Introduction
Ubiquitin protein ligase E3A (UBE3A), also known as
human papilloma virus E6-associated protein (E6-AP), is an
important member of ubiquitin proteasome systems. The E6/E6-AP
complex has previously been shown to target p53 for ubiquitination
and degradation through the ubiquitin-proteasome pathway (UPP) in
cervical cancer (1,2). It has been reported that abnormal
expression of UBE3A is associated with various types of tumor,
including prostate, cervical and breast cancer (3–5).
However, how the loss of function of UBE3A causes disease
pathogenesis remains to be elucidated. A previous study by our
group reported that UBE3A was overexpressed in breast cancer and
associated with its pathological mechanism (6). Therefore, it may be hypothesized that
UBE3A has an important role in breast cancer through its
ubiquitin-protein ligase activity.
Previously, the degradation of various proteins has
been shown to be mediated by UBE3A, including c-Myc (7), human telomerase reverse transcriptase
(8), p27 (9), progesterone receptor-B protein and
annexin A1 protein (10,11). To date, these cellular substrates
of E6-AP have been identified and characterized; however, the
significance of these interactions in disease remains to be fully
elucidated.
Annexin A2 is a member of the annexin family, which
has been reported to be abundantly expressed in numerous types of
malignant tissue, and is believed to have an important role in
tumorigenesis and cancer progression (12). A previous study demonstrated that
annexin A2 could be inhibited by RNA interference (RNAi), which
resulted in decreased metastasis and invasion of breast cancer
cells (13). Furthermore, a
previous study by our group showed that annexin A2 was
overexpressed and ubiquitinated in breast cancer tissues (14). These findings suggested that
annexin A2 may contribute to the pathogenesis of breast cancer, as
it is mediated by ubiquitination, and it may be a novel substrate
of E6-AP.
The function of UBE3A in the proliferation, invasion
and apoptosis of breast cancer have remained elusive; therefore,
the present study used RNAi to silence UBE3A expression in BT-549
breast cancer cells in order to determine its function. The results
indicated that UBE3A is involved in carcinogenesis by regulating
the levels of annexin A2 in breast cancer cells, which renders it a
potential molecular marker of breast cancer and a target of breast
cancer therapy.
Materials and methods
Reagents
The BT-549 human breast cancer cell line was
obtained from the Cell Bank of Chinese Academy of Sciences
(Beijing, China). RPMI-1640 cell culture medium was purchased from
HyClone Laboratories, Inc. (Logan, UT, USA).
Lipofectamine® 2000 and TRIzol® were obtained
from Invitrogen Life Technologies (Carlsbad, CA, USA). Fetal bovine
serum (FBS), penicillin and streptomycin were purchased from Gibco
Life Technologies (Carlsbad, CA, USA). First Strand cDNA Synthesis
kit and the polymerase chain reaction (PCR) kit were purchased from
Takara Bio, Inc. (Otsu, Japan). Rabbit monoclonal UBE3A (1:1,000;
no. 5571-1), rabbit polyclonal β-actin (1:2,000; no. 5779-1) and
rabbit polyclonal annexin A2 (1:1,000; no. S0555) primary
antibodies, as well as goat anti-rabbit horseradish
peroxidase-conjugated immunoglobulin G (1:5,000; no. 3053-1-1)
secondary antibody were purchased from Epitomics (Burlingame, CA,
USA). Enhanced Chemiluminescence (ECL) Reagent kit, fibronectin and
Transwell assay plates were obtained from EMD Millipore (Billerica,
MA, USA). Cell Counting Kit-8 (CCK-8) and Annexin V-Fluorescein
Isothiocyanate (FITC)/Propidium Iodide (PI) Cell Apoptosis
Detection kit were purchased from Nanjing Keygen Biotech., Co.,
Ltd., Nanjing, China). Negative siRNA and specific UBE3A siRNAs
were synthesized by Takara Bio, Inc. Matrigel® was
purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Cell culture
The BT-549 human breast cancer cells were cultured
in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin and 100
U/ml streptomycin at 37°C in an atmosphere containing 5%
CO2.
Design and synthesis of siRNA targeting
UBE3A
Using siDirect Version 2.0 design software
(http://sidirect2.rnai.jp/), three
specific siRNAs were designed to target the UBE3A gene, and the
negative siRNA fragments were synthesized by Takara Bio, Inc.
