Introduction
There are numerous types of B-cell lymphoma,
including: Burkitt lymphoma, chronic lymphocytic leukemia, diffuse
large B-cell lymphoma, mantle cell lymphoma, follicular lymphoma,
marginal zone lymphoma/mucosa-associated lymphoid tissue lymphoma,
nodal marginal zone B-cell lymphoma, splenic marginal zone lymphoma
and other less frequent malignancies (1,2). The
pathogenic mechanisms of B-cell lymphoma remain largely unknown;
therefore, further investigation may aid in improving the accuracy
of lymphoma diagnosis and gene therapy.
Previous studies have demonstrated the importance of
microRNAs (miRNA), a type of non-coding small RNA, in cell
proliferation, apoptosis and differentiation (3,4).
Altered miRNA expression has also been suggested in lymphoma
development and progression (5–7). A
transgenic mouse model with conditional overexpression of miR-155
in B-lymphocytes, exhibited polyclonal pre-leukemic pre-B-cell
proliferation, followed by mature B-cell malignancy (8). In addition, miR-155 has been shown to
have prognostic implications in a subtype of human Non-Hodgkin
lymphoma and diffuse large B-cell lymphoma (9). These findings suggest that aberrant
expression of certain miRNAs may be a potential biomarker for the
diagnosis of lymphoma.
In the present study a miRNA expression screen was
conducted using B-cell lymphoma and non-tumoral samples, in order
to observe the expression of different miRNAs and determine whether
any are involved in the development of B-cell lymphoma.
Materials and methods
Human samples
A total of 22 fresh-frozen samples of B-cell
lymphoma and 16 non-tumoral samples (reactive lymph nodes) were
collected. A total of 38 samples were collected at the People’s
Hospital of Guizhou Province (Guiyang, China) between 2009 and
2012. The non-tumoral samples were collected from different
patients. All lymphoma samples were obtained patients who had been
diagnosed and were obtained prior to the patients receiving
therapy. The project was approved by the Ethics Committee of the
People’s Hospital of Guizhou Province.
Cell culture
The Daudi and Raji B-cell lymphoma cell lines were
obtained from American Type Culture Collection (Manassas, VA, USA).
The cells were cultured in Dulbecco’s modified Eagle’s medium
(Gibco Life Technologies, Beijing, China) supplemented with 10%
fetal bovine serum (Gibco Life Technologies). The cultures were
maintained at 37°C in a humidified atmosphere containing 5%
CO2.
Analysis of miRNA expression using
TaqMan® reverse transcription-quantitative polymerase
chain reaction (RT- qPCR)
Total RNA from the tissue samples and cell lines was
extracted using the miRNA Isolation kit (Ambion Life Technologies,
Carlsbad, CA, USA). The expression levels of mature miRNAs were
assayed using a TaqMan® MicroRNA assay, specific for
hsa-miR-204 (Applied Biosystems Life Technologies, Foster City, NJ,
USA). Briefly, 10 ng total RNA was reverse transcribed into cDNA
using specific stem-loop RT primers. The primer sequences were as
follows: p21, forward 5′-TGTCCGTCAGAACCCATGC-3′ and reverse
5′-AAAGTCGAAGTTCCATCGCTC-3′; p27, forward
5′-AGGAGGAGATAGAAGCGCAGA-3′ and reverse 5′-GTGCGGACTTGGTACAGGT-3′;
Cyclin A, forward 5′-CGCTGGCGGTACTGAAGTC-3′ and reverse
5′-GAGGAACGGTGACATGCTCAT-3′; Cyclin B1, forward
5′-AATAAGGCGAAGATCAACATGG-3′ and reverse
5′-TTTGTTACCAATGTCCCCAAGAG-3′; Cyclin D1, forward:
5′-GCTGCGAAGTGGAAACCATC-3′ and reverse
5′-CCTCCTTCTGCACACATTTGAA-3′. All primers were provided by Biosune
(Shanghai, China). The qPCR was performed using an Applied
Biosystems 7900 Real-time PCR system (Life Technologies, Grand
Island, NY, USA) and a TaqMan® Universal PCR Master mix
(TaKaRa Bio Inc., Dalian, China). All of the primers were obtained
from the TaqMan miRNA assays. Small nuclear U6 snRNA (Applied
Biosystems Life Technologies) was used as an internal control. PCR
conditions included an initial holding period at 94°C for 5 min,
followed by a two-step PCR program consisting of 94°C for 5 sec and
60°C for 30 sec for 45 cycles. Relative quantification analysis of
gene expression analysis was performed according to the
2−ΔΔ method.
