Introduction
Malignant colorectal cancer or carcinoma (CRC)
originates from the epithelial cells of the colon or rectum. CRC is
associated with a high risk of cancer morbidity and mortality and a
previous genomic analysis demonstrated that colorectal and rectal
cancers have considerably similar patterns (1). CRC is responsible for ~500,000 deaths
annually (2), and in developed
countries, one out of three cases of CRC are fatal (3). Genetic studies have resolved the
regulation of cellular metabolism, proliferation, differentiation
and survival in CRC (4). However,
further research is required to fully understand the molecular
changes associated with the pathophysiology of CRC. The effector
molecules and signal transduction pathways responsible for the
development of CRC remain to be elucidated.
Previous studies have indicated that conserved
intra-membrane proteolytic mechanisms are associated with the
regulation of cellular processes, including transcriptional
control, growth factor secretion and apoptosis (5,6). It
has been well demonstrated that intramembrane proteases are
involved in critical cellular processes, including apoptosis and
signaling transduction (6–10). The rhomboid serine proteases are
expressed in most species, and have been observed in both bacteria
and humans (11), they are
considered to be one of the most-widely conserved membrane
proteases (12). The first
rhomboid serine protease was initially identified in a Drosophila
genetics study, where it was shown to activate the upstream
epidermal growth factor receptor (EGFR) signaling pathway (13). Previous research has also
demonstrated that numerous yeast rhomboid proteases have a role in
mitochondrial membrane remodeling (14).
Rhomboid domain containing 1 (RHBDD1), a mammalian
rhomboid protease highly expressed in testis, has previously been
identified as a pro-apoptotic member of the B-cell lymphoma 2
family (15). RHBDD1 has been
shown to be highly expressed in chronic myeloid leukemia patients,
as compared with healthy controls (16). RHBDD1 exhibited a proteolytic
activity in the tumor suppressor activated pathway-6, in both
HCT116 and RKO colon cancer cells (15). Downregulation of RHBDD1 also
demonstrated the ability to suppress proliferation and colony
formation capability of HepG2 hepatocellular carcinoma cells
(17). However, the precise
function of RHBDD1 in CRC progression remains unclear. In the
present study, RHBDD1 was confirmed to be highly expressed in
numerous CRC cell lines. To determine the role of RHBDD1 in human
CRC, a lentivirus-mediated short hairpin RNA (shRNA) was used to
knockdown RHBDD1 expression in RKO CRC cells. The effects of RHBDD1
expression knockdown on CRC cell growth were then investigated.
Materials and methods
Cell Culture
SW480, SW620, RKO, DLD-1, HCT116 and HT-29 human CRC
cell lines and HEK293T human embryonic kidney cells were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). SW480, SW620, RKO and DLD-1 cells were cultured in
RPMI-1640, supplemented with 10% fetal bovine serum (FBS; HyClone
Laboratories Inc., Logan, UT, USA). HCT116 and HT-29 cells were
cultured in McCoy’s 5A media, supplemented with 10% FBS
(Sigma-Aldrich, St. Louis, MO, USA). HEK293T cells were cultured in
Dulbecco’s modified Eagle’s medium, supplemented with 10% FBS
(HyClone Laboratories Inc.).
Construction of recombinant
lentivirus
The small interfering RNA (siRNA) sequence for
RHBDD1 (NM_032276) (5′-GCTGGGATTCTTGTTGGACTA-3′) was screened and
validated, to confirm its use as the candidate siRNA. A
non-silencing siRNA sequence (5′-CCAAGGAAGTGCAATTGCATA-3′) was used
as a control. shRNAs corresponding to both the RHBDD1 and control
siRNA sequences, were synthesized as 21-nt inverse repeats,
separated by a 9-nt loop for each sequence and inserted downstream
of the U6 promoter in the pFH-L lentiviral vector (Shanghai
Hollybio Co., Ltd., Shanghai, China). The lentiviruses were
generated by triple transfection of 80% confluent HEK293T cells,
with modified pFH-L plasmid, and helper vectors pVSVG-I and
pCMVΔR8.92 (Shanghai Hollybio Co., Ltd.), using
Lipofectamine® 2000 (Invitrogen Life Technologies,
Carlsbad, CA, USA). The lentiviruses were harvested in serum-free
medium after three days, filtered and concentrated using primed
Centricon® Plus-20 filter devices (Millipore, Billerica,
MA, USA).
