (http://cran.r-project.org/web/packages/plotrix/index.html)
in R.
Pathway analysis
Pathway enrichment analysis of all of the DEGs was
performed using the KEGG database (http://www.kegg.jp/) (20). The KEGG maps of biological
functions, and the corresponding DEGs were obtained.
Results
Data processing and identification of
DEGs
Following normalization, a differential comparison
between the expression profiles was performed, with the cut-off
values set at FDR<0.05 and |logFC|>1. A total of 127 DEGs
were identified, of which 58 were downregulated and 69 were
upregulated (Table I).
 | Table IDifferentially expressed genes in
pediatric allergic asthma. |
Table I
Differentially expressed genes in
pediatric allergic asthma.
| Genes | ID | adj.P-val | logFC |
|---|
| Downregulated
genes |
| GPI | 208308_s_at | 0.023 | −1.44282 |
| MLXIP | 202519_at | 0.023 | −1.32908 |
| TPP1 | 200742_s_at | 0.023 | −1.26717 |
| NEU1 | 208926_at | 0.023 | −1.21158 |
| ACTN1 | 208636_at | 0.023 | −1.20938 |
| LAMP1 | 201551_s_at | 0.040 | −1.14585 |
| MYOF | 211864_s_at | 0.027 | −1.14455 |
| GLB1 | 201576_s_at | 0.037 | −1.13821 |
| NFKB1 | 209239_at | 0.044 | −1.13729 |
| ACP2 | 202767_at | 0.043 | −1.08902 |
| M6PR | 200900_s_at | 0.047 | −1.06069 |