Transfection
miR-204 mimics, antisense oligonucleotides and
negative controls were obtained from GenePharma, Co., Ltd.
(Shanghai, China). For the transient transfections, a complex of
Lipofectamine® 2000 (Invitrogen Life Technologies) and
25 nm miRs was prepared, according to the manufacturer’s
instructions. For the STAT5 re-introduction experiments, Daudi or
Raji cells were pre-transfected with miR-204 mimics or negative
control (NC) for 24 h, and then transfected with STAT5 expression
plasmids or empty vector (EV) for another 30 h.
Bromodeoxyuridine (BrdU) assays
A cell proliferation ELISA (BrdU kit; Beyotime
Institute of Biotechnology, Haimen, China) was used to analyze the
incorporation of BrdU during DNA synthesis, according to the
manufacturer’s instructions. All of the experiments were performed
in triplicate. Absorbance was measured at 450 nm using a Spectra
Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA, USA).
Western blotting
The cells or tissues were harvested and lysed with
ice-cold lysis buffer (50 mm Tris-HCl, pH 6.8, 100 mm 2-ME, 2% w/v
SDS and 10% glycerol). Following centrifugation at 20,000 × g for
10 min, the proteins in the supernatants were quantified. The
proteins were separated by 10% SDS PAGE and transferred to
nitrocellulose membranes (GE Healthcare Life Sciences, Chalfont,
UK). The membranes were then blocked with 10% nonfat milk in
phosphate-buffered saline, followed by an incubation with the
primary antibodies indicated. The membranes were then further
incubated with horseradish peroxidase-linked secondary antibodies
(Cell Signaling Technology, Inc., Danvers, MA, USA). The signals
were detected using SuperSignal West Pico Chemiluminescent
Substrate kit (Pierce Biotechnology, Inc., Rockford, IL, USA),
according to the manufacturer’s instructions. Anti-signal
transducer and activator of transcription 5 (STAT5; rabbit
polyclonal antibody targeting human STAT5, cat no. ab7969, 1:2,000)
and β-actin (mouse monoclonal antibody targeting human β-actin, cat
no. ab6276, 1:1,000) were purchased from Abcam (Cambridge, MA,
USA). The protein expression levels were normalized to total
β-actin. The potential target genes of miR-204 were predicted by
miRWalk software (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
Luciferase reporter assay
Total cDNA from the Daudi cells was used to amplify
the 3′-untranslated region (3′-UTR) of STAT5 by PCR. The STAT5
3′-UTR was cloned into the pMIR-Report vector carrying firefly
luciferase (Ambion Life Technologies), yielding pMIR-Report-STAT5.
Mutations were introduced into the potential miR-204 binding sites,
using the QuikChange Site-Directed Mutagenesis kit (Agilent
Technologies, Inc., Santa Clara, CA, USA). The cells were
transfected with the pMIR-Report vectors containing the 3′-UTR
variants and miR-100 precursor control plasmids for 36 h. The
pRL-TK vector (Promega Corporation, Madison, WI, USA) carrying the
Renilla luciferase gene was used as an internal control to
normalize the transfection efficiency. Luciferase values were
determined using the Dual-Luciferase Reporter Assay system (Promega
Corporation).
miRNA microarray
miRNA microarrays were performed by Kangchen
(Shanghai, China) using Human miRNA microarray chips (Release 16.0,
8×60K; Agilent Technologies).
Cell cycle analysis
For cell cycle analysis, propidium iodide (PI) was
used to identify the proportion of cells in each of the three
interphase stages of the cell cycle. Cells were harvested and fixed
in 1 ml cold 70% ethanol overnight at −20°C. DNA was stained in
PI/RNaseA solution and the DNA content was detected using flow
cytometry. Data was analyzed using WinMDI 2.8 software (freeware
from Joe Trotter, La Jolla, CA, USA).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean from ≥4 separate experiments. Differences between the
groups were analyzed using Student’s t-test or one-way analysis of
variance. Statistical analyses were performed with Prism GraphPad
software (Version 5.01) (GraphPad Software Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-204 expression levels are decreased
in B-cell lymphoma
To identify novel miRNAs that are dysregulated in
B-cell lymphoma, a ‘miRNome’ expression profile was performed,
using miRNA microarrays, on RNA isolated from freshly frozen B-cell
lymphoma and non-tumoral samples. The screen revealed a pronounced
downregulation in the expression levels of miR-204 in B-cell
lymphoma tissues (P<0.01; Fig.