RNA extraction and quantitative
polymerase chain reaction (qPCR)
RKO cells were pre-cultured and infected with the
recombinant lentivirus for five days. Total RNA was extracted using
TRIzol® reagent (Gibco Life Technologies, Carlsbad, CA,
USA), according to the manufacturer’s instructions. Total RNA (5
mg) was reverse transcribed to produce the first strand of cDNA,
using 200 U/ml SuperScript II RT (Invitrogen, Carlsbad, CA, USA).
RHBDD1 mRNA expression levels were evaluated by qPCR using a
Bio-Rad Connect Real-Time PCR platform (Bio-Rad Laboratories,
Hercules, CA, USA) with SYBR Green PCR core reagents (Bio-Rad
Laboraties); β-actin was used as an internal reference. The
following primers were used: RHBDD1 forward,
5′-GCAGGACTGAGTGAAGAAGAAC-3′, and reverse,
5′-GTGAGAGATGAAACCCGTAGG-3′; and β-actin forward,
5′-GTGGACATCCGCAAAGAC-3′ and reverse, 5′-AAAGGGTGTAACGCAACTA-3′.
The RT-qPCR analysis was performed with the following amplification
steps: initial denaturation at 95°C for 1 min, followed by 40
cycles of denaturation at 95°C for 5 sec and annealing extension at
60°C for 20 sec. The results are presented as cycle threshold (Ct)
values, the threshold PCR cycle number at which the amplified
product is first detected. The average Ct was calculated for both
RHBDD1 and β-actin, and the ΔCT was determined as the mean of the
triplicate Ct values for RHBDD1, minus the mean of the triplicate
Ct values for β-actin.
Western blot analysis
RKO cells, five days after lentiviral infection,
were lysed in 2X SDS sample buffer [100 mM Tris-Hcl (pH 6.8), 10 mM
EDTA, 4% SDS, 10% glycine]. The proteins (3 μg) were loaded
onto a polyacrylamide gel and were separated by SDS-PAGE, followed
by transfer to polyvinylidene difluoride membranes (Millipore). The
blots were blocked with TBST containing 5% non-fat dry milk at room
temperature for 1 h and then incubated with primary antibodies:
rabbit anti-RHBDD1 (1:500 dilution; Sigma-Aldrich) and mouse
anti-GAPDH (1:3,000 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) at 4°C overnight. The blots were then probed with
antibodies against RHBDD1 (1:500 dilution; Sigma-Aldrich) and mouse
anti-GAPDH, (1:3,000 dilution; Santa Cruz Biotechnology, Inc.).
Following washing three times (5 min each) with TBST, the blots
were incubated with the corresponding horseradish
peroxidase-conjugated secondary antibodies: goat anti-rabbit IgG
(1:5,000 dilution; Santa Cruz Biotechnology, Inc.) and goat
anti-mouse IgG (1:5,000 dilution; Santa Cruz Biotechnology, Inc.)
at room temperature for 2 h. The blots were then visualized using a
Super Enhanced Chemiluminescence Detection Reagent (Applygen
Technologies Inc., Beijing, China).
Colony formation assay
RKO cells were cultured in 24-well plates and
treated with the recombinant lentiviruses at a multiplicity of
infection (MOI) of 20. Following a 72 h incubation, the cells were
washed, re-cultured in the prepared 6-well plates (400 cells/well)
and allowed to form natural colonies. Eight days later the cells in
both groups were subjected to Giemsa staining. Briefly, the cells
were washed and fixed using paraformaldehyde, the fixed cells were
then washed twice with phosphate-buffered saline (PBS), treated
with Giemsa (Sigma-Aldrich) for 10 min, washed three times with
double-distilled H2O, and then photographed using a
digital camera. The number of colonies (>50 cells/colony) were
counted.
MTT Viability Assay
RKO cells were cultured in 24-well plates and
inoculated with either RHBDD1-shRNA or control-shRNA lentiviruses,
at a MOI of 20. Following a 72 h incubation, the cells were washed,
re-cultured in the prepared 96-well plates (2,000 cells/well) and
the cell viability was analyzed using MTT reagent (Sigma-Aldrich)
and acidic isopropanol (10% SDS, 5% isopropanol and 0.01 mol/l
HCl).