| HGSNAT | 218017_s_at | 0.023 | −1.05619 |
| Upregulated
genes |
| UTP14A | 221098_x_at | 0.026 | 1.00982 |
| KLHL23 | 213610_s_at | 0.023 | 1.02244 |
| EFCAB11 | 210525_x_at | 0.023 | 1.02378 |
| NUCKS1 | 217802_s_at | 0.025 | 1.03147 |
| FIP1L1 | 221007_s_at | 0.023 | 1.03372 |
| MBD4 | 214048_at | 0.023 | 1.03480 |
| PPFIBP1 | 203735_x_at | 0.023 | 1.03706 |
| INHBC | 207688_s_at | 0.026 | 1.04053 |
| PPARA | 210771_at | 0.023 | 1.04131 |
| EZR | 217234_s_at | 0.038 | 1.04663 |
| CDC42BPA | 214464_at | 0.023 | 1.05413 |
| INVS | 211054_at | 0.023 | 1.06234 |
| BMP2K | 37170_at | 0.026 | 1.06702 |
| RREB1 | 203704_s_at | 0.045 | 1.07460 |
| DMP1 | 217067_s_at | 0.023 | 1.07594 |
Hierarchical clustering of DEGs
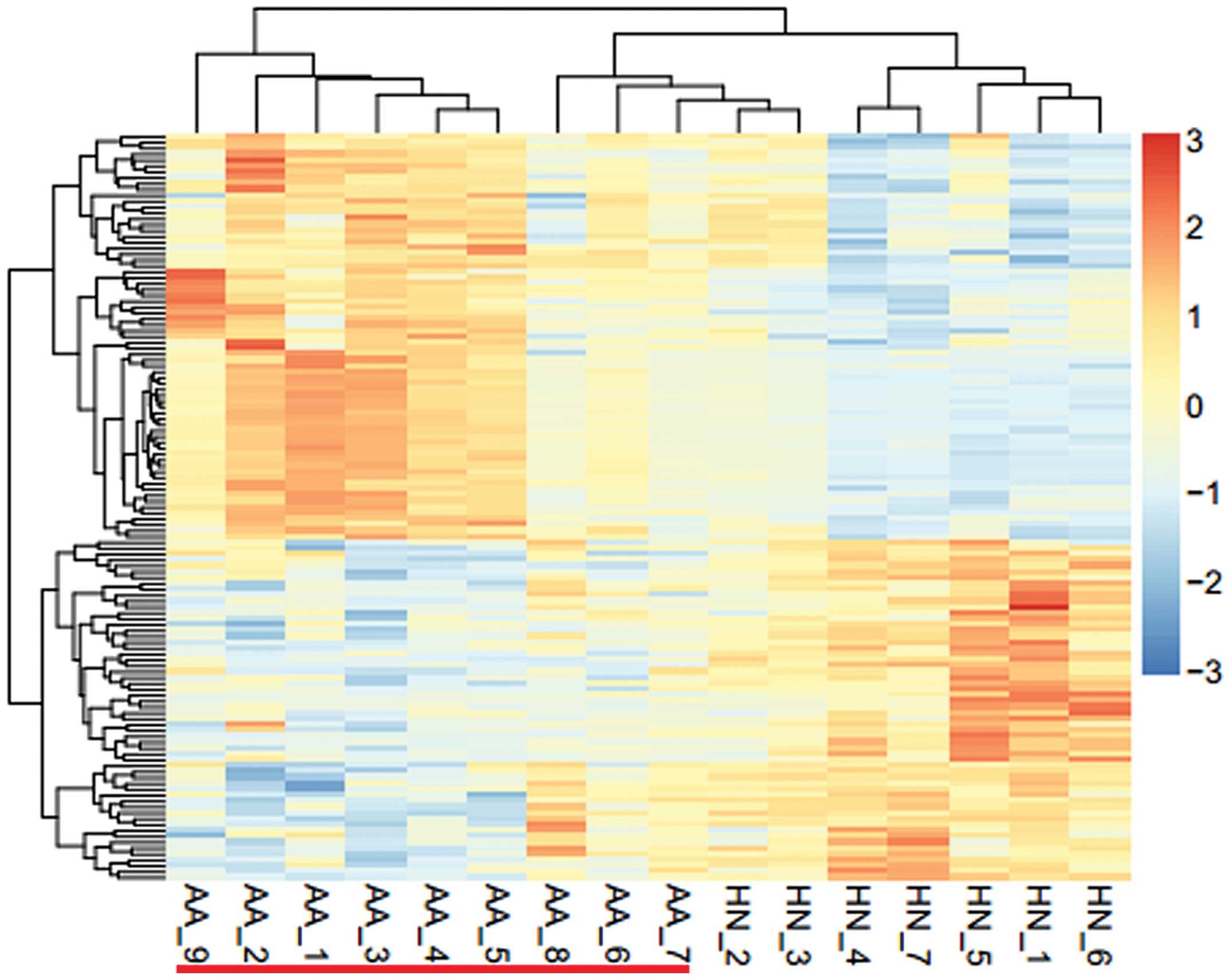
Hierarchical clustering of the identified genes is
presented in Fig. 1. The logFC
values of the DEGs ranged between three times upregulated and three
times downregulated. The samples from the patients with pediatric
allergic asthma could easily be distinguished from the samples of
the healthy control group. These results suggested that the
identified DEGs were significantly characteristic of allergic
asthma and may be used to distinguish between normal samples and
those from patients with asthma.