1A). Decreased miR-204 expression levels were further confirmed
by qPCR (Fig. 1B). These results
are the first, to the best of our knowledge, to demonstrate that
the expression levels of miR-204 are significantly reduced in
B-cell lymphoma tissue.
Transfection with miR-204 mimics inhibits
cell proliferation
To assess the effects of miR-204 on B-cell lymphoma
growth, miR-204 mimics or NCs were transfected into Daudi and Raji
cells. Transfection with the miR-204 mimics inhibited the
proliferative ability of the cells, as evidenced by BrdU
incorporation assays (Fig. 2A and
B). Furthermore, miR-204 mimics significantly increased the
percentage of cells in the G0/G1 phase and
decreased the percentage of cells in the S phase (Fig. 2C and D).
The present study also determined whether the
inhibition of cell proliferation was associated with the expression
of the genes that regulate the G1/S transition. These
include the cyclin-dependent kinase (CDK) inhibitors p21 and p27,
and the CDK regulators cyclins A, B1 and D1. The expression levels
of p21 and p27 were increased, whereas the expression levels of
cyclins A, B1 and D1 were decreased in the Daudi and Raji cells
transfected with the miR-204 mimics, as compared with the
NC-transfected cells (Fig. 2E and
F).
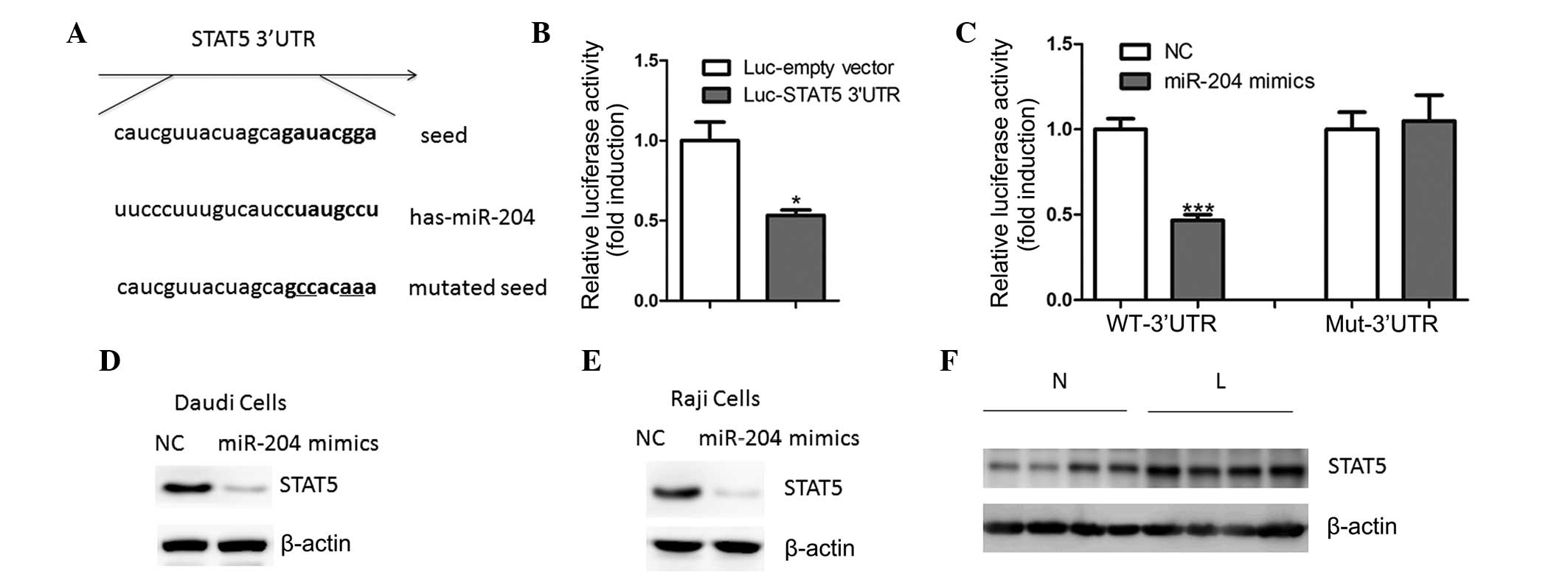
miR-204 directly regulates STAT5
expression in B-cell lymphoma cells
An aim of the present study was to explore the
molecular mechanisms of miR-204 regulation of cell proliferation. A
bioinformatics approach (miRWalk) was used to identify numerous
putative human miR-204 target genes (data not shown). The gene
encoding STAT5 was identified as harboring a potential miR-204
binding site (Fig. 3A) in its
3′-UTR. In Daudi cells, transfection with the luciferase reporter
containing the STAT5 3′-UTR resulted in reduced luciferase
activity, as compared with the cells transfected with the reporter
lacking the STAT5 3′-UTR (Fig.