Specific siRNA sequences are shown in Table I.
 | Table IsiRNA sequences. |
Table I
siRNA sequences.
| Name | Sequences |
|---|
| siRNA1 | Sense strand:
5′-CUGUAUACUGGAUGUACAUTT-3′
Anti-sense strand: 5′-AUGUACAUCCAGUAUACAGTT-3′ |
| siRNA2 | Sense strand:
5′-CUCAAAUCAGGAACUGUAUTT-3′
Anti-sense strand: 5′-AUACAGUUCCUGAUUUGAGTT-3′ |
| siRNA3 | Sense strand:
5′-GAAGACAAUGCUUUCCAUATT-3′
Anti-sense strand: 5′-UAUGGAAAGCAUUGUCUUCTT-3′ |
| Negative control
siRNA | Sense strand:
5′-AGGUGACUAGCACUGUUAGTT-3′
Anti-sense strand: 5′-GUAACAGUGCUAGUCACCUTT-3′ |
Cell transfection
One day prior to transfection, the BT-549 cells were
seeded in a six-well plate (2×105 cells). A total of 5
μl siRNA, at 20 μM stock, was added to 250 μl
RPMI-1640. In addition, 5 μl Lipofectamine® 2000
was diluted in the same amount of RPMI-1640 medium. Following an
incubation for 5 min at room temperature, the diluted siRNAs were
gently added to the diluted Lipofectamine® 2000 medium
and incubated for 20 min at room temperature. Subsequently, the
mixture was added to the cells, alongside 1.5 ml serum-free and
antibiotic-free RPMI-1640 medium. Following a 6-h incubation, the
mixture was removed and 2 ml RPMI-1640 supplemented with 10% FBS
was added to the plate. Cells treated with
Lipofectamine® 2000 only were considered the control
group.
Semi-quantitative reverse transcription
(RT)-PCR
The BT-549 cells transfected with siRNAs were
harvested 72 h post-transfection. Total RNA was extracted using
TRIzol® reagent according to the manufacturer’s
instructions. The cDNA was synthesized using the PrimeScript RT
reagent kit. PCR amplification was conducted using the Taq
polymerase enzyme (Takara Bio, Inc.). The PCR reaction program was
set as follows: 95°C for 1 min, followed by 30 cycles of 94°C for
30 sec, 55°C for 30 sec and 72°C for 1 min, then a final extension
step of 72°C for 5 min and then stored at 4°C for further use.
β-actin was used as the internal control. The PCR products were
then separated by 1.5% agarose gel electrophoresis (Takara Bio,
Inc.) at 105 V for 30 min. Images were captured using the GelDoc XR
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and the
images were analyzed using PD QUEST 8.0 software (Bio-Rad
Laboratories, Inc.). The sequences of the specific primers are
shown in Table II. The experiment
was repeated three times.
 | Table IIPolymerase chain reaction primers. |
Table II
Polymerase chain reaction primers.
| Name | Sequences | Product length
(bp) |
|---|
| UBE3A | Forward:
5′-CACTTGTCCGGCTAGAGATGAT-3′
Reverse: 5′-TCCCCATTAGCTTCCTGTAGAC-3′ | 324 |
| Annexin A2 | Forward:
5′-TGACGCTGGAGTGAAGAGGAA-3′
Reverse: 5′-GCCCTTAGTGTCTTGCTGGATA-3 | 379 |
| β-actin | Forward:
5′-GACCCAGATCATGTTTGAGACC-3
Reverse: 5′-ATCTCCTTCTGCATCCTGTCG-3′ | 594 |
Western blot analysis
Total protein was extracted from the cells using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China), 72 h post-transfection. Total
protein was quantified using the bicinchoninic acid method
(Beyotime Institute of Biotechnology). According to the size of the
molecular weight of the protein extracts, the appropriate
concentration of SDS-PAGE gel (UBE3A, 8%; annexin A2 and β-actin,
12%). Equal amounts of total protein (30 μg) were separated
by SDS-PAGE. The proteins were then transferred to polyvinylidene
fluoride (PVDF) membranes (0.45 μm; EMD Millipore) in
transfer buffer (Beyotime Institute of Biotechnology) by
electroblotting using a Mini-Trans-Blot Electrophoretic Transfer
Cell (Bio-Rad Laboratories, Inc.) at 200 mA for 1–2 h. The PVDF
membranes were blocked for 1 h at room temperature using blocking
solution (Beyotime Institute of Biotechnology), and were then
incubated with the specified dilutions of the primary antibodies
for UBE3A, annexin A2 and β-actin overnight at 4°C. The membranes
were then washed three times with Tris-buffered saline-Tween (15
min/wash), prior to an incubation with the recommended dilution of
the labeled secondary antibody at 37°C for 1 h. The goat
anti-rabbit alkaline phosphatase-conjugated antibody was detected
colorimetrically using ECL Plus. Images were captured using the
GelDoc XR system and were analyzed using Image Lab™ 3.0 software
(Bio-Rad Laboratories, Inc.). β-actin was used as the internal
control. The experiment was repeated three times.
