Introduction
Myocarditis is an inflammation of the heart muscle
that is most often due to enteroviral infection. Coxsackievirus B3
(CVB3) is hypothesized to be the most common causative agent
underlying myocarditis in humans. Viral myocarditis (VM) affects
5–20% of the human population, and is potentially fatal among
infants as well as children (1,2).
Although VM is frequently resolved among older individuals, it may
develop into chronic myocarditis and/or dilated cardiomyopathy,
leading to cardiac failure (3–5). The
pathogenesis of VM is based on adverse immune responses evoked by
infection of the cardiac muscle with cardiotropic viruses, which
leads to viral elimination as well as cardiac myocyte destruction,
reparative fibrosis and heart failure (6). Due to the current dearth of effective
therapies for the treatment of myocarditis, further studies of the
molecular mechanisms underlying these autodestructive inflammatory
signaling pathways within the immune system are required (7).
MicroRNAs (miRNAs) are short, noncoding RNA
sequences that function as key gene regulators, modulating gene
expression at the post-transcriptional level by targeting the
3′-untranslated region (3′UTR) of messenger RNA sequences (8). Gene expression studies have
demonstrated that miRNAs are differentially expressed in heart
disease, and loss-of-function studies in mice have confirmed that
miRNAs are able to regulate multiple cell processes essential to
heart function (9,10). miRNAs have significant functions in
the maintenance of normal human physiological conditions, and
abnormal miRNA expression has been associated with numerous human
diseases, varying from psychiatric disorders to types of malignant
cancer (11–13).
Recently, a group reported that disturbed miRNA
expression, including that of miR-155, -21 and -146b, was
associated with VM pathogenesis (10). It has also been hypothesized that
miRNAs, for example miR-208b and miR-499-5p, may be used as novel
biomarkers for VM diagnosis (14).
In the present study, the expression levels of eight candidate
miRNAs which may be involved in the pathogenesis of VM were
evaluated, in order to determine which were significantly altered.
Subsequently, bioinformatics tools were used to predict target
genes of the identified dysregulated miRNAs, and the results were
verified by dual-luciferase assay and western blot analysis. The
biological functions of the identified miRNAs were also studied in
mouse cardiac myocytes. NF-κB signaling was upregulated only by
overexpressed miR-214, but not miR-146a. Thus, miR-214 is
associated with myocardial injury during VM.
Materials and methods
Patients
All human material was obtained from the First
Affiliated Hospital of Xinxiang Medical University (Weihui, China).
Samples were stored at -80°C immediately following collection and
were available for research purposes in accordance with the
Declaration of Helsinki. The study was approved by the ethics
committee of The First Affiliated Hospital of Xinxiang Medical
University (Weihui, China). Informed consent was obtained from the
patients or their families. miRNA expression was evaluated in right
ventricular septal specimens from patients with myocarditis at the
acute stage (n=8), within 3 months after the disease had been
diagnosed, as well as a clinical history of myocarditis and
confirmed viral presence in cardiac biopsies. The control subjects
(n=8) comprised age-matched patients presenting with unexplained
ventricular tachy-arrhythmias, but with normal ejection fractions
and no identified systemic or cardiac inflammation or viral
presence at the time of biopsy.
Cell culture
HeLa cells were cultured in Dulbecco’s modified
Eagle medium supplemented with 10% fetal bovine serum (HyClone,
Thermo Fisher Scientific, Waltham, MA, USA), 100 IU/ml penicillin
and 10 mg/ml streptomycin. Cells were maintained at 37°C in a 5%
CO2 atmosphere.
