Introduction
Lung cancer remains the leading cause of
cancer-associated mortality in China and numerous other countries
in the world (1). Non-small cell
lung cancer (NSCLC) accounts for ~85% of all types of lung cancer
(2), with <15% of patients
surviving beyond 5 years (3,4).
Metastasis is a major cause of morbidity and mortality in NSCLC
cancer. Surgical resection of primary lung cancer is frequently
followed by tumor recurrence at distant sites, including the lymph
nodes (5), bone (6) and brain (7). Therefore, in depth investigations
into the mechanisms underlying NSCLC invasion and metastasis are
required to gain insights into disease progression and to identify
potential therapeutic targets and treatment strategies.
A disintegrin and metalloproteinases (ADAMs)
comprise a family of type I transmembrane proteins containing a
metalloproteinase and disintegrin extracellular domain, and are
involved in cell adhesion, migration, cell signal transduction and
the proteolytic processing of multiple transmembrane proteins
(8). In addition, ADAMs are
important in a range of human diseases, including inflammatory
diseases and asthma (9,10). ADAM family proteins, including
ADAM9, ADAM12, ADAM15 and ADAM17 have also been associated with
human cancer metastasis, formation and progression (11,12).
Among these members of the ADAM protein family,
ADAM9 in particular has been associated with tumorigenesis and
tumor cell metastasis (13). It
has been demonstrated that ADAM9 expression is frequently
upregulated in various types of cancer, including breast cancer
(14), hepatocellular carcinoma
(15), gastric cancer (16), pancreatic ductal adenocarcinoma
(17), prostate cancer (18), renal cell carcinoma (19) and cervical squamous carcinoma
(20), correlating with cancer
progression, metastasis and predicting a shortened survival time in
patients (14–20). Consistent with these results, a
previous study demonstrated that ADAM9 is highly expressed in NSCLC
and highly expressed ADAM9 correlates with a shortened survival
time (21). Notably, Shintani
et al found that ADAM9 overexpression enhances cell adhesion
and invasion of NSCLC cells via modulation of other adhesion
molecules and altering sensitivity to growth factors (22). A previous study demonstrated that
RNA interference (RNAi)-mediated downregulation of endogenous
ADAM9 could inhibit adenoid cystic carcinoma cell growth and
metastasis in vitro and in vivo (23). The generation of MDA-MB-231
knockdown clones lacking ADAM9 expression inhibited breast
cancer cell invasion in vitro (24). The above studies imply that ADAM9
is important in tumor metastasis.
However, relatively little is known about the role
of ADAM9 in NSCLC cells. Thus, in the present study, the
feasibility of lentiviral vector delivered small hairpin RNA
(shRNA) against ADAM9 for the treatment of NSCLC in vitro
and in vivo was assessed and the molecular pathways involved
were analyzed.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM) and fetal
bovine serum (FBS) were purchased from HyClone (Logan, UT, USA).
Lipofectamine™ 2000 and TRIzol® reagent was obtained
from Invitrogen Life Technologies (Carlsbad, CA, USA). Moloney
murine leukemia virus (M-MLV) reverse transcriptase was purchased
from Promega (Madison, WI, USA). All other chemicals were obtained
from Sigma-Aldrich (St. Louis, MO, USA). For western blot analysis,
the following primary antibodies were used: Mouse monoclonal
anti-human β-actin (1:5,000 dilution; cat. no. A2228;
Sigma-Aldrich) and mouse monoclonal anti-human ADAM9 (1;2,000
dilution; cat. no. sc-23290) purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA).
Cell culture
The human non-small-cell lung cancer cell lines A549
and human embryonic kidney 293T cell lines were purchased from the
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences, Shanghai Institute of Cell Biology, Chinese Academy of
Sciences (Shanghai, China). A549 cells and 293T cells were cultured
in DMEM supplemented with heat-inactivated 10% FBS at 37°C in a
humidified atmosphere containing 5% CO2.
Construction of the ADAM9 shRNA
lentivirus vector and cell infection
The following oligonucleotides were synthesized. The
sequence for the negative control small interfering RNA (siRNA) was
5′-AATTCTCCGAACGTGTCACGT-3′, which did not target any genes in
humans, mice or rats and was essential for determining the effects
of siRNA delivery. The ADAM9 siRNA sequence was
5′-GGCGGGATTAATGTGTTTG-3′. The stem-loop-stem oligos (shRNAs) were
synthesized, annealed and ligated into the NheI/Pac
I-linearized pFH-Lvector. The lentiviral-based shRNA-expressing
vectors were confirmed by DNA sequencing. The generated plasmids
were referred to as pFH-LshADAM9 or pFH-LshCon, respectively.
Recombinant lentiviral vectors and packaging vectors were then
transfected into 293T cells via Lipofectamine™ 2000 (Invitrogen
Life Technologies) to generate the lentivirus. Supernatants
containing either the lentivirus expressing the ADAM9 shRNA or the
control shRNA were harvested 72 h after transfection. The
lentiviruses were purified by ultra-centrifugation at 17,000 × g
for 30 min and the titer of the lentiviruses was determined.