Cell proliferation assay
The CCK-8 assay was performed to examine cell
proliferation. The experiment was repeated five times. Cells
(2×103/well) in the logarithmic phase were cultured in
96-well culture plates. The cells were divided into the following
three groups: siRNA1 group, negative control group and blank
control group. The methods of cell transfection were the same as
stated above. A total of 10 μl CKK-8 reagent was added at
four time-points (24, 48, 72 and 96 h post-transfection) and the
plates were incubated at 37°C for 2 h. The absorbance was then
measured at a wavelength of 450 nm using a microplate reader
(Biochrom Anthos 2010; Autobio Diagnostics Co., Ltd., Zhengzhou,
China).
Flow cytometric assay
The Annexin V-FITC/PI Apoptosis Detection kit was
used to detect the rate of cell apoptosis according to the
manufacturer’s instructions. The experiment was repeated five
times. Cells (2×105/ml) in the logarithmic phase were
cultured in six-well culture plates. The cells were divided into
the same three groups as for the cell proliferation assay. Briefly,
the cells were collected 72 h post-transfection, after being washed
twice with cold phosphate-buffered saline (PBS). The cells were
then suspended in 500 μl 1× binding buffer and subsequently,
5 μl Annexin V and PI were added to the cells. The cells
were incubated for 15 min at room temperature in the dark, followed
by flow cytometric analysis (FACSCalibur; Becton Dickinson,
Franklin Lakes, NJ, USA). The apoptotic rate was calculated as the
percentage of Annexin V-positive and PI-negative cells out of the
total number of cells. The assay was conducted using a flow
cytometer and was analyzed using WinMDI 2.9 software (www.bioon.com).
Cell invasion assay
Transwell chambers were used to perform invasion
experiments. Briefly, Matrigel® was thawed at 4°C
overnight, and was then diluted with an equal amount of PBS. A
total of 80 μl diluted Matrigel® was added to the
upper chamber, and 30 μl fibronectin was added to the lower
chamber of a 24-well Transwell plate (24-well insert; pore size, 8
μm; EMD Millipore). The Transwell chamber was then incubated
at 37°C for ≥4 h. The cells were harvested from the cell culture
plates 72 h post-transfection, and were washed three times with
RPMI-1640 supplemented with 1% FBS. The cells (4×104
cells) were seeded into the upper chamber, and 600 μl
RPMI-1640 containing adhesive stubstrate (diluted
Matrigel®) was added to the lower chamber. The Transwell
chamber was incubated at 37°C for 24 h. The cells that did not
invade through the pores were removed using a cotton swab, whereas
the cells on the lower surface of the membrane were stained with
crystal violet (Beyotime Institute of Biotechnology) and counted
using an IX53-F32PH microscope (Olympus Corp., Tokyo, Japan). The
experiment was repeated tree times.
Statistical analysis
The results are presented as the mean ± standard
deviation. Statistical analyses were performed by one-way analysis
of variance using SPSS version 13.0 software (SPSS Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
mRNA expression levels of UBE3A and
annexin A2 in BT-549 cells
The relative mRNA expression levels of UBE3A and
annexin A2 were calculated as UBU3A/β-actin and annexin A2/β-actin,
respectively. The UBE3A specific siRNAs effectively silenced the
mRNA expression levels of UBE3A, as compared with levels in the
control groups (n=3, P≤0.01). In addition, following UBE3A
knockdown with siRNA1, the relative mRNA expression levels of
annexin A2 were down-regulated, as compared with those in the
control groups (n=3, P≤0.05) (Fig.
1A and Table III).
