Introduction
Cervical cancer (CC) has become the second most
common female cancer, particularly in developing countries
(1). In mainland China, it remains
an important public health problem (2,3).
Therefore, identification of novel mechanisms of CC may aid the
development of strategies for the diagnosis, treatment and
determination of prognosis.
A previous study has shown that oncogenes and tumor
suppressors exhibit critical roles in the development of CC
(4). Recent studies also indicate
that expression of these genes is tightly regulated by microRNAs
(miRNAs), a class of small endogenous RNA molecules (5,6). For
example, FoxO1, a tumor suppressor in several types of human
cancer, is controlled by miR-96, miR-223 and miR-370 (7–9).
Whilst the reason why these distinct miRNAs regulate the same
target gene remains unknown, dysregulation of certain miRNAs may
contribute to tumor initiation and/or progression. The present
study analyzed the expression levels of miR-181 in cervical cancer
tissues. Furthermore, the roles and mechanisms of miR-181 in the
development of cervical cancer were determined.
Materials and methods
Tissue samples and cell culture
In total, 30 pairs of samples from CC tissues and
the adjacent normal tissues were obtained from patients who
underwent surgery in the Department of Obstetrics and Gynecology.
This study was approved by the Institutional Review Board of
Changhai Hospital, The Second Military Medical University
(Shanghai, China). Written informed consent was obtained from each
patient. The CC cell lines (HeLa, HeLa-229, SiHa and C33A) and
normal cervical epithelial cells (End1/E6E7) were obtained from the
Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were
maintained in Dulbecco’s modified Eagle’s medium (Invitrogen,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum
(Invitrogen). TNF-α (10 ng/ml), IL-1β (20 ng/ml) and IL-6 (20
ng/ml) were obtained from Bioyare Company (Shanghai, China).
Quantitative PCR
miRNA Isolation kits (Ambion, Austin, TX, USA) were
used to harvest RNA from tissues or cells. Expression of miR-181
was determined by Taqman® MicroRNA assay (Applied
Biosystems, Foster City, CA). Quantitative PCR was performed using
a 7300 Real-Time PCR system (Applied Biosystems). The PCR
conditions included an initial incubation period at 94°C for 5 min,
followed by a two-step PCR program consisting of 94°C for 5 sec and
60°C for 30 sec, for 40 cycles. The primer sequences for miR-181
were: Forward 5′-AACATTCAACGCTGTCGGTGAAGT-3′, and reverse
5′-ACTTCACCGACAGCGTTGAATGTT-3′. All of the samples were normalized
to the internal control (U6 small nuclear RNA).
Cell proliferation assay
Cell counts were estimated by trypsinizing the cells
and performing analysis with a Coulter counter (Beckman Coulter,
Fullerton, CA, USA). For BrdU incorporation assays, a cell
proliferation enzyme-linked immunosorbent assay (Beyotime,
Shanghai, China) was used to analyze the incorporation of BrdU
during DNA synthesis following the manufacturer’s instructions.
Absorbance was measured at 450 nm in the SpectraMax 190 ELISA
reader (Molecular Devices, Sunnyvale, CA, USA).
Cell cycle analysis
The cells were labeled for 15 min with propidium
iodide and immediately analyzed by flow cytometry (Becton
Dickinson, San Jose, CA, USA). A total of ~10,000 cells were
acquired and analyzed for DNA content. All of the data were
collected, stored and analyzed by ModFit software (http://www.vsh.com/products/mflt/). Histograms
represent the percentage of cells in each phase of the cell cycle
(G0/G1, S and G2/M).
MicroRNA mimics and transfection
Human miR-181 mimics were purchased from Ambion. The
negative controls consist of a scrambled sequence (Ambion). All
transfection was performed using Lipofectamine 2000 (Invitrogen)
following the manufacturer’s instructions.