For lentivirus infection, A549 cells were cultured
in 6-well plates. Subsequently, ADAM9 shRNA-expressing lentivirus
or non-targeting shRNA-expressing lentivirus (negative control) was
added, with a multiplicity of infection of 10 in A549 cells, and
mock-infected cells were used as negative controls. After 3 days of
infection, cells were observed under a fluorescence microscope
(MicroPublisher 3.3 RTV; Olympus, Tokyo, Japan).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from A549 cells 5 days after
infection with the lentivirus constructs using TRIzol®
reagent (Invitrogen Life Technologies). cDNA was synthesized from
total RNA using M-MLV reverse transcriptase with random primers
according to the manufacturer’s instructions (Promega). qPCR
analysis was performed using a SYBR Green Master mix kit (Applied
Biosystems, Foster City, CA, USA) on a Bio-Rad connect Real-Time
PCR platform (Bio-Rad Laboratories, Inc., Hercules, CA, USA). In
brief, each PCR reaction mixture, containing 10 μl of 2X
SYBR Green Master mix, 1 μl of sense and antisense primers
(5 μmol/μl) and 1 μl of cDNA (10 ng), was run
for 40 cycles with denaturation at 95°C for 15 sec, annealing at
58°C for 10 sec and extension at 72°C for 30 sec in a total volume
of 20 μl. For relative quantification, 2−ΔΔCT was
calculated and used as an indication of the relative expression
levels, which was calculated by subtracting the CT values of the
control gene from the CT values of ADAM9. The primer sequences for
the PCR amplification of the ADAM9 gene were: ADAM9, sense
5′-TGTGGGAACAGTGTGTTCAAGGA-3′ and antisense
5′-CCAATTCATGAGCAACAATGGAAG-3′. β-actin was used as an internal
control. The primer sequences for β-actin were: β-actin, sense
5′-GTGGACATCCGCAAAGAC-3′ and antisense
5′-AAAGGGTGTAACGCAACTA-3′.
Western blot analysis
A549 cells were collected 5 days after infection
with the lentivirus constructs and were then washed twice with
phosphate-buffered saline (PBS; Sigma-Aldrich) and lysed in RIPA
lysis buffer (Sigma-Aldrich). Protein concentrations were
determined using Protein Assay reagent (Bio-Rad Laboratories,
Inc.). Protein samples (30 mg) were loaded on 8% sodium dodecyl
sulfate-polyacrylamide gels. Following electrophoresis, the
proteins were transferred onto polyvinylidene difluoride membranes
(Bio-Rad Laboratories, Inc.). Blocking was performed using 5%
non-fat milk in a 1X mixture of Tris-buffered saline and Tween 20
(Sigma-Aldrich). The membranes were then incubated overnight at 4°C
with the primary antibodies. Following washing and incubation with
sheep anti-mouse horseradish peroxidase-conjugated polyclonal IgG
secondary antibody (1:10,000; cat no. RPN4201; Amersham
Biosciences, Uppsala, Sweden) for 2 h at room temperature, blotted
proteins were detected using an enhanced chemiluminescence system
(Millipore Corporation, Billerica, MA, USA) with the BioSpectrum
Imaging System (UVP, Upland, CA, USA).
Cell proliferation assay
To measure the effect of downregulation of ADAM9 by
RNAi on cell proliferation, a cell counting kit-8 (CCK-8) assay
(Dojindo Laboratories, Kumamoto, Japan) was performed. In brief,
A549 cells infected with the ADAM9 shRNA lentivirus (Lv/sh-ADAM9)
or the non-silencing shRNA lentivirus (Lv/sh-NC), along with
untreated cells, were seeded in 96-well plates at a density of
5×103 cells per well. The proliferative activity was
determined at the end of different experimental periods (24, 48,
72, 96 and 120 h) using the CCK-8 assay according to the
manufacturer’s instructions. When the media changed from red to
yellow, the absorbance value at a wavelength of 450 nm was detected
using an enzyme-linked immunosorbent assay reader (Multiskan EX;
Thermo Labsystems, Vantaa, Finland). The experiment was performed
at least three times with similar results.
The proliferation rate of cells was determined by
measuring the incorporation of 5-bromodeoxyuridine (BrdU) into the
genomic DNA. In brief, A549 cells infected with Lv/sh-ADAM9 or
Lv/sh-NC, along with untreated cells, were seeded in a 96-well
plates at a density of 2×103 cells per well. A BrdU
incorporation assay was performed using a BrdU Cell Proliferation
Assay kit (Chemicon, Temecula, CA, USA) according to the
manufacturer’s instructions. Briefly, 20 μl of 1/500 diluted
BrdU was added and the assay was incubated for 6 h. Subsequently,
100 μl of 1/200 diluted anti-BrdU provided by the BrdU Cell
Proliferation Assay kit (Chemicon) and peroxidase-conjugated goat
anti-mouse polyclonal IgG antibodies (1:1,000 dilution; cat. no.
Ig-0296G; Sigma-Aldrich) were used according to the manufacturer’s
instructions. The plates were washed twice with PBS containing 0.1%
Triton X-100 (Sigma-Aldrich) and then 100 μl
3,3′,5,5′-tetramethylbenzidine peroxidase substrate (Sigma-Aldrich)
was added. Plates were read at a dual wavelength of 450/550 nm and
the growth rate of cells was calculated as described previously
(25).
Apoptosis analysis
To measure the effect of lentivirus-mediated siRNA
targeting ADAM9 on the cell apoptosis of A549 cells, a terminal
deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
was performed. In brief, cellular DNA fragmentation was measured
with the ApoTag Red in situ Apoptosis detection kit
(Chemicon International, Billerica, MA, USA) according to the
manufacturer’s instructions when A549 cells were infected with
Lv/sh-ADAM9 or Lv/sh-NC for 48 h. To quantify the apoptotic cells,
the TUNEL-positive cells were counted using a confocal microscope
(BX53; Olympus).
In addition, at the molecular level, caspase-3,
caspase-8 and caspase-9 activity was subsequently detected by ELISA
as an additional indicator of apoptosis.
Caspase activity
The activity of caspase-3, caspase-8 and caspase-9
was determined using caspase colorimetric protease assay kits
(Millipore Corporation) according to the manufacturer’s
instructions. In brief, A549 cells were infected with Lv/sh-ADAM9
or Lv/sh-NC for 24 h. Following treatment, cells were washed twice
with ice-cold PBS and harvested by centrifugation at 1,000 × g for
5 min. The cell pellets were then lysed in 150 μl buffer
provided in the kit. Protein concentrations of lysates were
measured by the Lowry method. An aliquot of lysates (80 μl)
was incubated with 10 μl substrate of each caspase at 37°C
for 2 h. Samples were analyzed at 405 nm in a microplate reader
(Thermo Fisher Scientific Inc., Waltham, MA, USA). The relative
caspase activity of the control group was referred to as 100.