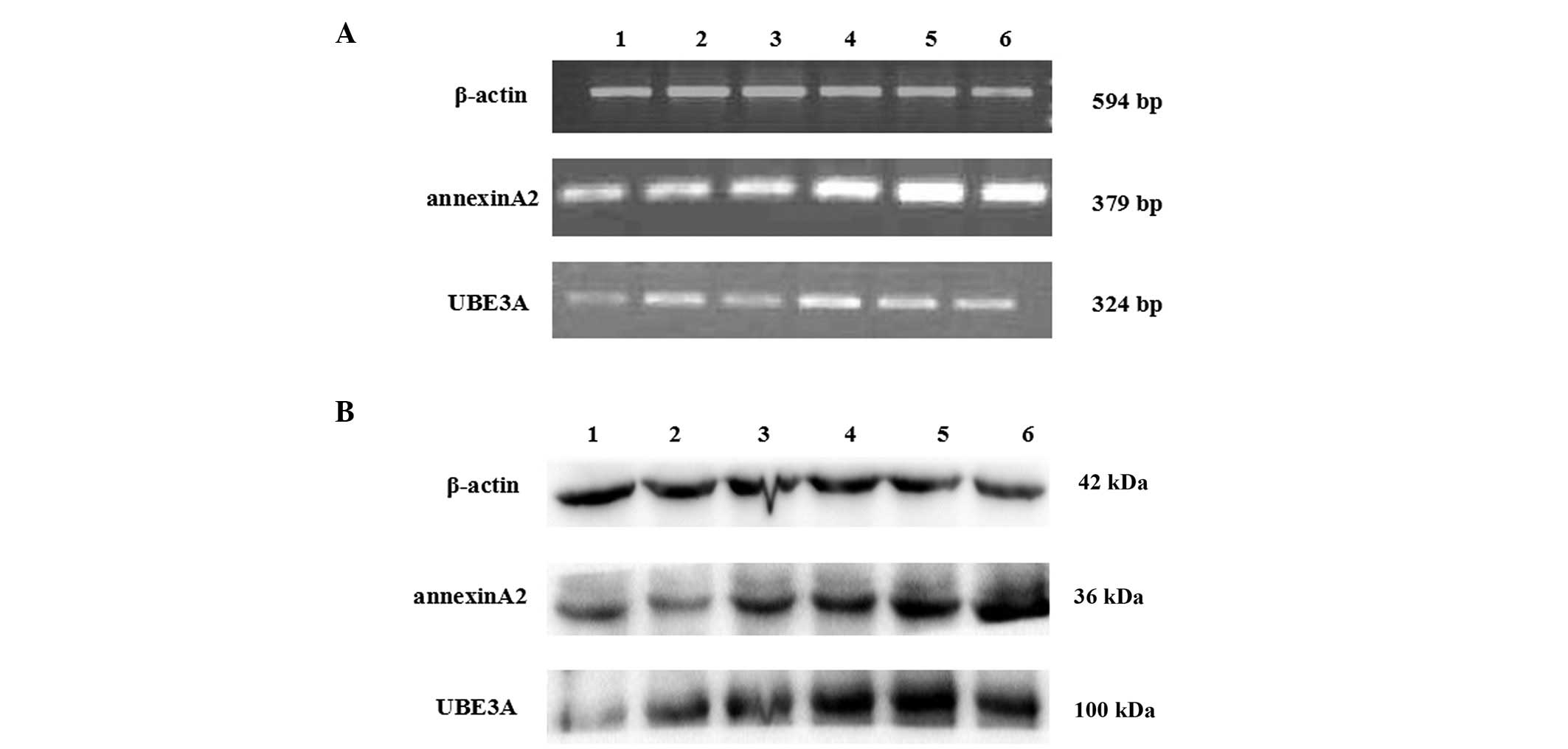 | Figure 1BT-549 human breast cancer cells were
transfected with siRNA1, siRNA2, siRNA3 and negative control siRNA,
using Lipofectamine® 2000. The cells treated with
Lipofectamine® 2000 only were considered the control.
Lane 1, siRNA1 group; lane 2, siRNA2 group; lane 3, siRNA3 group;
lane 4, negative control group; lane 5, Lipofectamine®
2000 group; lane 6, blank control group. (A) Total RNA was
extracted from the cells 72 h post-transfection. Reverse
transcription polymerase chain reaction was performed to measure
the relative mRNA expression levels of UBE3A and annexin A2. The
PCR products were seperated by 1.5% agarose gel electrophoresis.
The mRNA expression levels of β-actin served as a loading and
normalization control. (B) Protein expression levels of UBE3A and
annexin A2 protein expression levels were determined by western
blotting. The protein expression levels of β-actin served as a
loading and normalization control. Data were averaged from three
independent experiments. siRNA, small interfering RNA; UBE3A,
ubiquitin protein ligase E3A. |
 | Table IIIRelative mRNA expression levels of
UBE3A and annexin A2 in BT-549 human breast cancer cells. |
Table III
Relative mRNA expression levels of
UBE3A and annexin A2 in BT-549 human breast cancer cells.
| Group | n | UBE3A/β-actin | Annexin
A2/β-actin |
|---|
| siRNA1 group | 3 | 0.15±0.016 | 0.942±0.032 |
| siRNA2 group | 3 | 0.32±0.026 | 1.009±0.014 |
| siRNA3 group | 3 | 0.23±0.016 | 0.99±0.02 |
| Negative siRNA
group | 3 | 0.59±0.014 | 1.333±0.022 |
| Lipofectamine 2000
group | 3 | 0.587±0.021 | 1.326±0.018 |
| Control group | 3 | 0.592±0.027 | 1.336±0.026 |
| F | | 285.197 | 219.939 |
| P-value | | ≤0.05 | ≤0.05 |
Protein expression levels of UBE3A and
annexin A2 in BT-549 cells
The relative protein expression levels of UBE3A and
annexin A2 were calculated as UBU3A/β-actin and annexin A2/β-actin,
respectively. The protein expression levels were concordant with
the mRNA expression levels of UBE3A and annexin A2. siRNA1 was
shown to be the most effective siRNA for silencing UBE3A mRNA as
well as protein expression; therefore, the siRNA1 group was
selected as the experimental group for subsequent assays (Fig. 1B and Table IV).