Western blot analysis
Cells were harvested and lysed with ice-cold lysis
buffer (50 mm Tris-HCl, pH 6.8, 181 mm 2-mercaptoethanol, 2% w/v
SDS, 10% glycerol; Biosune Biotechnology Co., Ltd., Shanghai,
China). Following centrifugation at 10,000 x g and 4°C for 15 min,
proteins in the supernatants were quantified and separated by 10%
SDS-PAGE and transferred to a nitrocellulose membrane (Amersham
Bioscience, Amersham, UK). After blocking with 10% nonfat milk in
phosphate-buffered saline, membranes were immunoblotted with
antibodies as indicated, followed by horseradish peroxidase-linked
anti-mouse or anti-rabbit IgG secondary antibodies (Cell Signaling
Technology, Inc., Beverly, MA, USA). The signals were detected by
SuperSignal West Pico Chemiluminescent Substrate kit (Pierce,
Rockford, IL, USA) according to the manufacturer’s instructions.
Polyclonal rabbit anti-YY1, polyclonal rabbit p53 and polyclonal
mouse C-myc antibodies were purchased from Abcam (Cambridge, MA,
USA). Protein levels were normalized to GAPDH (polyclonal mouse;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Luciferase reporter assay
The wild-type and mutant 3′-UTR fragments of the YY1
gene were cloned into pMir-Report (Ambion). Mutations were
introduced in potential miR-181 binding sites using the QuikChange
Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA).
Luciferase values were determined using the Dual-Luciferase
Reporter Assay system (Promega, Madison, WI, USA).
Tumor growth assay
Male BALB/c nude mice aged 4 weeks old were
purchased from Shanghai Laboratory Animal Company (Shanghai,
China). HeLa cells (2×105), either stably expressing
miR-181 or negative controls, were injected subcutaneously into the
front legs of the mouse. The mice were observed over 5 weeks for
tumor formation. The mice were then sacrificed by cervical
dislocation, the tumors were recovered and the wet weight of each
tumor was determined.
Statistical analysis
Differences between groups were analyzed using
Student’s t-test analysis and expressed as the mean ± standard
error from at least three separate experiments. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-181 is downregulated in CC tissues
and cells
Expression of miR-181 in CC tissues was compared to
that in the adjacent normal tissues using quantitative PCR. As a
result, it was revealed that miR-181 expression levels were greatly
reduced in CC tissues (Fig. 1A).
The circulating levels of miR-181 were quantified in the serum of
CC patients and healthy controls, and a significant difference was
also observed between the two groups (Fig. 1B).
Inflammatory cytokines inhibit miR-181
expression
To investigate the reasons for the downregulation of
miR-181, its expression was determined in CC cell lines and normal
cervical cells. The expression levels of miR-181 in four CC cell
lines (HeLa, HeLa-229, SiHa and C33A), was observed to be lower
compared with those in the normal cervical cell line (End1/E6E7)
(Fig. 2A).
Previous studies have demonstrated that inflammatory
responses are responsible for the high rate of CC development
(10,11). Therefore, End1/E6E7 cells were
treated with several pro-inflammatory cytokines. In the present
study TNFα, IL-1β and IL-6 significantly repressed miR-181
expression in a dose-dependent manner (Fig. 2B). Furthermore, HeLa cells were
transfected with plasmids expressing a dominant negative (Dn) IκBα
(Fig. 2C), which could bind with
NF-κB and inhibit NF-κB-mediated inflammatory signaling (12). As expected, Dn-IκBα significantly
upregu-lated miR-181 expression levels in HeLa cells compared with
those in the controls (Fig. 2D).
These results suggest that the downregulation of miR-181 in CC
tissues and cell lines may be due to, at least in part, the
persistent activation of inflammatory response.