Wound healing assay
To assess the effect of downregulation of ADAM9 on
cell migration, a wound healing assay was performed. In brief, A549
cells were infected with Lv/sh-ADAM9 or Lv/sh-NC, along with
untreated cells, seeded in a 6 cm dish with 1.5×106
wells per dish and cultured for 24 h. A linear wound of the
cellular monolayer was created by scratching the confluent cell
monolayer using a plastic pipette tip. The monolayer of scratched
cells was washed with PBS to remove debris. Following incubation at
37°C with 5% CO2 for 24 h, images of the area of
migration were captured under a light microscope (CX41; Olympus)
for evaluation. All experiments were performed in triplicate.
Transwell migration assay
The migration assay of A549 cells was performed
in vitro using Transwell chambers (Corning Life Sciences,
Tewksbury, MA, USA) in which the two chambers were separated by a
Matrigel-coated polycarbonate membrane (8 μm pore size). In
brief, A549 cells infected with Lv/sh-ADAM9 or Lv/sh-NC, along with
untreated cells, were seeded into the cell culture insert (8
μm pore size; Falcon, BD Biosciences, Franklin Lakes, NJ,
USA), precoated with 25 μl of 20% Matrigel (2–3 mg⁄ml
protein) and then placed in a 24-well plate (Falcon) with
1×105 wells per well. Following culturing cells at 37°C
for 40 h, they were fixed and stained with 0.5% crystal violet. The
cells on the top of the cell culture insert were removed by wiping
with a cotton swab and cell invasion was observed using an
immunofluorescence microscope (CKX41; Olympus) by counting the
cells that had invaded into the bottom of the cell culture insert.
All experiments were performed in triplicate.
Tumor xenograft assay
All animal experiments were performed following the
standards of animal care as outlined in the Guide for the Care and
Use of Experimental Animals of Jilin University (Changchun, China),
following a protocol approved by the Ethics Committees of the
Disease Model Research Center, The First Hospital of Jilin
University. Approximately 6 week-old male BALB/c nude mice were
maintained under specific pathogen-free conditions and provided
with food and water ad libitum.
In vitro cultured A549 cells were harvested
and a tumorigenic dose of 2×106 cells was injected
intraperitoneally into BALB/c mice. Tumor volume was calculated
using the following formula: Tumor volume = length ×
width2 / 2. When tumors grew to an average volume of 75
mm3, mice were randomly divided into the Lv/sh-ADAM9
group, control group (untreated group) and the Lv/sh-NC group (n=10
in each group). The Lv/sh-ADAM9 group and Lv/sh-NC group were
administered with Lv/sh-ADAM9 or Lv/sh-NC plus PBS in a total
volume of 20 μl (10 μl virus plus 10 μl PBS)
once a week for 3 weeks, respectively. The control group were
administered 20 μl PBS once a week for 3 weeks. Following
sacrificing the mice, the tumor weight was measured 21 days after
treatment. Tumor volume was measured prior to administration of the
treatment injections and on days 7, 14 and 21 of treatment. The
primary tumors were measured and western blot analysis was
performed for ADAM9 protein expression. Subsequently, TUNEL
staining was performed on 5 μm sections of the excised
tumors using an ApoTag Red in situ Apoptosis detection kit
(Chemicon International) according to the manufacturer’s
instructions.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Statistical analysis between two samples was performed
using Student’s t-test. Statistical comparison of more than two
groups was performed using one-way analysis of variance followed by
Tukey’s post hoc test. Graphpad Prism 6.0 software (GraphPad
Software, San Diego, CA, USA) and SPSS® 19.0 (SPSS,
Inc., Chicago, IL, USA) for Windows® were used for
statistical analyses. P<0.05 was considered to indicate a
statistically significant difference.
Results
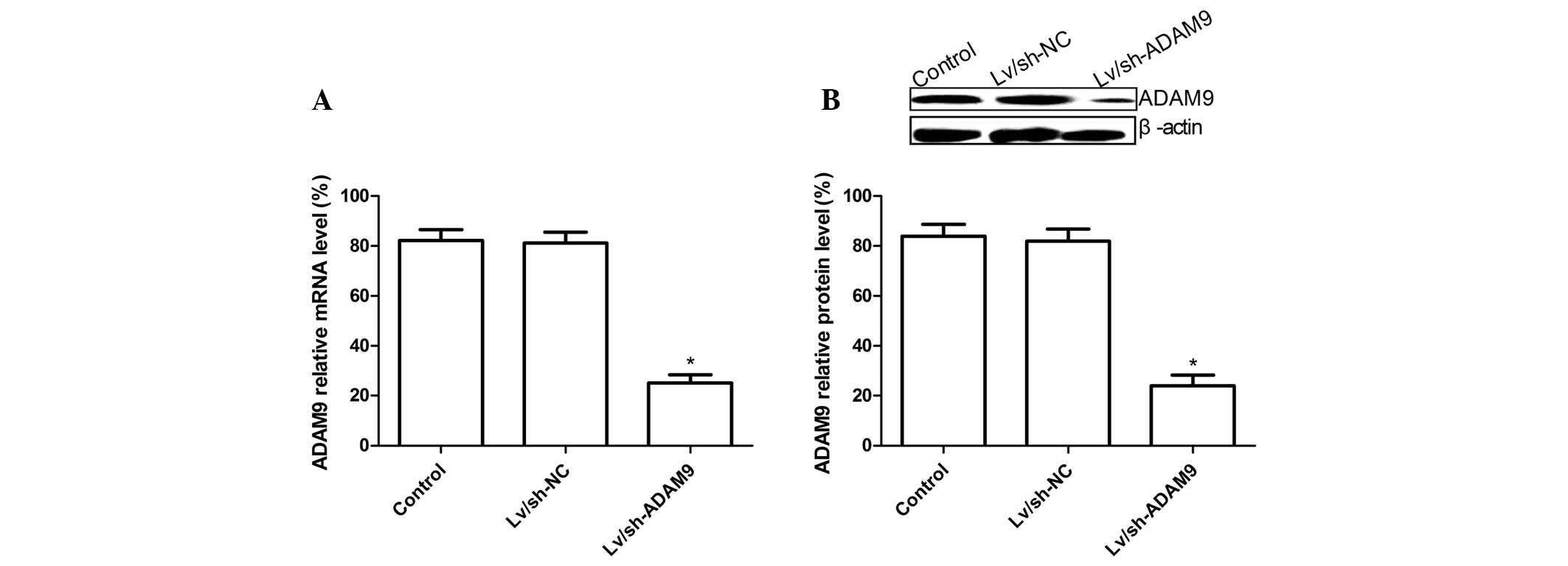
ADAM9 silencing inhibits ADAM9 expression
in A549 cells
The lentivirus carrying the ADAM9 siRNA or negative
control siRNA was infected into A549 cells for 5 days and then
ADAM9 expression at the mRNA and protein levels was
determined by RT-qPCR and western blot analysis, respectively.