Purification of neonatal murine cardiac
myocytes
Six specific pathogen free BALB/c mice (8–10 weeks
old) in their first day of pregnancy were were purchased from
Shanghai Laboratory Animal Centre (Chinese Academy of Science,
Shanghai, China). All of the mice were housed under pathogen free
conditions. The mice were maintained in a controlled environment at
a temperature of 25±1°C and relative humidity of 50–60%. The
environment was artificially illuminated (fluorescent lights), and
the mice underwent a 12:12 h light/dark cycle and were fed a
standard chow diet. The neonatal mice (postnatal 0–3 days) were
sacrificed by cervical dislocation. Cardiac myocytes obtained from
neonatal mice were prepared as previously described (15). Briefly, the mouse hearts were
removed, minced finely and stepwise enzymatic digestion was
performed using 0.25% trypsin-0.02% EDTA. The dissociated cells
were subsequently washed with complete basal Eagle’s medium and the
endothe-lial cells and fibroblasts were removed by two sequential 1
h adsorptions to plastic flasks at 37°C. Non-adherent myocytes were
removed, washed once, resuspended in complete basal medium and
plated into tissue culture wells. Following 48 h of incubation, the
myocytes had attached to the plastic tissue culture wells, and
>95% of the cells were identified as cardiac myocytes according
to observations regarding their shape and beating activity. The
cultured cells were subsequently evaluated as described below. The
experiment was approved by the ethical committee of The First
Affiliated Hospital of Xinxiang Medical University.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR analysis was used to examine the relative
expression levels of eight candidate miRNAs. Total RNA was
extracted from the cell or tissue samples with TRIzol (Invitrogen
Life Technologies, Carlsbad, CA, USA), according to the
manufacturer’s instructions. Eight candidate miRNAs (miR-214,
miR-155, miR-146b, miR-208b, miR-21, miR-499, miR-375 and miR-125b)
were selected, which had previously been reported to be associated
with VM pathogenesis (10,14,16,17),
and their expression levels were evaluated using TaqMan miRNA
RT-Real Time PCR (Applied Biosystems, Foster City, CA, USA).
Single-stranded complementary DNA was synthesized using the TaqMan
MicroRNA Reverse Transcription kit (Applied Biosystems) and
subsequently amplified with the TaqMan Universal PCR Master mix
(Applied Biosystems), alongside the miRNA-specific TaqMan MGB
probes (Applied Biosystems). The reaction mixture was incubated at
95°C for 30 sec, followed by 40 cycles of 95°C for 8 sec and 60°C
for 30 sec. The U6 small nuclear RNA was used for normalization.
Each sample, within each group, was evaluated in triplicate and
each experiment was repeated ≥3 times.
Target gene prediction
Potential target genes were searched for using the
online bioinformatics tool TargetScan Release 6.2 (http://www.targetscan.org/) (18). The results suggested that ITCH mRNA
may be directly targeted by miR-214.
Dual luciferase assay
The full length of ITCH 3′UTR was cloned into a pGL3
vector (Promega Corp., Madison, WI, USA), downstream of the firefly
luciferase coding region in order to produce the luciferase
reporter vector. For the luciferase reporter assays, HeLa cells
were seeded into 48-well plates. miRNA mimics, miRNA antagonists or
their corresponding controls and luciferase reporter vectors were
co-transfected using Lipofectamine® 2000 (Invitrogen
Life Technologies). The pRT-TK vector, which expresses
Renilla luciferase was used as a transfection control. Cells
were harvested following two days of cotransfection and evaluated
with the Dual-Luciferase assay (Promega Corp.). Each transfection
was performed in triplicate in three independent experiments. The
results are expressed as relative luciferase activity: (Firefly
luciferase/Renilla luciferase). A reporter vector containing
mutant ITCH 3′UTR was constructed using Fast Mutagenesis system
(Beijing TransGen Biotech Co., Ltd, Beijing, China), to identify
the binding region of miR-214. The binding region sequence of
miR-214 (CCUGCUG) was replaced with CAUACUA. The wild type or
mutant ITCH 3′UTR reporter vector was then transfected into HEK293T
cells (China Infrastruture of Cell Line Resources, Beijing, China)
alongside the miR-214 mimic or inhibitor using lipofectamine 2000.
A total of 48 h post-transfection, the cells were lysed by
incubation in Passive Lysis buffer on ice for 15 min and luciferase
activity was detected using the Dual-Luciferase Assay kit.
Western blot analysis
Protein extracts were boiled in
SDS/β-mercaptoethanol sample buffer, and 20 μg samples were loaded
onto 8% polyacrylamide gels. The proteins were separated by
electrophoresis, and blotted onto poly-vinylidene difluoride
membranes (Amersham Pharmacia Biotech, St. Albans, Herts, UK) via
electrophoretic transfer. The membrane was incubated with rabbit
polyclonal anti-human ITCH antibody (1:500; cat. no. ab31097;
Abcam, Cambridge, MA, USA) and mouse monoclonal anti-human β-actin
antibody (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology
Inc., Dallas, TX, USA) for 1 h at 37°C. The membrane was
subsequently incubated with horseradish peroxidase-conjugated goat
anti-rabbit (1:5,000; cat. no. sc-2004) or goat anti-mouse
(1:10,000; cat. no. sc-2060) secondary antibodies (Santa Cruz
Biotechnology, Inc.) for 1 h at 37°C. Detection was performed via
enhanced chemiluminescence using an ECL kit (Pierce Biotechnology
Inc., Thermo Fisher Scientific, Rockford, IL, USA). β-actin was
used as a loading control.