 | Table IVRelative protein expression levels of
UBE3A and annexin A2 in BT-549 human breast cancer cells. |
Table IV
Relative protein expression levels of
UBE3A and annexin A2 in BT-549 human breast cancer cells.
| Groups | n | UBE3A/β-actin | Annexin
A2/β-actin |
|---|
| siRNA1 group | 3 | 0.276±0.019 | 0.452±0.033 |
| siRNA2 group | 3 | 0.332±0.025 | 0.547±0.036 |
| siRNA3 group | 3 | 0.312±0.022 | 0.543±0.048 |
| Negative siRNA
group | 3 | 0.471±0.031 | 0.833±0.03 |
| Lipofectamine 2000
group | 3 | 0.507±0.021 | 0.846±0.041 |
| Control group | 3 | 0.478±0.031 | 0.887±0.027 |
| F | | 47.655 | 81.604 |
| P-value | | ≤0.05 | ≤0.05 |
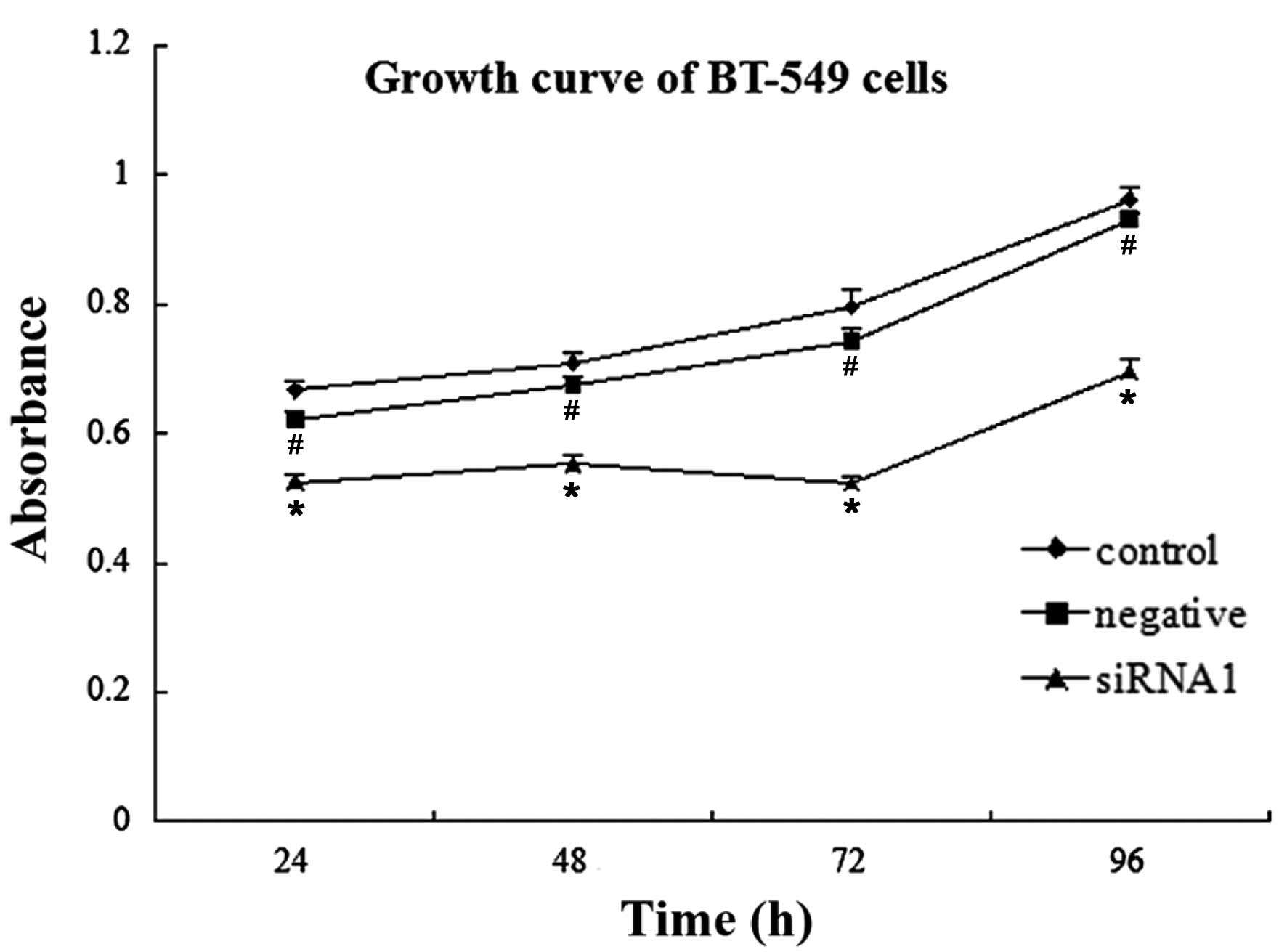
Cell proliferation
Proliferation of the BT-549 cells in the siRNA1
group was significantly inhibited 24, 48, 72 and 96 h
post-transfection, as compared with that of the blank control group
(n=5, P<0.01). However, there were no statistically significant
differences detected between the negative siRNA group and the blank
control group at the four post-transfection time-points (n=5,
P>0.05) (Fig. 2 and Table V).