RT-qPCR analysis demonstrated that the expression level of ADAM9
mRNA in Lv/sh-ADAM9-infected A549 cells was decreased compared with
that in untreated cells or Ad/sh-NC-infected cells (P<0.05;
Fig. 1A). At the protein level,
similar results were obtained by western blotting using anti-ADAM9
antibody (P<0.05; Fig. 1B).
These data suggested that lentivirus-mediated siRNA targeting ADAM9
could specifically and significantly inhibit the gene expression of
ADAM9 in A549 cells.
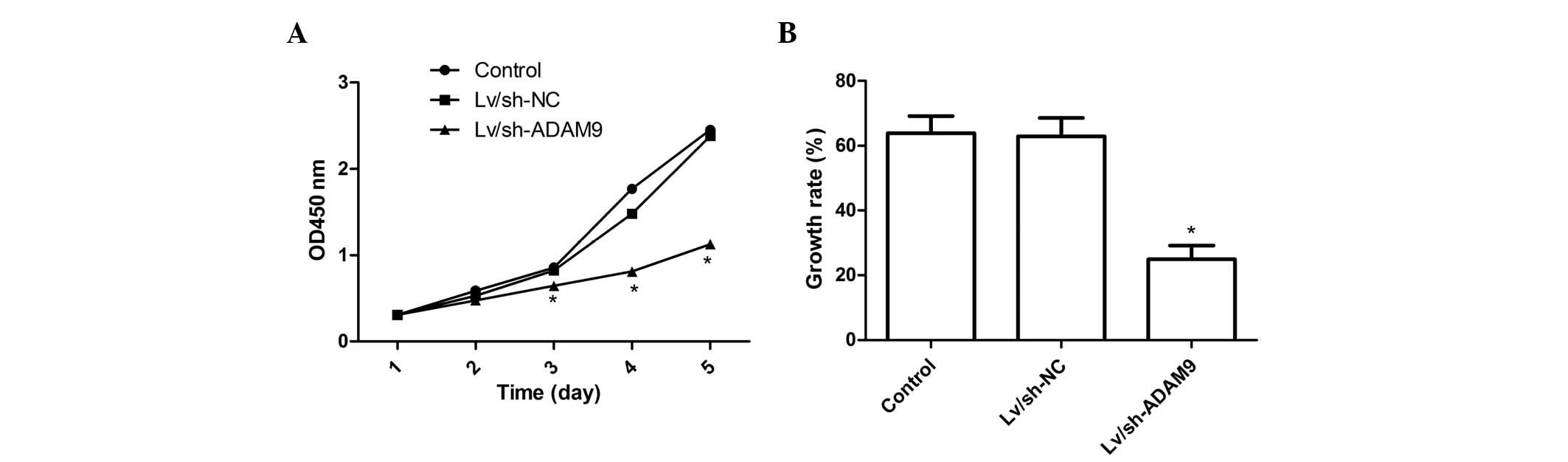
ADAM9 silencing inhibits cell
proliferation in A549 cells
To further assess the role of ADAM9 in regulating
A549 cell proliferation, CCK-8 assays were performed on A549 cells
following adenovirus infection for 5 days. Fig. 2A shows that no statistically
significant differences were observed in cell viability between
uninfected cells and cells infected with Lv/sh-NC, indicating that
the adenoviral system itself had no cytotoxic effect on cells,
while the viability of A549 cells was markedly inhibited by ADAM9
silencing (P<0.05, compared with the control). The inhibitory
effect of Lv/sh-ADAM9 on cell proliferation can be observed
beginning on day 2 and became more prominent on days 4 and 5
(P<0.05; Fig. 2A). In addition,
BrdU incorporation assays also demonstrated that the inhibition of
ADAM9 expression significantly reduced the growth rate of A549
cells during the 48 h incubation period (P<0.05; Fig. 2B). These findings suggest that
ADAM9 silencing markedly decreased the proliferative ability of
A549 cells.