Homology analysis
The sequences of human and mouse ITCH gene were
downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and analyzed
using CLUSTAL X, version 1.83 (http://www.clustal.org/) (19).
Cytokine assays
The levels of tumor necrosis factor (TNF)-α,
interleukin (IL)-1β, IL-6 and monocyte chemoattractant protein
(MCP)-1 of cell culture supernatants were determined with ELISA
(eBioscience Inc., San Diego, CA, USA) according to the
manufacturer’s instructions.
Statistical analysis
Data were analyzed with SPSS Statistical Package
version 16 (SPSS, Inc., Chicago, IL, USA). Independent two group’s
analyses are used t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-214 and miR-146b are downregulated in
the heart tissue of patients with VM
In the present study, the expression levels of eight
miRNAs, which were previously reported to exhibit altered
expression levels during CVB3 infection, were evaluated in heart
tissue samples from patients with CVB3-infected VM. miR-214 and
miR-146b were revealed to be significantly upregulated (Fig. 1A and B). Using bioinformatics
tools, ITCH, an E3 ubiquitin ligase which functions as an NF-κB
suppressor, was identified as a predicted target gene of miR-214
and miR-146b (Fig. 1C).
The expression of ITCH is suppressed by
miR-214, but not miR-146b, in HeLa cells
To confirm whether the expression of ITCH was
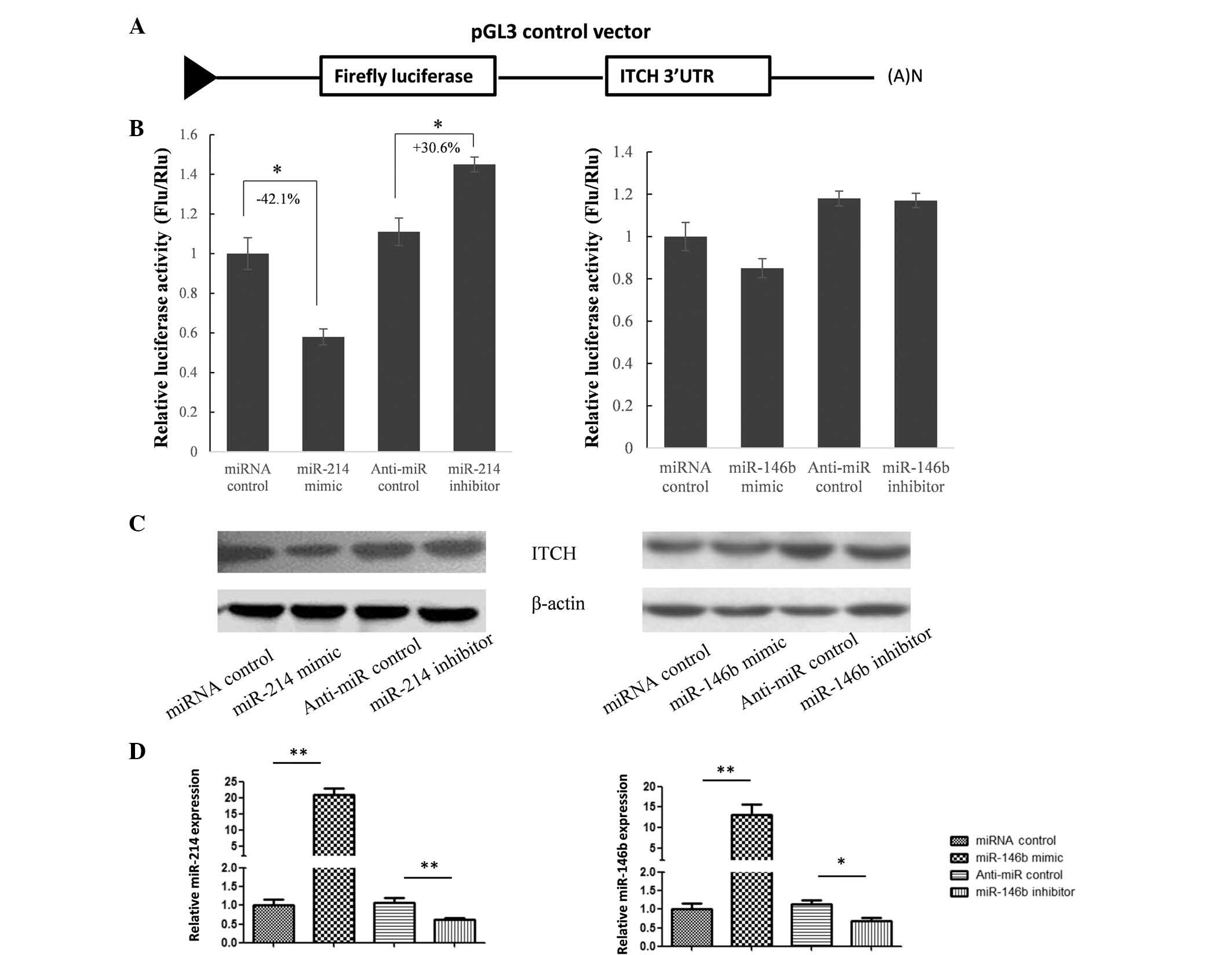
supressed by these miRNAs, the full length (3.6 kb) ITCH 3′UTR was
cloned into a pGL3 vector downstream of the luciferase coding
region (Fig. 2A). The results of a
dual-luciferase assay in HeLa cells indicated that miR-214
significantly supressed firefly luciferase expression by binding to
the 3′UTR of ITCH; however, miR-146b did not significantly
influence luciferase expression levels (Fig. 2B). To further confirm whether
miR-214 repressed ITCH expression, miR-214 was overexpressed and
knocked down in HeLa cells, in which ITCH expression was detectable
(Fig. 2C). Protein expression
levels of ITCH were evaluated by western blot analysis (Fig. 2C), and expression levels of miR-214
and miR-146b were detected by RT-qPCR (Fig. 2D). As shown in Fig. 2B and C, ITCH expression levels were
significantly reduced by the miR-214 mimic and slightly upregulated
by the miR-214 inhibitor, which indicated that ITCH was a target
gene of miR-214.
miR-214 upregulates TNF-α and IL-6