Flow cytometric analysis
RKO cells were harvested by centrifugation at 404 ×
g for 5 min, 72 h following infection. The pellets were washed
twice with cold PBS, fixed with cold 70% ethanol, centrifuged and
resuspended with PBS. The pellets were washed twice with cold PBS,
fixed with cold 70% ethanol at 4°C overnight and then resuspended
in propidium iodide/RNase/PBS for incubation in the dark (room
temperature for 30 min). The suspensions were filtered through a
400-mesh membrane and subjected to cell cycle analysis using a flow
cytometer (BD Biosciences, San Jose, CA, USA). (BD Biosciences, San
Jose, CA, USA).
Statistical analysis
All statistical analyses were performed using SPSS
version 13.0 (SPSS, Inc., Chicago, IL, USA)software. The
differences between the groups were compared using a Student’s
t-test, and data were expressed as the means ± standard deviation
of triplicate experiments. A P<0.05 was considered to indicate a
statistically significant difference.
Results
Knockdown of RHBDD1 expression by
siRNA
Preliminary studies were performed to determine the
prime candidate CRC cell line for further in vitro studies.
The mRNA and protein expression levels of RHBDD1 were analyzed in
six CRC cell lines: SW480, SW620, RKO, DLD-1, HCT116 and HT-29. As
shown in Fig. 1A and B, RHBDD1 was
widely expressed in all of the cell lines tested. The RKO cell
line, which had moderate RHBDD1 expression levels, is commonly used
in colon cancer studies due to its high proliferation rate and low
coefficient of MOI. Therefore, the following in-depth in