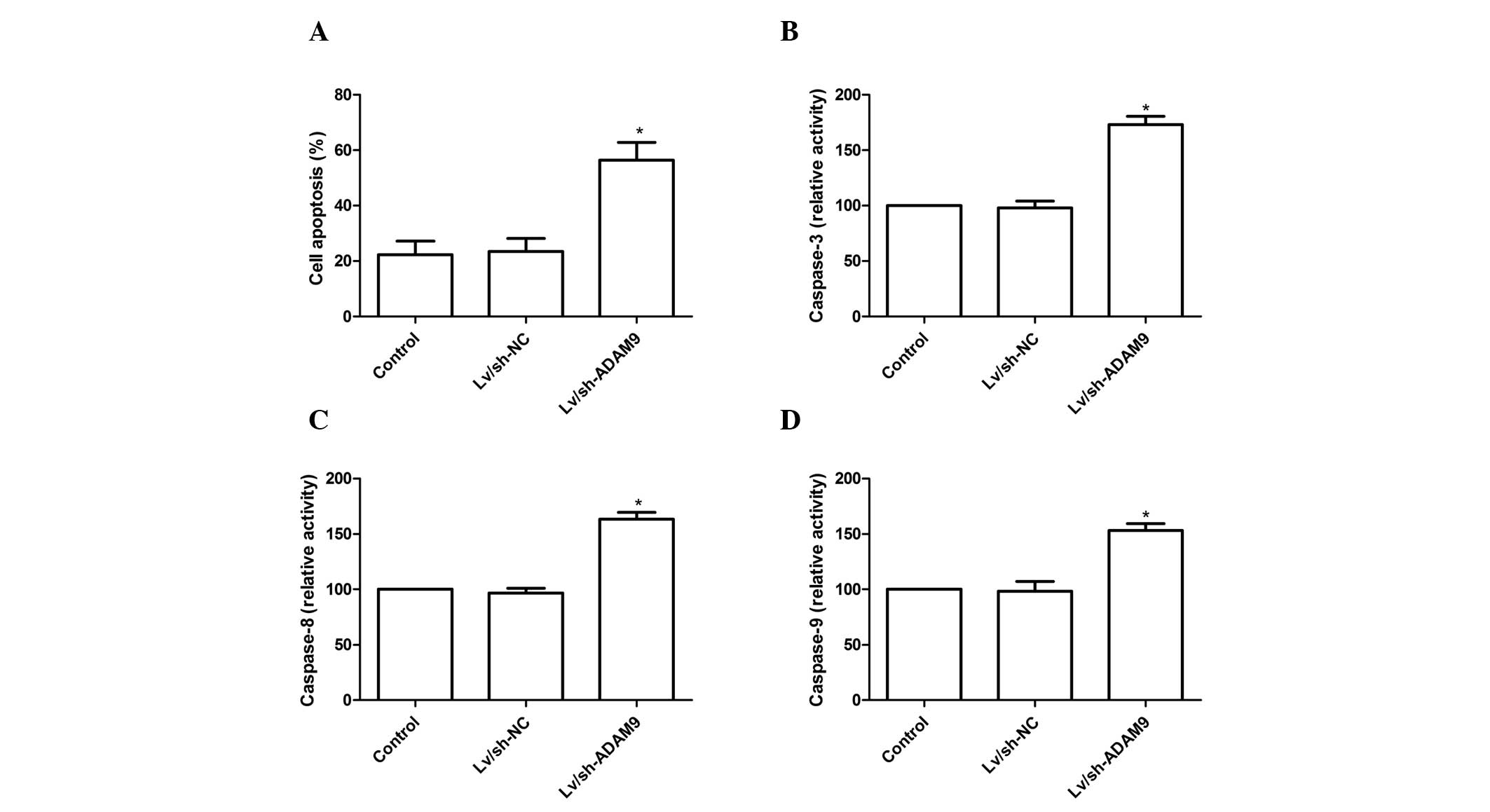
ADAM9 silencing induces cell apoptosis in
A549 cells
To investigate the effect of ADAM9 silencing on cell
apoptosis in A549 cells, a TUNEL assay was performed. Compared with
the control group and Lv/sh-NC group, cell apoptosis was
significantly increased in the Lv/sh-ADAM9 group (P<0.05;
Fig. 3A). Subsequently, the
effects of ADAM9 silencing on caspase-3, caspase-8 and caspase-9
activity were analyzed in A549 cells. As shown in Fig. 3B–D, caspase-3, caspase-8 and
caspase-9 activity in the Lv/sh-ADAM9 treatment group significantly
increased compared with that in the control group or Lv/sh-NC group
(P<0.05). These results suggest that ADAM9 silencing can induce
cell apoptosis in A549 cells.
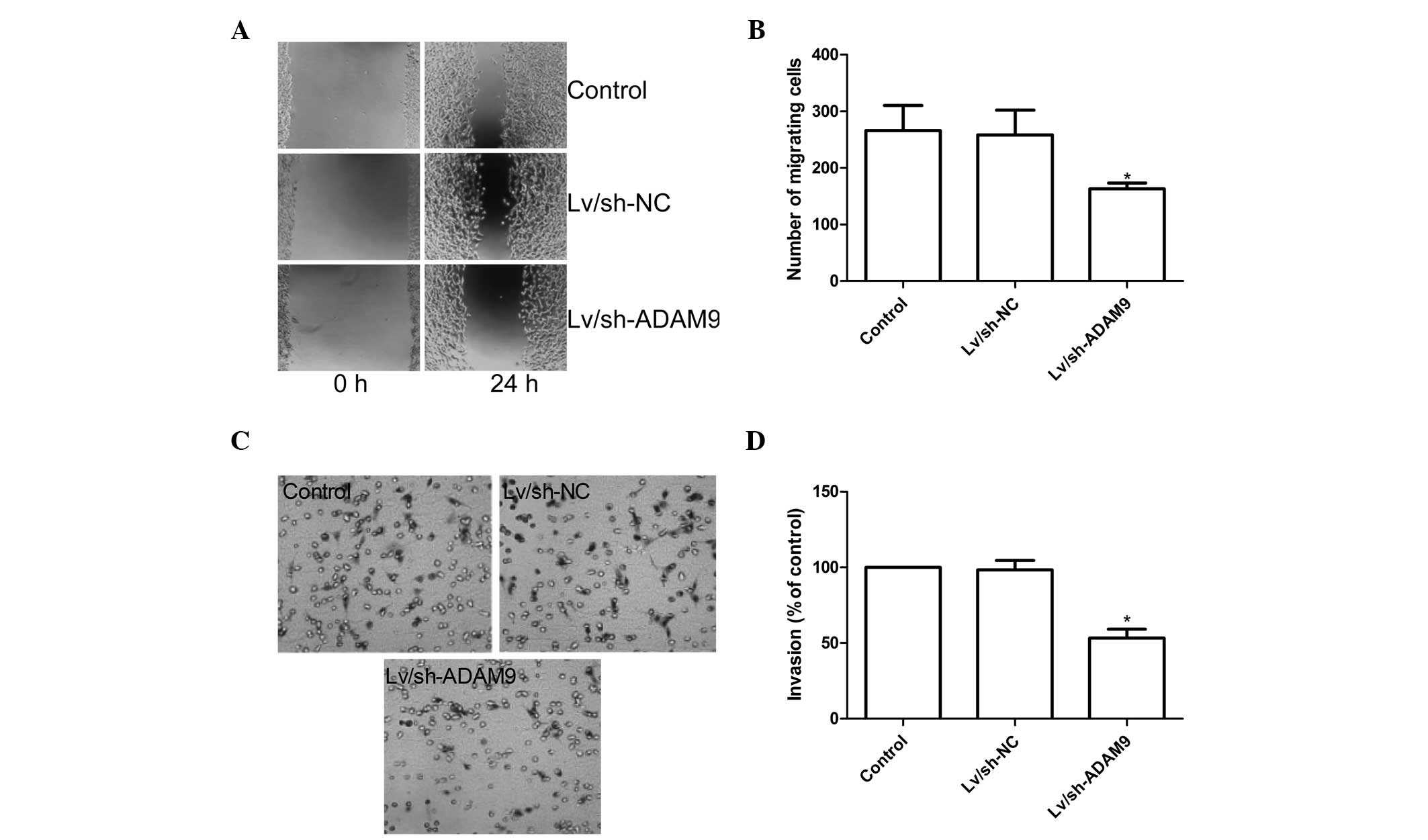
ADAM9 silencing inhibits cell migration
and invasion in A549 cells
To ascertain the inhibitory effect of ADAM9
silencing on A549 cell motility in vitro, a wound-healing
assay was performed to investigate the effects on the migration
potential of A549 cells. A scratch was introduced into confluent
monolayers of different treatment groups and the time-dependent
movement of cells into the injured area was monitored