expression by targeting ITCH
The NF-κB family of transcription factors is crucial
in the mediation of inflammatory responses. ITCH is a key component
of the ubiquitin-editing complex required for termination of the
pro-inflammatory activation of the c-Jun N-terminal kinases and
NF-κB. Following transfection with miR-214 mimic and CVB3
infection, ITCH expression levels were markedly downregulated,
compared with those of cells transfected with the miRNA control
(Fig. 3A). To further investigate
the biological functions of miR-214 overexpression, the expression
levels of TNF-α and IL-6 in cell culture supernatants were
examined. As indicated in Fig. 3B,
CVB3 infection induced upregulation of TNF-α and IL-6 levels, and
following transfection with miR-214 mimic, the concentration of
these cytokines was significantly enhanced (P<0.05). Following
cotransfection with ITCH expression vector, the expression levels
of TNF-α and IL-6 were significantly reduced (Fig. 3C), indicating that miR-214 promoted
cell inflammatory responses partially via the suppression of ITCH
expression.
miR-214 upregulates TNF-α, IL-1β, IL-6
and MCP-1 expression by targeting ITCH in murine cardiac
myocytes
To further investigate this overexpression function
in cardiac cells, cardiac myocytes were purified from neonatal
mice. Prior to the functional study, the miR-214 binding site in
ITCH 3′UTR was identified and the sequences of the human and mouse
ITCH 3′UTR were aligned. As exhibited in Fig. 4A, when three nucleotides were
mutated, luciferase activity was no longer repressed by miR-214,
indicating that the miR-214 binding site was CCUGCUG. Furthermore,
this region is highly conserved between human and mouse. Following
infection with CVB3, the expression of ITCH was upregulated in a
time-dependent manner in those cells transfected with the miRNA
control. An analogous effect on ITCH expression was observed in
those cells transfected with miR-214 mimic; however, the ITCH
protein levels were markedly lower in mouse cardiac myocytes
(Fig. 4B). Furthermore, the
concentrations of TNF-α, IL-6, IL-1β and MCP-1 in cell culture
supernatants were upregu-lated by miR-214 mimic transfection and
downregulated by miR-214 antagonist transfection (Fig. 4C).
 | Figure 4miR-214 upregulates TNF-α, IL-1β, IL-6
and MCP-1 expression by targeting ITCH in murine cardiac myocytes.