Functional enrichment analysis of
DEGs
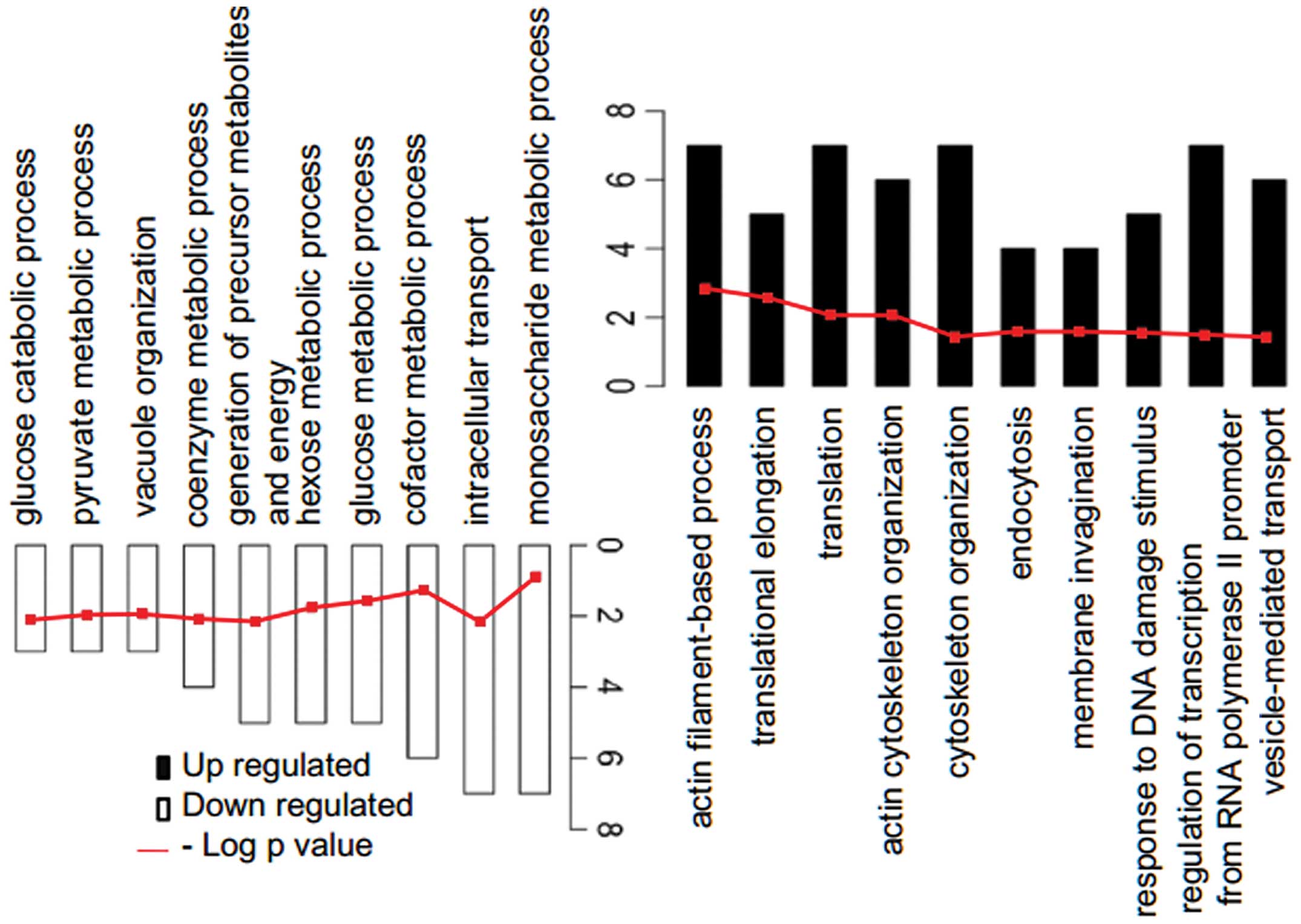
The identified DEGs were assembled into up- and
downregulated genes and mapped into DAVID for functional enrichment
analysis using the topological approach (Table I). The available functional
enrichments are presented in a bar chart, with the P-values
displayed in a line chart (Fig.
2). The up- and downregulated genes were significantly enriched
in the actin filament-based process and the monosaccharide
metabolic process, respectively.
Pathway analysis
For further detail regarding the biological
processes in which the identified DEGs participated in, part of
these pathways were analyzed using KEGG. A pathway shown to be
associated with the downregulated genes was the lysosomal pathway
(P=6.4×10−9), of which seven downregulated DEGs were
involved [M6PR, TPP1, GLB1, NEU1, ACP2, LAMP1 and HGSNAT].