microscopically. Cells in the control and Lv/sh-NC groups began
migrating 8 h after scratching. After 24 h, cells in the
Lv/sh-ADAM9 group migrated significantly less than those of the
control and Lv/sh-NC groups (P<0.05; Fig. 4A and B).
The ability of ADAM9 silencing to reduce the
invasiveness of A549 cells was further investigated by the
transwell system assay. The results demonstrated that invasion was
also decreased significantly in the Lv/sh-ADAM9 treatment group
compared with the control and the Lv/sh-NC groups (P<0.05;
Fig. 4C and D).
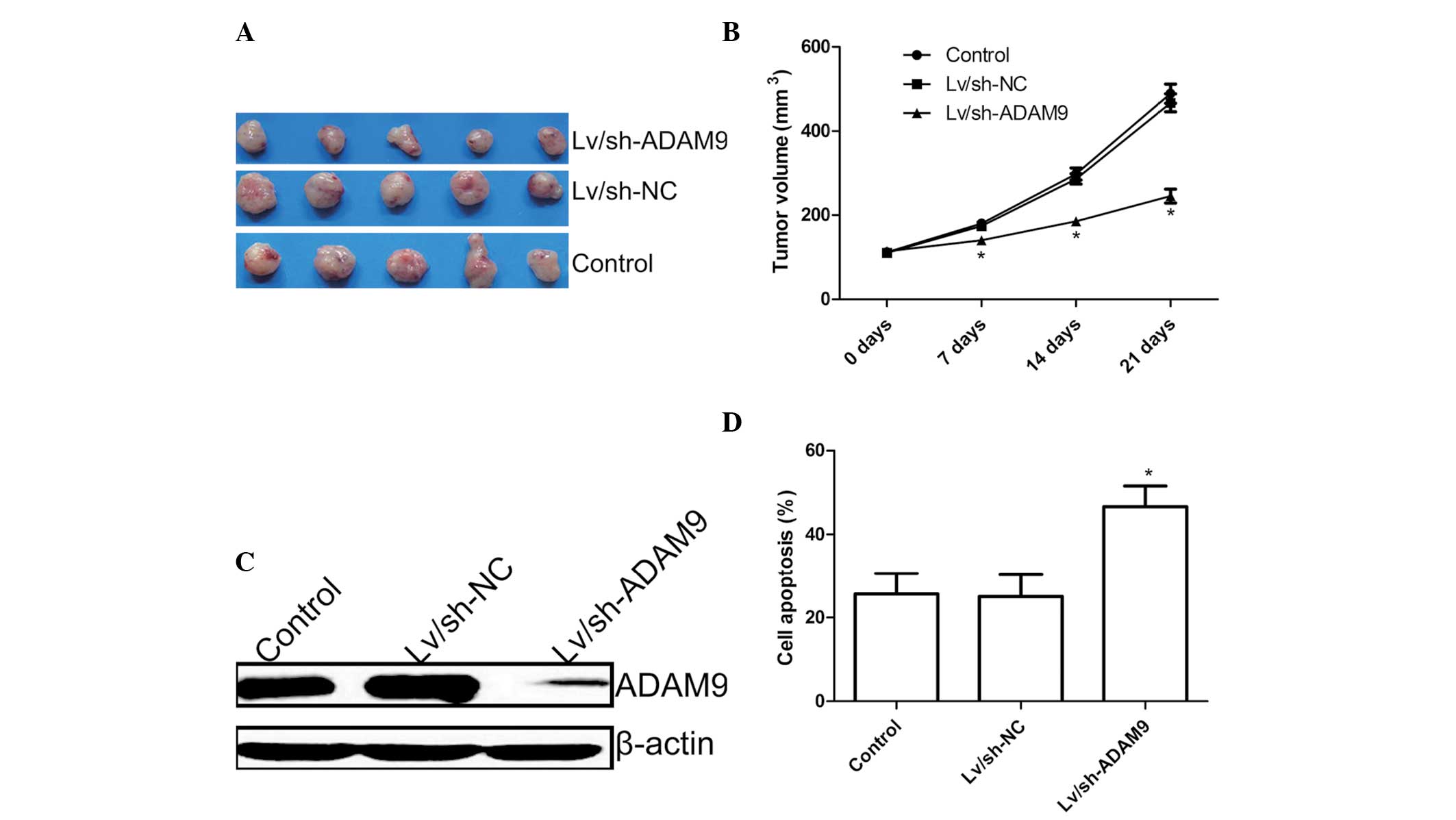
ADAM9 silencing suppresses tumor growth
in vivo
The in vivo therapeutic efficacy of
downregulation of ADAM9 was assessed in male BALB/c mice bearing
A549 tumor cells. Tumor growth was monitored for 21 days. At 1 day
after the end of treatment, animals were sacrificed and final tumor
weights and tumor volume were determined. It was found that the
tumor weight of the Lv/sh-ADAM9 group was significantly lower than
those of the control group and Lv/sh-NC group (Fig. 5A). In addition, the present study
also found that tumor volume following infection with Lv/sh-ADAM9
was significantly lower compared with the control group and
Lv/sh-NC group (P<0.05; Fig.