(A) Homology analysis of miR-214 binding site in human and mouse
ITCH 3′UTR. Dual luciferase assay was used to confirm the location
of the miR-214 binding site in ITCH 3′UTR. (B) Murine cardiac
myocytes were transfected with miR-214 mimic, and 24 h after
transfection, cells were infected with CVB3. ITCH protein levels
were evaluated by western blot analysis at five time-points
following CVB3 infection. (C) Levels of TNF-α, IL-1β, IL-6 and
MCP-1 expression in cell culture supernatants were determined by
enzyme-linked immunosorbent assay. The results of paired groups
were analyzed using t-test. *P<0.05,
**P<0.01. TNF-α, tumor necrosis factor-α; IL,
interleukin; MCP-1, monocyte chemoattractant protein-1; 3′UTR, 3′
untranslated region; CVB3, coxsackievirus B3; WT, wild-type; MU,
mutant. |
Discussion
Myocarditis most frequently occurs as a result of
infection by enteroviruses, and CVB3 is hypothesized to be the most
common causative factor in human myocarditis (6). Despite decades of extensive research,
the pathogenesis of VM has remained elusive, and there is currently
no effective therapy available for the treatment of this disease
(20). Experimental studies have
revealed that although CVB3 is able to directly destroy the
myocardium (21,22), it is the associated aberrant
inflammatory response that is primarily responsible for the
induction of myocyte damage (23,24).
Additionally, clinical studies have identified enhanced levels of
circulating TNF-α, IL-1β, IL-6 and other pro-inflammatory cytokines
in patients with myocarditis (25,26).
Furthermore, specific immunosup-pressive agents may be used to
control the inflammatory response in clinical therapies (27). Therefore, modulation of the
inflammatory response represents a potential therapeutic strategy
for the treatment of viral myocarditis.
The expression of miRNAs, which function as key gene
regulators, was found to be disturbed during CVB3 infection, and
certain miRNAs were revealed to have promotional and/or inhibitory
roles in the mediation of CVB3 infection. In the present study,
eight candidate miRNAs, the functions of which are associated with
myocarditis, were selected (10,14,28,29),
and their expression was evaluated by RT-qPCR. The results revealed
that miR-146b and miR-214 expression levels were significantly
upregulated in VM tissues compared with those of the control
tissues. Online bioinformatics tools were used to predict ITCH, an
NF-κB signaling suppressor, as a target gene of miR-214. This
prediction was verified by dual-luciferase assay and western blot
analysis. To investigate the biological function of miR-214, TNF-α
and IL-6 expression levels were evaluated in HeLa cell culture
supernatants. It was demonstrated that the expression levels of
TNF-α and IL-6 were upregulated by miR-214 mimic transfection.
There was no significant difference in TNF-α or IL-6 expression
between the miR-214 knockdown group and that of the control group,
although this may have been a result of the low expression levels
of miR-214 in HeLa cells. This hypothesis was indirectly confirmed
by the experiments performed in murine cardiac myocytes, where the
expression of cytokines was significantly reduced by transfection
with the miR-214 antagonist.
In conclusion, to the best of our knowledge, the
present study was the first to report that the upregulation of
miR-214 expression was associated with VM pathogenesis by targeting
ITCH. Overexpression of miR-214 upregulated the expression of
cytokines and chemokines that enhance myocardial inflammation by
weakening NF-κB pathway feedback signaling.
References
|
1
|
Duncan BW, Bohn DJ, Atz AM, French JW,
Laussen PC and Wessel DL: Mechanical circulatory support for the
treatment of children with acute fulminant myocarditis. J Thorac
Cardiovasc Surg. 122:440–448. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woodruff JF: Viral myocarditis. A review.
Am J Pathol. 101:425–484. 1980.PubMed/NCBI
|
|
3
|
Bowles NE, Richardson PJ, Olsen EG and
Archard LC: Detection of Coxsackie-B-virus-specific RNA sequences
in myocardial biopsy samples from patients with myocarditis and
dilated cardiomyopathy. Lancet. 1:1120–1123. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kandolf R, Klingel K, Mertsching H, et al:
Molecular studies on enteroviral heart disease: patterns of acute
and persistent infections. Eur Heart J. 12(Suppl D): 49–55. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morimoto S, Hiramitsu S, Yamada K, et al:
Clinical and pathologic features of chronic myocarditis: four
autopsy cases presenting as dilated cardiomyopathy in life. Am J
Cardiovasc Pathol. 4:181–191. 1992.PubMed/NCBI
|
|
6
|
Maier R, Krebs P and Ludewig B:
Immunopathological basis of virus-induced myocarditis. Clin Dev
Immunol. 11:1–5. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shauer A, Gotsman I, Keren A, et al: Acute
viral myocarditis: current concepts in diagnosis and treatment. Isr
Med Assoc J. 15:180–185. 2013.PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Rosa S, Curcio A and Indolfi C:
Emerging role of microRNAs in cardiovascular diseases. Circ J.