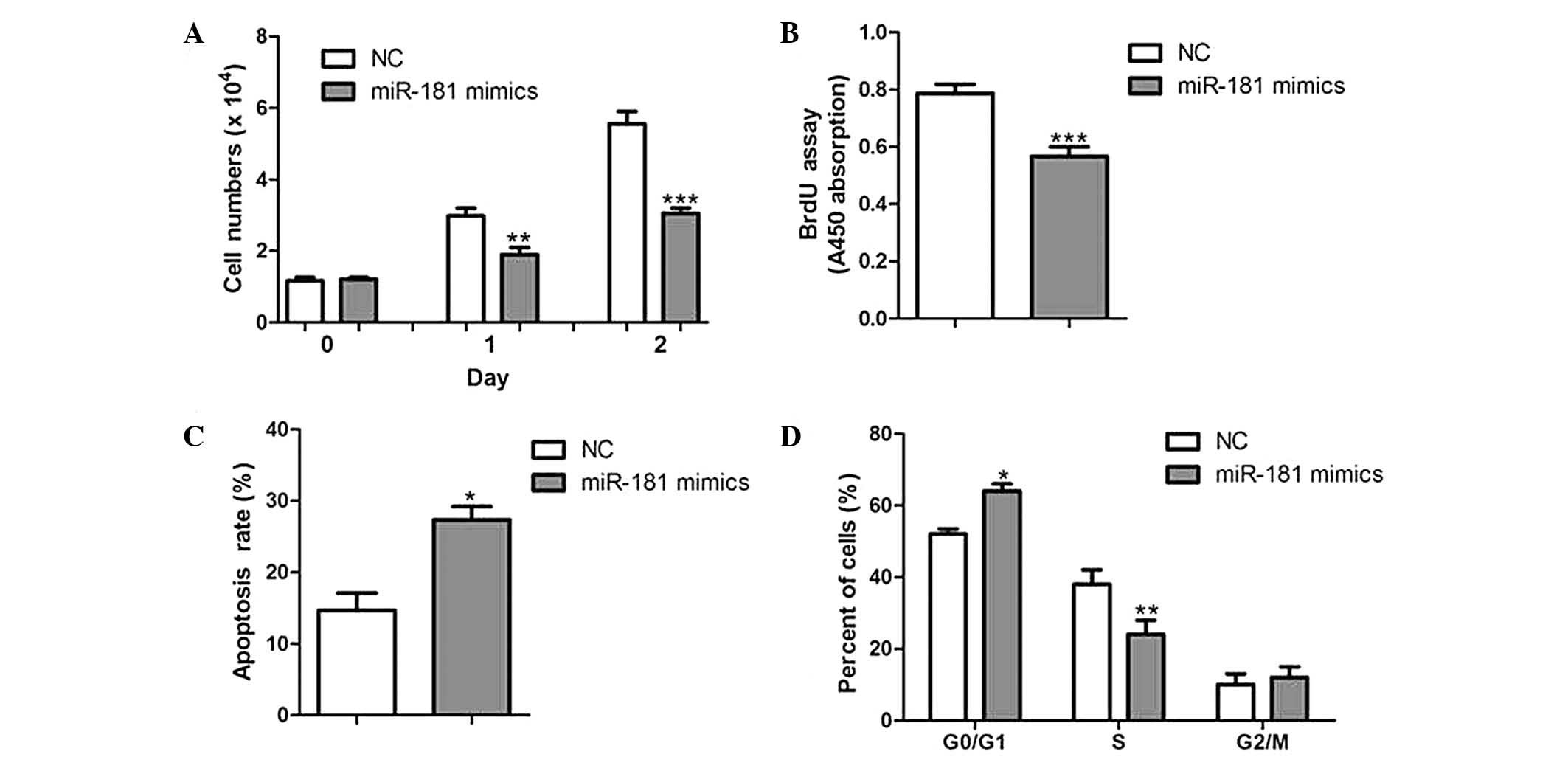
miR-181 overexpression inhibits CC cell
proliferation in vitro
To determine the effect of miR-181 on CC cell
growth, HeLa cells were transfected with either miR-181 mimics or
negative controls. As a result, cell growth was significantly
reduced in miR-181-overexpressed cells compared with that of their
corresponding controls (Fig. 3A).