5B).
In addition, in the present study, the expression of
ADAM9 in grafted tumor tissues was also examined by western blot
analysis. It was found that ADAM9 protein expression levels were
decreased in the Lv/sh-ADAM9 treatment group compared with the
control group and Lv/sh-NC group (Fig.
5C).
The ADAM9 silencing effects on tumor tissue cell
apoptosis were also determined in vivo by TUNEL. The results
demonstrated that ADAM9 silencing could significantly induce cell
apoptosis compared with the control group and Lv/sh-NC group
(P<0.05; Fig. 5D). These
results indicate that ADAM9 silencing could suppress the tumor
growth of NSCLC in a mouse model.
Discussion
ADAMs are membrane-anchored enzymes with critical
roles in the proteolytic processing of numerous membrane-bound
molecules that include growth factors, receptors and adhesion
molecules, which affect multiple cellular processes, including cell
proliferation, differentiation, migration and invasion (26). Due to their ability to rapidly
affect signaling activities between cells and their environment,
ADAMs have been involved in a range of human diseases, including
cancer (27,28) and represent attractive therapeutic
targets for the treatment of various types of cancer. ADAM9 an
important member of the ADAM family, is frequently upregulated in
various types of cancer, including NSCLC and is involved in cancer
progression and metastasis (14–21).
In line with this opinion, our results demonstrated that
downregulation of ADAM9 by RNAi inhibited A549 cell proliferation,
migration and invasion in vitro. The results further
demonstrated that ADAM9 is important in tumor progression.
Although ADAM9 has not been identified as the
principal sheddase of any given substrate so far, it is an active
proteinase capable of shedding various molecules with important
roles in cancer development and angiogenesis. For example, Peduto
et al reported that ADAM9 promotes the proliferation of
prostate cancer cells through hydrolyzing and releasing ligands for
the epidermal growth factor receptor and fibroblast growth factor
receptor on the surface of exfoliative cells (29). Furthermore, Shintani et al
suggested that ADAM9 may enhance the adhesion and invasive
abilities of tumor cells through its regulation of cell adhesion
molecules and altering the sensitivity of tumor cells to growth
factors, thereby promoting tumor cell metastasis to the brain
(22). A previous study
demonstrated that downregulation of ADAM9 by RNAi in prostate
cancer cells results in the impairment of proliferation and the
osteolytic reaction, as well as causing G1/G0 cell cycle arrest.
The G1-to-S phase negative regulators p21Cip1/WAF1 and
p27Kip1 acted as downstream targets of the ADAM9-REG4
signaling pathway (30). Hamada
et al demonstrated that re-expression of miR-126 and
siRNA-based knockdown of ADAM9 in pancreatic cancer cells resulted
in reduced cellular migration, invasion and induction of an
epithelial marker, and that the miR-126/ADAM9 axis is important in
the inhibition of invasive growth of pancreatic cancer cells
(31). The present study
demonstrated that ADAM9 silencing significantly inhibited the
migration and invasion capacity of A549 human lung cancer cells.
These studies, in combination with the results reported in the
present study, support the possibility that downregulating ADAM9
may contribute to the inhibition of cancer cell invasion and
metastasis.
Previously, the successful use of siRNA in
downregulating gene expression in several model systems has led to
numerous attempts to examine this methodology in a potentially
therapeutic setting and this method has been widely used to
investigate gene function for cancer therapy (32,33).
To examine the possibility of ADAM9 as an effective therapeutic
target, endogenous ADAM9 expression was silenced using
lentivirus-mediated siRNA targeting the ADAM9 gene in A549
cells and analyzed its anti-cancer effect. Our results demonstrated
that downregulation of ADAM9 expression by RNAi inhibits
proliferation, migration and invasion, induces apoptosis and
decreases tumor growth in a nude mouse model.
In conclusion, in the present study, the results
demonstrated that downregulation of ADAM9 expression using
lentivirus-mediated siRNA targeting ADAM9 significantly inhibited
cell proliferation, migration and cell invasion, induced cell
apoptosis in vitro and suppressed tumor growth in a nude
mouse model. Considering the significance of cell invasion in
metastatic progression, ADAM9 is thus a potential therapeutic
target for the treatment of NSCLC.
Acknowledgments
This study was supported by the Science and
Technology Research and innovation team funded by Jilin province
(grant no. JL2013528).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reungwetwattana T, Weroha SJ and Molina
JR: Oncogenic pathways, molecularly targeted therapies and
highlighted clinical trials in non-small-cell lung cancer (NSCLC).
Clin Lung Cancer. 13:252–266. 2012. View Article : Google Scholar
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayama M, Chida M, Karube Y, et al:
One-step nucleic acid amplification for detection of lymph node
metastasis in lung cancer. Ann Thorac Cardiovasc Surg. 20:181–184.