Discussion
Allergic asthma is characterized by airway
hyperresponsiveness, inflammation and a cellular infiltration,
which is dominated by eosinophils (21). Numerous epidemiological studies
have linked the exacerbation of allergic asthma with an increase in
ambient inhalable particulate matter from air pollutants (22,23).
Furthermore, the majority of cases of allergic asthma have been
attributed to infection with respiratory viruses, as well as other
allergens (24). These infectious
and allergic stimuli induce airway hyper-responsiveness by
stimulating T lymphocytes and chemotaxis of acidophilic leukocytes
(25), which results in the
production of various pro-inflammatory cytokines and mediators to
induce inflammation (26). The
present study identified the up- and downregulated DEGs in allergic
asthma, which were significantly enriched in the actin
filament-based process and the monosaccharide metabolic process,
respectively. Concordant with the findings of the present study,
Wang et al (5) also
demonstrated that the downregulated genes were associated with the
monosaccharide metabolic process. Husain et al (27) previously reported that the actin
filament-based process [GO:0030029] is enriched in food allergy,
thus suggesting a possible involvement of certain DEGs with smooth
muscle contraction, bronchoconstriction and vasodilation, which are
common characteristics associated with type I allergic responses
and anaphylaxis (27). One of the
DEGs enriched in this process is scinderin (Scin), an
actin-filament severing and capping protein that is activated by
calcium (27). It has been
suggested that Scin may be a potential biomarker of type I
allergies, such as asthma (28).
Furthermore, the upregulated genes (PDPK1, EZR, MYO6, CDC42BPA,
OPHN1, ARF6 and WASL) identified in the present study that were
enriched in the actin filament-based process require further study
in order to determine whether they may be used as potential
biomarkers of allergic asthma.
Viral transcription-associated proteins have
previously been identified as DEGs in asthma (5). The present study identified seven
downregulated DEGs (M6PR, TPP1, GLB1, NEU1, ACP2, LAMP1 and HGSNAT)
that are associated with lysosomal function, which are associated
with the autolysis of cells. A previous study demonstrated that the
absence of MPRs and recycling cell surface receptors may lead to
distinguishment of lysosomes, membrane-bound organelles that
contain numerous hydrolytic enzymes from endosomes (29). TPP1 has previously been established
as a shared or restricted regulatory dendritic cell (DC) marker
(30), which has been suggested to
have an important role in the development of atopic asthma
(31). GLB1 gives rise to the GLB1
lysosomal enzyme and the elastin binding protein (EBP), which are
involved in elastic fiber deposition (32). GLB1 forms a complex with protective
protein cathepsin A (PPCA), NEU1 and galactosamine 6-sulphate
sulfatase inside lysosomes, whereas EBP binds PPCA and NEU1 on the
cell surface. ACP is present in the lysosomes inside DCs and was
previously reported as a key enzyme that is able to digest antigens
(33), thus indicating that it may
have a similar role in allergic asthma. The specific functions of
LAMP-1 and -2, which belong to the N-glycosylated proteins present
in lysosomal membranes, have only recently begun to be recognized
(34). The normal functions of
LAMP-1 can be substituted by the structurally-associated LAMP-2;
however, LAMP-2 has more specific tasks. Knockout of LAMP-2
expression in mice has revealed roles for LAMP-2 in lysosomal
enzyme targeting, autophagy and lysosomal biogenesis (35). Furthermore, LAMP-2 deficiency in
humans leads to Danon disease, which is associated with fatal
cardiomyopathy and myopathy (36).
A previous study demonstrated that loss of HGSNAT activity leads to
mucopolysaccharidosis IIIC (MPSIIIC), a lysosomal disease (37). Subsequent ana lysis of this novel
lysosomal protein revealed mutations in MPSIIIC and confirmed that
it encoded HGSNAT (38). Among the
genes identified in the present study (M6PR, TPP1, GLB1, NEU1,
ACP2, LAMP1 and HGSNAT) that were significantly associated with
lysosomal function, only TPP1 has been confirmed as being
associated with atopic asthma (29). Further analyses of the specific
functions of these identified DEGs in atopic asthma is
required.