3B). Furthermore, overexpression of miR-204 led to a reduction
in luciferase activity (Fig. 3C).
Conversely, mutation of the miR-204 binding motif abrogated the
reduced luciferase expression (Fig.
3C). Transfection of the cells with miR-204 mimics led to a
reduction in the endogenous protein expression levels of STAT5 in
the Daudi and Raji cells (Fig. 3D and
E), whereas its mRNA expression levels remained unchanged (data
not shown). These results suggest that miR-204 may regulate STAT5
expression at the translational level. Concordantly, the protein
expression levels of STAT5 were also upregulated in B-cell lymphoma
tissue samples, as compared with the non-tumoral tissues (Fig. 3F).
To verify the functional association between miR-204
and STAT5, the Daudi and Raji cells were transfected with STAT5
expression plasmids, following transfection with miR-204 mimics
(Fig. 4A and B). Re-introduction
of STAT5 reversed the antiproliferative effects of miR-204,
confirming the specific importance of STAT5 for miR-204 action in
cell proliferation (Fig. 4C and
D). These results suggest that STAT5 is an important target
gene of miR-204 in the B-cell lymphoma.
Inhibition of miR-204 promotes the
proliferation of B-cell lymphoma
As described above, miR-204 has a negative role on
the proliferation of B-cell lymphoma. However, it remains unknown
whether inhibiting miR-204 promotes cell proliferation. Therefore,
the Daudi and Raji cells were transfected with miR-204 antisense
oligonucleotides. The ectopic expression of hsa-miR-204 antisense
enhanced the proliferative ability of the two cell lines, as
compared with the NC-transfected cells (Fig. 5A and B). Furthermore,
overexpression of antisense miR-204 resulted in a significantly
lower percentage of cells in the G1/G0 phase
and an increased percentage of cells in the S phase, as compared
with the NC-transfected cells (Fig. 5C
and D). In concordance with these findings, the expression
levels of p21 and p27 were downregulated, whereas the expression
levels of cyclins A, B1 and D1 were upregulated in the miR-204
antisense-transfected cells (Fig. 5E
and F). Furthermore, the protein expression levels of STAT5
were increased in response to miR-204 inhibition (Fig. 5G and H), further validating STAT5
as a miR-204 target.
Discussion
The present study demonstrated that miR-204
expression is downregulated in B-cell lymphoma tissue. Forced
overexpression of miR-204 was able to inhibit cell proliferation in
Daudi and Raji cells, whereas the inhibition of miR-204 by
antisense oligonucleotides enhanced cell proliferation. The present
study is the first, to the best of our knowledge, to identify
miR-204 as a potential tumor suppressor in the progression of
B-cell lymphoma. However, in vivo studies are required to
further establish this notion.
Initial studies demonstrated that miR-204 has an
important physiological role in normal cells, particularly in
developmental events and autophagy miR-204 was previously shown to
regulate mesenchymal progenitor cell differentiation, by targeting
the 3′-UTR of Runt-related transcription factor 2 (10). Furthermore, miR-204 is essential
for the maintenance of the epithelial barrier function, through
regulation of transforming growth factor-β receptor 2 and SNAIL2
(11). miR-204 may also regulate
numerous aspects of eye development in the medaka fish (12). Ablation of miR-204 expression
resulted in an eye phenotype characterized by microphthalmia,
abnormal lens formation and altered dorsoventral patterning of the
retina (12). Subsequent reports
have indicated that miR-204 expression is downregulated in numerous
types of human cancer (13).