 | Table VAbsorbance of BT-549 human breast
cancer cells at various time-points. |
Table V
Absorbance of BT-549 human breast
cancer cells at various time-points.
| Groups | 24 h | 48 h | 72 h | 96 h |
|---|
| siRNA1 |
0.5244±0.0130a | 0.5528±0.0147 | 0.523±0.0120 | 0.695±0.0190 |
| Negative siRNA |
0.6218±0.0110b | 0.6744±0.0143 | 0.7432±0.0184 | 0.9318±0.0094 |
| Control | 0.6668±0.0163 | 0.7098±0.0143 | 0.796±0.0268 | 0.9622±0.02 |
Cell apoptosis
The rate of apoptosis of the cells in the siRNA1
group, negative control group and blank control group was
11.104±1.1935%, 2.852±0.7213% and 2.788±0.3954%, respectively, 72 h
post-transfection. The rate of apoptosis was significantly higher
in the siRNA1 group, as compared with the control groups (n=5,
P<0.05) (Fig. 3).
Cell invasion
The number of invasive cells in the siRNA1 group,
negative control group and blank control group were 53.96±4.55,
93.16±3.91 and 94.43±3.71, respectively, 72 h post-transfection.
The number of invasive cells in the siRNA group was significantly
lower as compared with that in the blank control group (n=10,
P<0.01) (Fig. 4).
Discussion
The UPP is an important pathway of protein
degradation for numerous proteins vital to cellular regulation and
function. Furthermore, the ubiquitin-proteasome system is a crucial
regulator of the cell cycle, and it is well-known that abnormal
cell cycle control may lead to oncogenesis (15–17).
A previous study demonstrated that UPP dysregulation has an
important role in mammary tumorigenesis (18). UBE3A is an important member of the
ubiquitin-ligase family and is one of the key enzymes associated
with the maintenance of normal cellular physiological functions
(15). Abnormal alterations of
UBE3A may cause the development of various diseases.
RNAi has recently been identified as being capable
of gene silencing. RNAi is mediated by double-stranded RNA, which
can degrade specific target mRNAs (4). The present study used chemically
synthesized siRNAs to suppress the expression of UBE3A. Chemical
synthesis of siRNA has previously been shown to effectively block
gene expression (19). According
to the principle of RNAi, specific siRNAs were synthesized in
vitro, and were then transfected into BT-549 human breast
cancer cells. The interference effects were detected using
semi-quantitative RT-PCR and western blotting. The results of the
present study demonstrated that three specific siRNAs were able to
effectively inhibit the expression of UBE3A. siRNA1 was shown to
exhibit the best interference effect. Following effective UBE3A
gene knockdown, the expression levels of annexin A2 were
downregulated. This may have an adverse effect on certain proteins
that are degraded in an E6-AP-dependent manner. Shimoji et
al (10) previously reported
that RNAi-mediated downregulation of endogenous E6-AP increased the
levels of endogenous annexin A1 protein, but had no effect on the
accumulation of endogenous annexin A2 protein.
The present study aimed to determine why the
expression of annexin A2 was downregulated following knockdown of
UBE3A expression. The results suggested that UBE3A may have a role
in controlling the functions of annexin A2 in breast cancer cells.