In conclusion, the results of the present study
provide information on the underlying molecular mechanism of
allergic asthma and provide a basis for future research. It was
hypothesized that the identified downregulation of M6PR, TPP1,
GLB1, NEU1, ACP2, LAMP1 and HGSNAT leads to disorders of lysosome
function, which results in asthma by causing T-cell dysfunction. To
date, the discovery of drugs for the treatment of pediatric asthma
has been limited as its pathogenesis has yet to be fully
elucidated. Therefore, the DEGs identified in the present study may
provide a basis for the development of future medication used to
treat this disease.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81170034) and the
Key Fund Project of Sichuan Provincial Department of Education
(grant no. 13ZA0210).
References
|
1
|
Tomita Y, Tomida S, Hasegawa Y, et al:
Artificial neural network approach for selection of susceptible
single nucleotide polymorphisms and construction of prediction
model on childhood allergic asthma. BMC Bioinformatics. 5:1202004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mannino DM, Homa DM, Akinbami LJ, Moorman
JE, Gwynn C and Redd SC: Surveillance for asthma - United States,
1980–1999. MMWR Surveill Summ. 51:1–13. 2002.PubMed/NCBI
|
|
3
|
Dietert RR and Zelikoff JT: Early-life
environment, developmental immunotoxicology, and the risk of
pediatric allergic disease including asthma. Birth Defects Res B
Dev Reprod Toxicol. 83:547–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmid-Ott G, Jaeger B, Adamek C, et al:
Levels of circulating CD8(+) T lymphocytes, natural killer cells,
and eosinophils increase upon acute psychosocial stress in patients
with atopic dermatitis. J Allergy Clin Immunol. 107:171–177. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang XQ, Wang XM, Zhou TF and Dong LQ:
Screening of differentially expressed genes and small molecule
drugs of pediatric allergic asthma with DNA microarray. Eur Rev Med
Pharmacol Sci. 16:1961–1966. 2012.PubMed/NCBI
|
|
6
|
Bender B, Milgrom H and Rand C:
Nonadherence in asthmatic patients: is there a solution to the
problem? Ann Allergy Asthma Immunol. 79:177–185. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Milgrom H, Bender B, Ackerson L, Bowry P,
Smith B and Rand C: Noncompliance and treatment failure in children
with asthma. J Allergy Clin Immunol. 98:1051–1057. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laird PW: Principles and challenges of
genomewide DNA methylation analysis. Nat Rev Genet. 11:191–203.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Creer TL and Wigal JK: Self-efficacy.
CHEST. 103:1316–1317. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heller MJ: DNA microarray technology:
devices, systems, and applications. Annu Rev Biomed Eng. 4:129–153.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kicic A, Hallstrand TS, Sutanto EN, et al:
Decreased fibronectin production significantly contributes to
dysregulated repair of asthmatic epithelium. Am J Respir Crit Care
Med. 181:889–898. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujita A, Sato JR, Rodrigues LO, Ferreira
CE and Sogayar MC: Evaluating different methods of microarray data
normalization. BMC Bioinformatics. 7:4692006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smyth GK: limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor Statistics for Biology and Health.
Gentleman R, Carey VJ, Huber W, Irizarry RA and Dudoit S: Springer;
New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
14
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
15
|
Deza MM and Deza E: Encyclopedia of
Distances. Springer; Berlin Heidelberg: 2009, View Article : Google Scholar
|
|
16
|
Szekely GJ and Rizzo ML: Hierarchical
clustering via joint between-within distances: Extending Ward’s
minimum variance method. J Classif. 22:151–183. 2005. View Article : Google Scholar
|
|
17
|
Huson DH, Richter DC, Rausch C, Dezulian
T, Franz M and Rupp R: Dendroscope: An interactive viewer for large
phylo-genetic trees. BMC Bioinformatics. 8:4602007. View Article : Google Scholar
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
20
|
Kanehisa M, Goto S, Hattori M, et al: From
genomics to chemical genomics: new developments in KEGG. Nucleic
Acids Res. 34:D354–D357. 2006. View Article : Google Scholar :
|
|
21
|
Peden DB: The epidemiology and genetics of
asthma risk associated with air pollution. J Allergy Clin Immun.