2014. View Article : Google Scholar
|
|
6
|
Coleman RE: Clinical features of
metastatic bone disease and risk of skeletal morbidity. Clin Cancer
Res. 12:6243–6249. 2006. View Article : Google Scholar
|
|
7
|
Sun DS, Hu LK, Cai Y, et al: A systematic
review of risk factors for brain metastases and value of
prophylactic cranial irradiation in non-small cell lung cancer.
Asian Pac J Cancer Prev. 15:1233–1239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloproteases: multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roy M, Luo YH, Ye M and Liu J: Nonsmall
cell lung cancer therapy: insight into multitargeted small-molecule
growth factor receptor inhibitors. Biomed Res Int. 2013:9647432013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Becherer JD and Blobel CP: Biochemical
properties and functions of membrane-anchored
metalloprotease-disintegrin proteins (ADAMs). Curr Top Dev Biol.
54:101–123. 2003.PubMed/NCBI
|
|
12
|
Duffy MJ, McKiernan E, O’Donovan N and
McGowan PM: Role of ADAMs in cancer formation and progression. Clin
Cancer Res. 15:1140–1144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peduto L: ADAM9 as a potential target
molecule in cancer. Curr Pharm Des. 15:2282–2287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O’Shea C, McKie N, Buggy Y, Duggan C, Hill
AD, McDermott E, O’Higgins N and Duffy MJ: Expression of ADAM-9
mRNA and protein in human breast cancer. Int J Cancer. 105:754–761.
2003. View Article : Google Scholar
|
|
15
|
Tannapfel A, Anhalt K, Häusermann P,
Sommerer F, Benicke M, Uhlmann D, Witzigmann H, Hauss J and
Wittekind C: Identification of novel proteins associated with
hepatocellular carcinomas using protein microarrays. J Pathol.
201:238–249. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carl-McGrath S, Lendeckel U, Ebert M,
Roessner A and Röcken C: The disintegrin-metalloproteinases ADAM9,
ADAM12 and ADAM15 are upregulated in gastric cancer. Int J Oncol.
26:17–24. 2005.
|
|
17
|
Grützmann R, Lüttges J, Sipos B, Ammerpohl
O, Dobrowolski F, Alldinger I, Kersting S, Ockert D, Koch R,
Kalthoff H, Schackert HK, Saeger HD, et al: ADAM9 expression in
pancreatic cancer is associated with tumour type and is a
prognostic factor in ductal adenocarcinoma. Br J Cancer.
90:1053–1058. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fritzsche FR, Jung M, Tölle A, Wild P,
Hartmann A, Wassermann K, Rabien A, Lein M, Dietel M, Pilarsky C,
Calvano D, Grützmann R, et al: ADAM9 expression is a significant
and independent prognostic marker of PSA relapse in prostate
cancer. Eur Urol. 54:1097–1106. 2008. View Article : Google Scholar
|
|
19
|
Fritzsche FR, Wassermann K, Jung M, Tölle
A, Kristiansen I, Lein M, Johannsen M, Dietel M, Jung K and
Kristiansen G: ADAM9 is highly expressed in renal cell cancer and
is associated with tumour progression. BMC Cancer. 8:1792008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zubel A, Flechtenmacher C, Edler L and
Alonso A: Expression of ADAM9 in CIN3 lesions and squamous cell
carcinomas of the cervix. Gynecol Oncol. 114:332–336. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Qi J, Chen N, Fu W, Zhou B and He
A: High expression of a disintegrin and metalloproteinase-9
predicts a shortened survival time in completely resected stage I
non-small cell lung cancer. Oncol Lett. 5:1461–1466.
2013.PubMed/NCBI
|
|
22
|
Shintani Y, Higashiyama S, Ohta M,
Hirabayashi H, Yamamoto S, Yoshimasu T, Matsuda H and Matsuura N:
Overexpression of ADAM9 in non-small cell lung cancer correlates
with brain metastasis. Cancer Res. 64:4190–4196. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Q, Liu X, Cai Y, Yu Y and Chen W:
RNAi-mediated ADAM9 gene silencing inhibits metastasis of adenoid
cystic carcinoma cells. Tumour Biol. 31:217–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Micocci KC, Martin AC, Montenegro Cde F,
et al: ADAM9 silencing inhibits breast tumor cell invasion in
vitro. Biochimie. 95:1371–1378. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin Y, Peng S and Yu H: RNAi-mediated
downregulation of NOB1 suppresses the growth and colony-formation
ability of human ovarian cancer cells. Med Oncol. 29:311–317. 2012.
View Article : Google Scholar
|
|
26
|
Klein T and Bischoff R: Active
metalloproteases of the A Disintegrin and Metalloprotease (ADAM)
family: biological function and structure. J Proteome Res.
10:17–33. 2011. View Article : Google Scholar
|
|
27
|
Murphy G: The ADAMs: signalling scissors
in the tumour microenvironment. Nat Rev Cancer. 8:929–941. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rocks N, Paulissen G, El Hour M, et al:
Emerging roles of ADAM and ADAMTS metalloproteinases in cancer.
Biochimie. 90:369–379. 2008. View Article : Google Scholar
|
|
29
|
Peduto L, Reuter VE, Shaffer DR, Scher HI
and Blobel CP: Critical function for ADAM9 in mouse prostate
cancer. Cancer Res. 65:9312–9319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu CM, Hsieh CL, He YC, et al: In vivo
targeting of ADAM9 gene expression using lentivirus-delivered shRNA
suppresses prostate cancer growth by regulating REG4 dependent cell
cycle progression. PLoS One. 8:e537952013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hamada S, Satoh K, Fujibuchi W, et al:
MiR-126 acts as a tumor suppressor in pancreatic cancer cells via
the regulation of ADAM9. Mol Cancer Res. 10:3–10. 2012. View Article : Google Scholar
|
|
32
|
Zhang J and Hua ZC: Targeted gene
silencing by small interfering RNA-based knock-down technology.
Curr Pharm Biotechnol. 5:1–7. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Uprichard SL: The therapeutic potential of
RNA interference. FEBS Lett. 579:5996–6007. 2005. View Article : Google Scholar : PubMed/NCBI
|