vitro investigations were conducted using RKO cells. To
investigate the role of RHBDD1 in human CRC, control (Lv-shCon) and
RHBDD1-shRNA (Lv-shRHBDD1), lentiviruses were constructed. RKO
cells were cultured and infected with either the Lv-shCon or
Lv-shRHBDD1 lentivirus. The images were photographed following a 72
h incubation. An embedded green fluorescent proteintag was used to
visualize the transfection efficiency of the lentiviruses. As shown
in Fig. 1C, >80% of the cells
were successfully infected with the recombinant lentiviruses. The
specificity and efficiency of the RHBDD1 RNA interference (RNAi)
treatment was further verified as the control lentivirus, inserted
with one irrelevant sequence, had no impact on RHBDD1 translation
(Fig. 1D). The target lentivirus
Lv-shRHBDD1 markedly downregulated the endogenous RHBDD1 mRNA
expression levels, as compared with the control lentivirus
(P<0.01). Knockdown efficiency was further confirmed using
western blot analysis, there was little RHBDD1 protein expression
detected following RHBDD1 specific knockdown by RNAi (Fig. 1E).
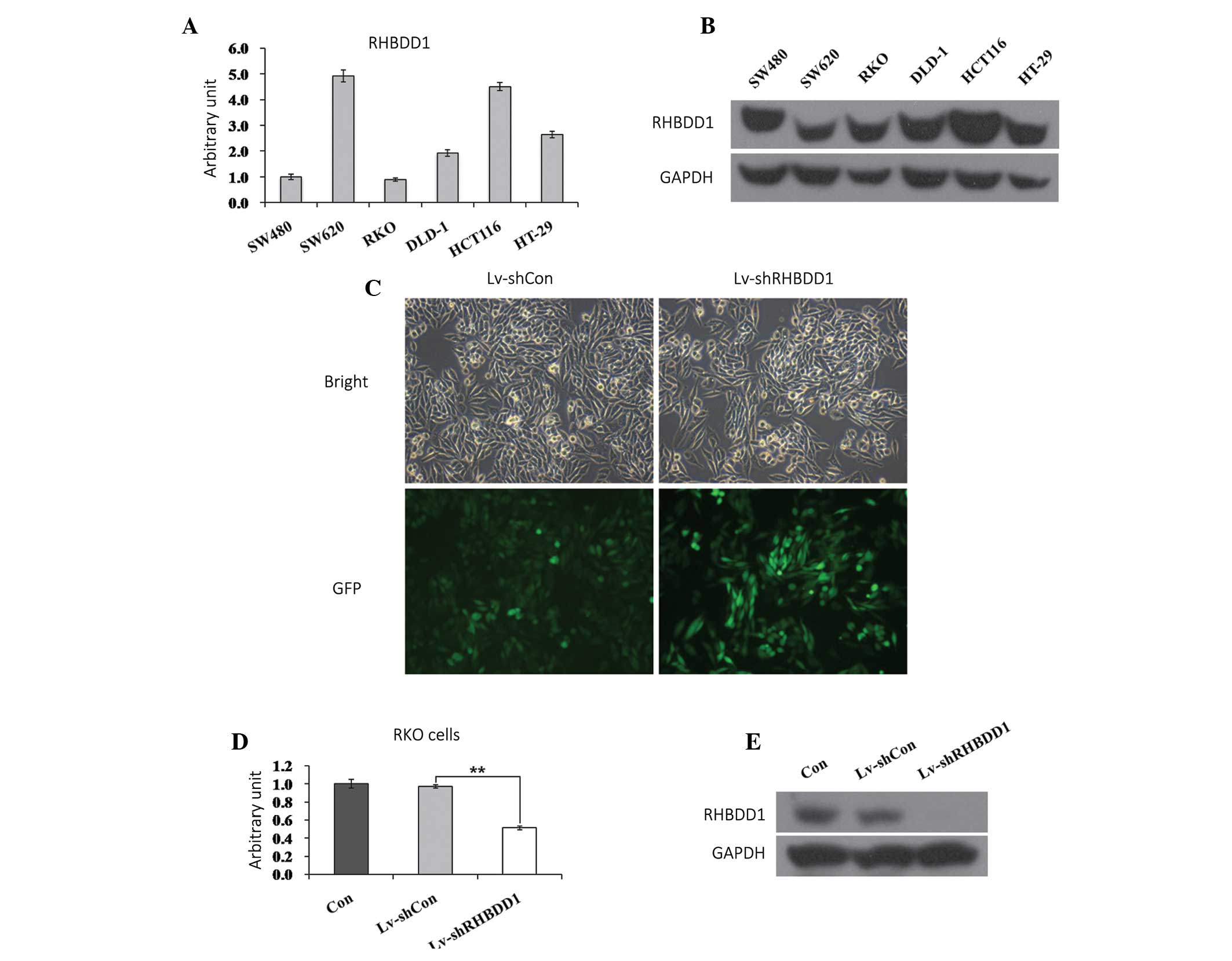 | Figure 1Downregulation of rhomboid domain
containing 1 (RHBDD1) by lentivirus-mediated small hairpin (sh)RNA.
(A) Quantitative polymerase chain reaction of RHBDD1 mRNA in six
colon cancer cell lines. (B) Western blot analysis of RHBDD1
protein in six colon cancer cell lines. (C) Representative graphs
of RKO cells infected with lentivirus at multiplicity of infection
of 20 (magnification, ×400). (D Expression analysis of RHBDD1 mRNA