Dysregulation of miR-204 has also been shown to mediate migration
and invasion of endometrial cancer, by regulating forkhead box C1
(14). In gastric cancer,
downregulation of miR-204 has been reported as having prognostic
value and correlates with increased staining of B-cell lymphoma 2
protein in tumoral specimens (15). Furthermore, ectopic expression of
miR-204 inhibits colony forming ability, migration and tumor
engraftment of gastric cancer cells (15). In glioma, loss of miR-204
expression has been demonstrated to enhance cell migration and stem
cell-like phenotype (16).
At the molecular level, the present study identified
the transcription factor STAT5 as a potential target of miR-204
function in B-cell lymphoma. This was supported by the observation
that miR-204 could bind and inhibit the activity of STAT5 3′-UTR;
miR-204 mimics reduced and antisense miR-204 increased the protein
expression levels of STAT5, respectively; and re-introduction of
STAT5 successfully reversed the antiproliferative roles of miR-204.
STAT5 is one of seven members of the STAT family of proteins and
has a key role in the generation of B-cell precursors (17). Persistent STAT5 activation is
oncogenic, leading to the late emergence of clonal B-cell
lymphoma/leukemia (18,19). Therefore, understanding the precise
role of the miR-204/STAT5 regulatory axis will advance our
knowledge of lymphoma biology, which may be beneficial for its
treatment.
Acknowledgments
The present study was supported by the Guizhou
Province Science and Technology Fund Projects [grant no. QKH J
(2009) 2214].
References
|
1
|
Seton-Rogers S: Lymphoma: Epigenetic
therapy gains momentum. Nat Rev Cancer. 12:7982012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young RM and Staudt LM: Targeting
pathological B cell receptor signalling in lymphoid malignancies.
Nat Rev Drug Discov. 12:229–243. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lawrie CH: MicroRNA expression in
lymphoma. Expert Opin Biol Ther. 7:1363–1374. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang LQ, Liang R and Chim CS: Methylation
of tumor suppressor microRNAs: lessons from lymphoid malignancies.
Expert Rev Mol Diagn. 12:755–765. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lawrie CH: MicroRNAs and lymphomagenesis:
a functional review. Br J Haematol. 160:571–581. 2013. View Article : Google Scholar
|
|
8
|
Sandhu SK, Volinia S, Costinean S, et al:
miR-155 targets histone deacetylase 4 (HDAC4) and impairs
transcriptional activity of B-cell lymphoma 6 (BCL6) in the
Eμ-miR-155 transgenic mouse model. Proc Natl Acad Sci USA.
109:20047–20052. 2012. View Article : Google Scholar
|
|
9
|
Ralfkiaer U, Hagedorn PH, Bangsgaard N, et
al: Diagnostic microRNA profiling in cutaneous T-cell lymphoma
(CTCL). Blood. 118:5891–5900. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364. 2010.
|
|
11
|
Wang FE, Zhang C, Maminishkis A, et al:
MicroRNA-204/211 alters epithelial physiology. FASEB J.
24:1552–1571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Conte I, Carrella S, Avellino R, et al:
miR-204 is required for lens and retinal development via Meis2
targeting. Proc Natl Acad Sci USA. 107:15491–15496. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paulin R, Courboulin A, Barrier M and
Bonnet S: From oncoproteins/tumor suppressors to microRNAs, the
newest therapeutic targets for pulmonary arterial hypertension. J
Mol Med (Berl). 89:1089–1101. 2011. View Article : Google Scholar
|
|
14
|
Chung TK, Lau TS, Cheung TH, et al:
Dysregulation of microRNA-204 mediates migration and invasion of
endometrial cancer by regulating FOXC1. Int J Cancer.
130:1036–1045. 2012. View Article : Google Scholar
|
|
15
|
Sacconi A, Biagioni F, Canu V, et al:
miR-204 targets Bcl-2 expression and enhances responsiveness of
gastric cancer. Cell Death Dis. 3:e4232012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ying Z, Li Y, Wu J, et al: Loss of miR-204
expression enhances glioma migration and stem cell-like phenotype.
Cancer Res. 73:990–999. 2013. View Article : Google Scholar :
|
|
17
|
Malin S, McManus S and Busslinger M: STAT5
in B cell development and leukemia. Curr Opin Immunol. 22:168–176.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blix ES, Irish JM, Husebekk A, et al:
Phospho-specific flow cytometry identifies aberrant signaling in
indolent B-cell lymphoma. BMC Cancer. 12:4782012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kirk R: Haematological cancer: Hit the
lymphoma, JAK. Nat Rev Clin Oncol. 9:6082012. View Article : Google Scholar : PubMed/NCBI
|