It may be hypothesized that ubiquitinated annexin A2 was not
degraded, but its function, activity or position may have been
altered, which was regulated by UBE3A in BT-549 breast cancer
cells. Another hypothesis may be that following knockdown of UBE3A,
annexin A2 may be degraded via UPP, or another pathway. Previous
studies have suggested that polyubiquitinated protein may be
generally degraded by the 26S proteasome; however, if
polyubiquitinated proteins are not connected to the target protein
48 or 63-bit lysines, but to the 6, 11, 27, 29 and 33 lysines, the
ubiquitinated proteins may not be degraded, but their funtions may
be altered (20,21). Furthermore, monoubiquitinated
proteins may not be degraded; however, their activity, positioning
or structure may be modified, thus regulating the endocytosis
pathway, histone modification, gene transcription and nuclear
protein localization (22). Future
studies may detect the ubiquitinated position or numbers of
ubiquitinated annexin A2 in the BT-549 breast cancer cells prior to
as well as following UBE3A RNAi, in order to further study the
association between annexin A2 and UBE3A. Exploring the role of
UBE3A in the transcriptional regulation of annexin A2 or other
genes may be an interesting area of research.
Treatment and prognosis of breast cancer are complex
and are associated with numerous factors in vivo and in
vitro. Cell proliferation, apoptosis and invasion are
considered the most important factors affecting the treatment and
prognosis of breast cancer. In the present study, a CCK-8 assay,
flow cytometry and a Transwell assay were used to detect the
proliferation, apoptosis and invasion of BT-549 cells following
silencing of UBE3A. The results demonstrated that the proliferation
of BT-549 cells was restrained following transfection withUBE3A
siRNA. The rate of apoptosis of the cells in the siRNA1 group was
increased, and the invasiveness of the cells was decreased. All of
these differences were statistically significant when comparing the
siRNA1 group with the control groups.
The present study aimed to determine how UBE3A could
influence proliferation, apoptosis and invasion of breast cancer
cells. A previous study demonstrated that UBE3A was able to impact
the function of cells, including transcription, signal
transduction, cell survival, cell cycle control and DNA repair
(23). In addition, it has been
reported that UBE3A can promote proliferation and inhibit apoptosis
of cervical cancer cells (4). In
prostate cancer cells, UBE3A has been shown to regulate the
phosphoinositide 3-kinase-Akt pathway, which results in increased
prostate cell growth, proliferation and decreased apoptosis
(24). Therefore, it was
hypothesized that in breast cancer cells, UBE3A may affect cell
signaling pathways and influence cell biological behavior.
The present study also demonstrated that following
knockdown of UBE3A, the expression of annexin A2 was downregulated.
Annexin A2 has previously been demonstrated to promote the
development, invasion and metastasis of breast cancer (13), which may explain the simultaneous
influence on proliferation, apoptosis and invasion of breast cancer
cells. Furthermore, UBE3A may regulate other cancer-associated
proteins, which have important roles in the pathogenesis of breast
cancer and directly or indirectly lead to cancer through regulating
cell proliferation, migration, invasion and apoptosis. E6-AP has
also been reported to promote the degradation of p53 (3,25,26).
Altered degradation of p53 may also affect cell cycle progression
and induce apoptosis. A future aim of our group is to use
two-dimensional polyacrylamide gel electrophoresis and
matrix-assisted laser desorption/ionization tandem time-of-flight
mass spectrometry incorporated with online database research to
identify the effect of knockdown of UBE3A expression on the levels
of various proteins.
In conclusion, the results of the present study
indicated that UBE3A has a role in regulating the functions of
annexin A2 and a positive role in the development of breast cancer.
These results suggested that UBE3A may be a potential marker for
treatment of breast cancer. However, the specific mechanisms
underlying the functions of UBE3A in breast cancer require further
research.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of China (grant no. 81172496), the
Sichuan Science and Technology Support Program (grant no.
2009SZ0116) and the Sichuan Department of Education (grant no.
10ZA075).
References
|
1
|
Yamamoto Y, Huibregtse JM and Howley PM:
The human E6-AP gene (UBE3A) encodes three potential protein
isoforms generated by differential splicing. Genomics. 41:263–266.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scheffner M, Huibregtse JM, Vierstra RD
and Howley PM: The HPV-16 E6 and E6-AP complex functions as a
ubiquitin-protein ligase in the ubiquitination of p53. Cell.
75:495–505. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khan OY, Fu G, Ismail A, et al:
Multifunction steroid receptor coactivator, E6-associated protein,
E6-AP is involved in development of the prostate gland. Mol
Endocrinol. 20:544–559. 2006. View Article : Google Scholar
|
|
4
|
Xiu XX, Zhang SL, Lu XY, Liang MY, Yu J
and Hou JP: SiRNA inhibition of E6AP expression in cervical cancer
cells. Zhonghua Bing Li Xue Za Zhi. 37:822–825. 2008.In
Chinese.
|
|
5
|
Ramamoorthy S, Tufail R, Hokayem JE, et
al: Overexpression of ligase defective E6-associated protein,
E6-AP, results in mammary tumorigenesis. Breast Cancer Res Treat.