Furthermore, cells overexpressing miR-181 had a lower rate of
proliferation and a higher rate of apoptosis (Fig. 3B and C). Cell cycle analysis of
miR-181-over-expressed cells showed a significant increase in the
percentage of cells in G0/G1 phase and a reduction of those in S
phase (Fig. 3D).
miR-181 overexpression inhibits tumor
growth in vivo
To further determine the roles of miR-181, HeLa
cells with stable overexpression of miR-181 were generated and
injected subcutaneously into the front legs of the nude mice. The
tumor growth was closely monitored for 5 weeks. After this time the
tumor size and weight were markedly reduced in
miR-181-overexpressed tumors in comparison with the control tumors
(Fig. 4A and B), suggesting that
miR-181 suppresses CC growth in vivo.
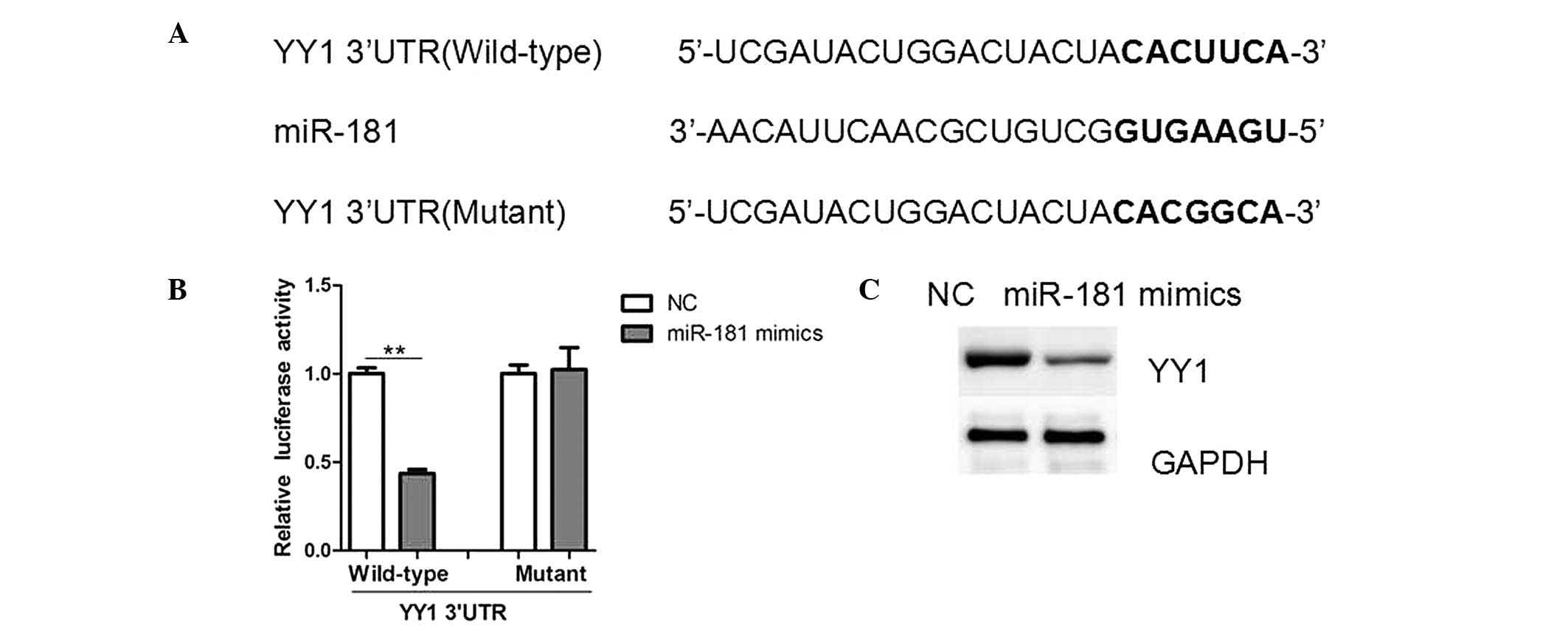
miR-181 targets YY1 3′-UTR and
downregulates its expression
To understand the underlying mechanism of growth
inhibition by miR-181, potential targets of miR-181 were searched
using TargetScan (www.targetscan.org) and miRWalk (www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
software. It was identified that YY1, an oncogene in human cancer,
harbored a potential miR-181 binding site (Fig. 5A). To verify whether YY1 is a
direct target of miR-181, luciferase report vectors that contained
the putative miR-181 binding sites within YY1 3′-UTR were
constructed. The overexpression of miR-181 significantly repressed
the luciferase activity when the reporter construct contained the
YY1 3′-UTR in HeLa cells (Fig.