78:567–575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Corsten MF, Papageorgiou A, Verhesen W, et
al: MicroRNA profiling identifies microRNA-155 as an adverse
mediator of cardiac injury and dysfunction during acute viral
myocarditis. Circ Res. 111:415–425. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: implications for alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu J, Li Y, Wang F, et al: Suppressed
miR-424 expression via upregulation of target gene Chk1 contributes
to the progression of cervical cancer. Oncogene. 32:976–987. 2013.
View Article : Google Scholar
|
|
13
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20.
2013.PubMed/NCBI
|
|
14
|
Corsten MF, Dennert R, Jochems S, et al:
Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial
damage in cardiovascular disease. Circ Cardiovasc Genet. 3:499–506.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen Y, Xu W, Chu YW, Wang Y, Liu QS and
Xiong SD: Coxsackievirus group B type 3 infection upregulates
expression of monocyte chemoattractant protein 1 in cardiac
myocytes, which leads to enhanced migration of mononuclear cells in
viral myocarditis. J Virol. 78:12548–12556. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu YL, Wu W, Xue Y, et al: MicroRNA-21
and -146b are involved in the pathogenesis of murine viral
myocarditis by regulating TH-17 differentiation. Arch Virol.
158:1953–1963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tijsen AJ, Pinto YM and Creemers EE:
Circulating microRNAs as diagnostic biomarkers for cardiovascular
diseases. Am J Physiol Heart Circ Physiol. 303:H1085–H1095. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thompson JD, Gibson TJ, Plewniak F,
Jeanmougin F and Higgins DG: The CLUSTAL_X windows interface:
Flexible strategies for multiple sequence alignment aided by
quality analysis tools. Nucleic Acids Res. 25:4876–4882. 1997.
View Article : Google Scholar
|
|
20
|
Esfandiarei M and McManus BM: Molecular
biology and pathogenesis of viral myocarditis. Annu Rev Pathol.
3:127–155. 2008. View Article : Google Scholar
|
|
21
|
Klingel K and Kandolf R: The role of
enterovirus replication in the development of acute and chronic
heart muscle disease in different immunocompetent mouse strains.
Scand J Infect Dis Suppl. 88:79–85. 1993.PubMed/NCBI
|
|
22
|
Fuse K, Chan G, Liu Y, et al: Myeloid
differentiation factor-88 plays a crucial role in the pathogenesis
of Coxsackievirus B3-induced myocarditis and influences type I
interferon production. Circulation. 112:2276–2285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leipner C, Grün K, Borchers M and Stelzner
A: The outcome of coxsackievirus B3-(CVB3-) induced myocarditis is
influenced by the cellular immune status. Herz. 25:245–248. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Calabrese F and Thiene G: Myocarditis and
inflammatory cardiomyopathy: microbiological and molecular
biological aspects. Cardiovasc Res. 60:11–25. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matsumori A, Yamada T, Suzuki H, Matoba Y
and Sasayama S: Increased circulating cytokines in patients with
myocarditis and cardiomyopathy. Br Heart J. 72:561–566. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levine B, Kalman J, Mayer L, Fillit HM and
Packer M: Elevated circulating levels of tumor necrosis factor in
severe chronic heart failure. N Engl J Med. 323:236–241. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schultz JC, Hilliard AA, Cooper LT Jr and
Rihal CS: Diagnosis and treatment of viral myocarditis. Mayo Clin
Proc. 84:1001–1009. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu HF, Ding YJ, Shen YW, et al: MicroRNA-1
represses Cx43 expression in viral myocarditis. Mol Cell Biochem.
362:141–148. 2012. View Article : Google Scholar
|
|
29
|
Hemida MG, Ye X, Zhang HM, et al:
MicroRNA-203 enhances coxsackievirus B3 replication through
targeting zinc finger protein-148. Cell Mol Life Sci. 70:277–291.
2013. View Article : Google Scholar
|