115:213–219. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
D’Amato G, Baena-Cagnani CE, Cecchi L, et
al: Climate change, air pollution and extreme events leading to
increasing prevalence of allergic respiratory diseases. Multidiscip
Respir Med. 8:122013. View Article : Google Scholar
|
|
23
|
Sugita M, Kuribayashi K, Nakagomi T,
Miyata S, Matsuyama T and Kitada O: Allergic bronchial asthma:
airway inflammation and hyperresponsiveness. Intern Med.
42:636–643. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaya M: Virus infection-induced
bronchial asthma exacerbation. Pulm Med. 2012:8348262012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Docherty SJ, Butcher LM, Schalkwyk LC and
Plomin R: Applicability of DNA pools on 500 K SNP microarrays for
cost-effective initial screens in genomewide association studies.
BMC Genomics. 8:2142007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yasuda H, Suzuki T, Zayasu K, et al:
Inflammatory and bronchospastic factors in asthma exacerbations
caused by upper respiratory tract infections. Tohoku J Exp Med.
207:109–118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Husain M, Boermans HJ and Karrow NA:
Mesenteric lymph node transcriptome profiles in BALB/c mice
sensitized to three common food allergens. BMC Genomics. 12:122011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Di Valentin E, Crahay C, Garbacki N, et
al: New asthma biomarkers: lessons from murine models of acute and
chronic asthma. Am J Physiol Lung Cell Mol Physiol. 296:L185–L197.
2009. View Article : Google Scholar
|
|
29
|
Luzio JP, Rous BA, Bright NA, Pryor PR,
Mullock BM and Piper RC: Lysosome-endosome fusion and lysosome
biogenesis. J Cell Sci. 113:1515–1524. 2000.PubMed/NCBI
|
|
30
|
Zimmer A, Bouley J, Le Mignon M, et al: A
regulatory dendritic cell signature correlates with the clinical
efficacy of allergen-specific sublingual immunotherapy. J Allergy
Clin Immunol. 129:1020–1030. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y and Liu S: Protein-protein
interaction network analysis of children atopic asthma. Eur Rev Med
Pharmacol Sci. 16:867–872. 2012.PubMed/NCBI
|
|
32
|
Caciotti A, Donati MA, Boneh A, et al:
Role of beta-galactosidase and elastin binding protein in lysosomal
and nonlysosomal complexes of patients with GM1-gangliosidosis. Hum
Mutat. 25:285–292. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hua H, Liang Z, Li W, et al: Phenotypic
and functional maturation of murine dendritic cells (DCs) induced
by purified Glycyrrhizin (GL). Int Immunopharmacol. 12:518–525.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eskelinen EL: Roles of LAMP-1 and LAMP-2
in lysosome biogenesis and autophagy. Mol Aspects Med. 27:495–502.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tanaka Y, Guhde G, Suter A, et al:
Accumulation of autophagic vacuoles and cardiomyopathy in
LAMP-2-deficient mice. Nature. 406:902–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eskelinen EL, Tanaka Y and Saftig P: At
the acidic edge: emerging functions for lysosomal membrane
proteins. Trends Cell Biol. 13:137–145. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ausseil J, Loredo-Osti JC, Verner A, et
al: Localisation of a gene for mucopolysaccharidosis IIIC to the
pericentromeric region of chromosome 8. J Med Genet. 41:941–945.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fan X, Zhang H, Zhang S, et al:
Identification of the gene encoding the enzyme deficient in
mucopolysaccharidosis IIIC (Sanfilippo disease type C). Am J Hum
Genet. 79:738–744. 2006. View
Article : Google Scholar : PubMed/NCBI
|