5B). However, mutation of the miR-181 binding site from the YY1
3′-UTR eliminated this effect of miR-181, suggesting that miR-181
directly inhibited YY1 expression by targeting its 3′-UTR (Fig. 5B). The miR-181 precursor inhibited
the protein expression of YY1 as indicated by western blot analysis
(Fig. 5C), whilst its mRNA levels
remained unchanged (data not shown). Therefore, these results
suggest that miR-181 negatively regulates YY1 expression at the
translational level.
YY1 has been reported to downregulate p53 expression
whilst upregulating C-myc, which are key regulators of tumor growth
and cell cycle progression (13,14).
In HeLa cells overexpressing miR-181 in the present study, an
increase in the expression levels of p53 was observed, as well as
the downregulation of C-myc, as compared with levels in controls
(Fig. 6A). Additionally, mRNA
levels of P21 and P27, downstream targets of P53, were increased,
while Cyclin D1, a target of C-myc, was repressed (Fig. 6B). The levels of YY1, p53 and C-myc
were also affected in tumors overexpressing miR-181, when compared
with the levels in negative controls (Fig. 6C). These results confirm that YY1
is an important target gene of miR-181 in CC.
Discussion
It has been demonstrated that several miRNAs are
dysregulated in CC tissues or cell lines, including miR-20, miR-196
and miR-203 (15–17). In the present study, through
experiments in vitro and in vivo, the role of miR-181
in CC was determined. It was found that miR-181 overexpression was
able to inhibit cell proliferation and induce apoptosis and cell
cycle arrest in G1 phase. Furthermore, miR-181 was able to inhibit
tumor growth in the nude mice. Therefore, miR-181 may be a tumor
suppressor in the development of CC; however further studies with
larger clinical samples are required.
Previous studies have shown that miR-181 also
functions as a tumor suppressor which triggers growth inhibition,
induces apoptosis and inhibits invasion in glioma cells (18). However, transforming growth factor
TGF-β-mediated upregulation of hepatic miR-181 promoted growth,
clonogenic survival, migration and invasion of hepatocellular
carcinoma cells (19). Although
the inconsistency in the roles of miR-181 remains unexplained, the
specificity of the roles of miR-181 may depend on its target genes
and/or cellular context.
YY1 was further experimentally validated as a
miR-181 target in CC in the present study. YY1, a transcription
factor of the polycomb group protein family, is widely expressed in
various tissues and participates in development, metabolism and
tumorigenesis (20–22). A study into YY1 expression in tumor
tissues revealed that YY1 overexpression may have an important role
in the regulation of sensitivity and resistance of tumor cells to
chemotherapy and immunotherapy (23). At the molecular level, a previous
study supports the role of YY1 in tumorigenesis via its association
with the tumor suppressor gene P53 (13). YY1 may interact with P53 and
promote its protein degradation by blocking p300-dependent p53
acetylation and stabilization (13). YY1 may also activate C-myc and
Cyclin D1 promoters to enhance their transcription (23). Therefore, inhibitors, antisense
therapies, and silencer RNAs that target YY1 may serve as potential
therapies for cancer progression.
In conclusion, the key finding of the present study
highlights the theory that overexpression of miR-181 inhibits CC
cell growth in vitro and in vivo by regulating YY1
expression. Therefore, the results regarding the functional
interaction of miR-181 and YY1 signaling in CC may aid the
development of novel gene therapies in the future.