132:97–108. 2012. View Article : Google Scholar
|
|
6
|
Deng S, Zhou H, Xiong R, et al:
Over-expression of genes and proteins of ubiquitin specific
peptidases (USPs) and proteasome subunits (PSs) in breast cancer
tissue observed by the methods of RFDD-PCR and proteomics. Breast
Cancer Res Treat. 104:21–30. 2007. View Article : Google Scholar
|
|
7
|
Liu X, Disbrow GL, Yuan H, Tomaic V and
Schlegel R: Myc and human papillomavirus type 16 E7 genes cooperate
to immortalize human keratinocytes. J Virol. 81:12689–12695. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Yuan H, Fu B, et al: The E6AP
ubiquitin ligase is required for transactivation of the hTERT
promoter by the human papillomavirus E6 oncoprotein. J Biol Chem.
280:10807–10816. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mishra A, Godavarthi SK and Jana NR:
UBE3A/E6-AP regulates cell proliferation by promoting proteasomal
degradation of p27. Neurobiol Dis. 36:26–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimoji T, Murakami K, Sugiyama Y, et al:
Identification of annexin A1 as a novel substrate for E6AP-mediated
ubiquitylation. J Cell Biochem. 106:1123–1135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramamoorthy S, Dhananjayan SC, Demayo FJ
and Nawaz Z: Isoform-specific degradation of PR-B by E6-AP is
critical for normal mammary gland development. Mol Endocrinol.
24:2099–2113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lokman NA, Ween MP, Oehler MK and
Ricciardelli C: The role of annexin A2 in tumorigenesis and cancer
progression. Cancer Microenviron. 4:199–208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma MR, Koltowski L, Ownbey RT,
Tuszynski G and Sharma MC: Angiogenesis-associated protein annexin
II in breast cancer: selective expression in invasive breast cancer
and contribution to tumor invasion and progression. Exp Mol Pathol.
81:146–156. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deng S, Jing B, Xing T, Hou L and Yang Z:
Overexpression of annexin A2 is associated with abnormal
ubiquitination in breast cancer. Genomics Proteomics
Bioinformatics. 10:153–157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramamoorthy S and Nawaz Z: E6-associated
protein (E6-AP) is a dual function coactivator of steroid hormone
receptors. Nucl Recept Signal. 6:e0062008.PubMed/NCBI
|
|
16
|
Yamasaki L and Pagano M: Cell cycle,
proteolysis and cancer. Curr Opin Cell Biol. 16:623–628. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Micel LN, Tentler JJ, Smith PG and
Eckhardt GS: Role of ubiquitin ligases and the proteasome in
oncogenesis: novel targets for anticancer therapies. J Clin Oncol.
31:1231–1238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dees EC and Orlowski RZ: Targeting the
ubiquitin-proteasome pathway in breast cancer therapy. Future
Oncol. 2:121–135. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Spänkuch-Schmitt B, Bereiter-Hahn J,
Kaufmann M and Stredhardt K: Effect of RNA sileneing of polo-like
kinase-1 (PLK1) on apoptosis and spindle formation in human cancer
cells. J Natl Cancer Inst. 94:1863–1877. 2002. View Article : Google Scholar
|
|
20
|
Liu S and Chen ZJ: Expanding role of
ubiquitination in NF-κB signaling. Cell Res. 21:6–21. 2011.
View Article : Google Scholar
|
|
21
|
Adhikari A and Chen ZJ: Diversity of
polyubiquitin chains. Dev Cell. 16:485–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sadowski M, Suryadinata R, Tan AR, Roesley
SN and Sarcevic B: Protein monoubiquitination and
polyubiquitination generate structural diversity to control
distinct biological processes. IUBMB Life. 64:136–142. 2012.
View Article : Google Scholar
|
|
23
|
Lochab S, Pal P, Kanaujiya JK, et al:
Proteomic identification of E6AP as a molecular target of tamoxifen
in MCF7 cells. Proteomics. 12:1363–1377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Srinivasan S and Nawaz Z: E3 ubiquitin
protein ligase, E6-associated protein (E6-AP) regulates PI3K-Akt
signaling and prostate cell growth. Biochim Biophys Acta.
1809:119–127. 2011. View Article : Google Scholar :
|
|
25
|
Jiang YH, Armstrong D, Albrecht U, et al:
Mutation of the Angelman ubiquitin ligase in mice causes increased
cytoplasmic p53 and deficits of contextual learning and long-term
potentiation. Neuron. 21:799–811. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mishra A and Jana NR: Regulation of
turnover of tumor suppressor p53 and cell growth by E6-AP, a
ubiquitin protein ligase mutated in Angelman mental retardation
syndrome. Cell Mol Life Sci. 65:656–666. 2008. View Article : Google Scholar : PubMed/NCBI
|