References
|
1
|
Meijer CJ and Berkhof J: Screening:
Cervical cancer - should we abandon cytology for screening? Nat Rev
Clin Oncol. 9:558–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li J, Kang LN and Qiao YL: Review of the
cervical cancer disease burden in mainland China. Asian Pac J
Cancer Prev. 12:1149–1153. 2011.PubMed/NCBI
|
|
3
|
Shi JF, Canfell K, Lew JB and Qiao YL: The
burden of cervical cancer in China: synthesis of the evidence. Int
J Cancer. 130:641–652. 2012. View Article : Google Scholar
|
|
4
|
Hung CF, Wu TC, Monie A and Roden R:
Antigen-specific immunotherapy of cervical and ovarian cancer.
Immunol Rev. 222:43–69. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haflidadóttir BS, Larne O, Martin M, et
al: Upregulation of miR-96 enhances cellular proliferation of
prostate cancer cells through FOXO1. PLoS One. 8:e724002013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu L, Li H, Jia CY, et al: MicroRNA-223
regulates FOXO1 expression and cell proliferation. FEBS Lett.
586:1038–1043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Z, Sun H, Zeng W, He J and Mao X:
Upregulation of MircoRNA-370 induces proliferation in human
prostate cancer cells by downregulating the transcription factor
FOXO1. PLoS One. 7:e458252012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mai CW, Kang YB and Pichika MR: Should a
Toll-like receptor 4 (TLR-4) agonist or antagonist be designed to
treat cancer? TLR-4: its expression and effects in the ten most
common cancers. Onco Targets Ther. 6:1573–1587. 2013.PubMed/NCBI
|
|
11
|
Deivendran S, Marzook KH and Radhakrishna
Pillai M: The role of inflammation in cervical cancer. Adv Exp Med
Biol. 816:377–399. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding G, Liu HD, Huang Q, et al: HDAC6
promotes hepatocellular carcinoma progression by inhibiting P53
transcriptional activity. FEBS Lett. 587:880–886. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grönroos E, Terentiev AA, Punga T and
Ericsson J: YY1 inhibits the activation of the p53 tumor suppressor
in response to genotoxic stress. Proc Natl Acad Sci USA.
101:12165–12170. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liao WR, Hsieh RH, Hsu KW, et al: The
CBF1-independent Notch1 signal pathway activates human C-myc
expression partially via transcription factor YY1. Carcinogenesis.
28:1867–1876. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening lymph node
metastasis in early stage cervical cancer. Genet Test Mol
Biomarkers. 17:631–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
How C, Hui AB, Alajez NM, et al:
MicroRNA-196b regulates the homeobox B7-vascular endothelial growth
factor axis in cervical cancer. PLoS One. 8:e678462013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu X, Er K, Mao C, et al: miR-203
suppresses tumor growth and angiogenesis by targeting VEGFA in
cervical cancer. Cell Physiol Biochem. 32:64–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi L, Cheng Z, Zhang J, et al:
hsa-mir-181a and hsa-mir-181b function as tumor suppressors in
human glioma cells. Brain Res. 1236:185–193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Hsu SH, Majumder S, et al:
TGFbeta-mediated upregulation of hepatic miR-181b promotes
hepatocarcinogenesis by targeting TIMP3. Oncogene. 29:1787–1797.
2010. View Article : Google Scholar :
|
|
20
|
Donohoe ME, Zhang X, McGinnis L, et al:
Targeted disruption of mouse Yin Yang 1 transcription factor
results in peri-implantation lethality. Mol Cell Biol.
19:7237–7244. 1999.PubMed/NCBI
|
|
21
|
Lu Y, Xiong X, Wang X, et al: Yin Yang 1
promotes hepatic gluconeogenesis through upregulation of
glucocorticoid receptor. Diabetes. 62:1064–1073. 2013. View Article : Google Scholar :
|
|
22
|
Wang H, Garzon R, Sun H, et al:
NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis
and rhabdomyosarcoma. Cancer Cell. 14:369–381. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gordon S, Akopyan G, Garban H and Bonavida
B: Transcription factor YY1: structure, function, and therapeutic
implications in cancer biology. Oncogene. 25:1125–1142. 2006.
View Article : Google Scholar
|