in RKO colorectal cancer cells with three treatments (control,
Lv-shCon, Lv-shRHBDD1) by qPCR. (E) Expression analysis of RHBDD1
protein in RKO cells with three treatments (control, Lv-shCon,
Lv-shRHBDD1) by western blotting. Con, uninfected; Lv-shCon,
control lentivirus with non-silencing; Lv-shRHBDD1,
RHBDD1-silencing lentivirus; GFP, green fluorescent protein.
**P<0.01, as compared to treatment with Lv-shCon. |
Knockdown of RHBDD1 inhibits RKO cell
proliferation
To explore the role of RHBDD1 in CRC tumorigenesis,
the proliferation of RKO cells following RHBDD1 expression
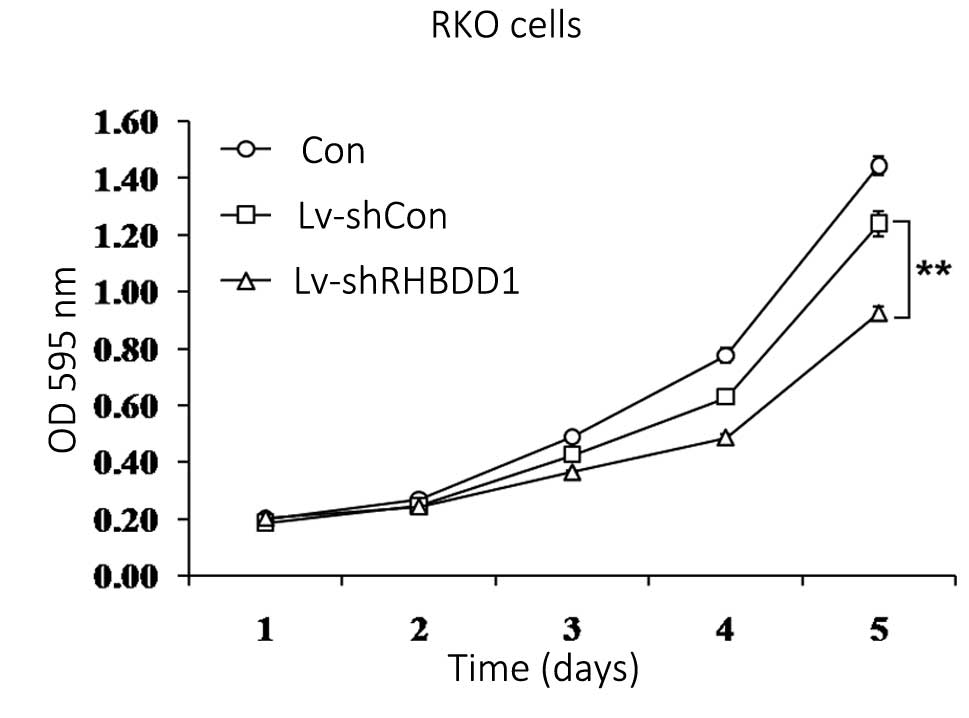
knockdown, was analyzed using an MTT assay. As shown in Fig. 2, the proliferation rate of
Lv-shRHBDD1 infected cells was markedly lower, as compared with the
Lv-shCon infected and uninfected cells (P<0.01). These results
indicate that knockdown of RHBDD1 expression may inhibit RKO cell
proliferation.
Knockdown of RHBDD1 suppresses colony
formation of RKO cells
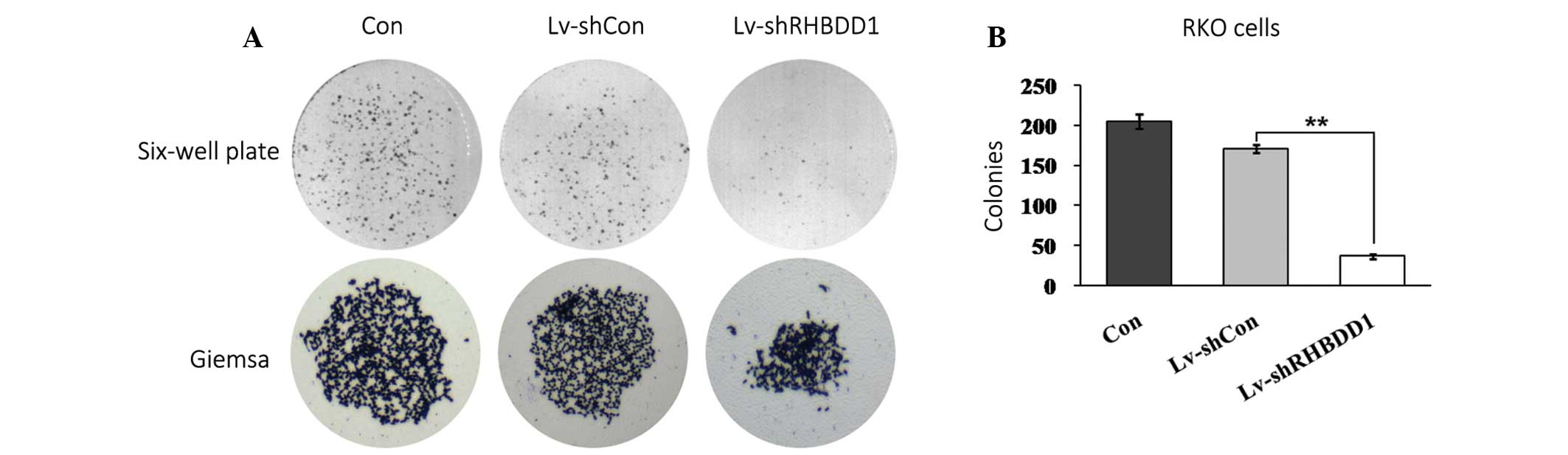
The colony forming capacity of RKO cells was
determined using a monolayer cell culture. As shown in Fig. 3A, knockdown of RHBDD1 expression
markedly suppressed colony formation in RKO cells, as determined by
a colony formation assay. The number of colonies formed in RKO
cells infected with Lv-shRHBDD1, was significantly decreased, as
compared with the Lv-shCon infected and uninfected cells (Fig. 3B, p<0.01). These results
indicate that RHBDD1 may have an important role in the cell growth
and tumorigenesis of CRC.
Knockdown of RHBDD1 induces G0/G1 phase
arrest in RKO cells
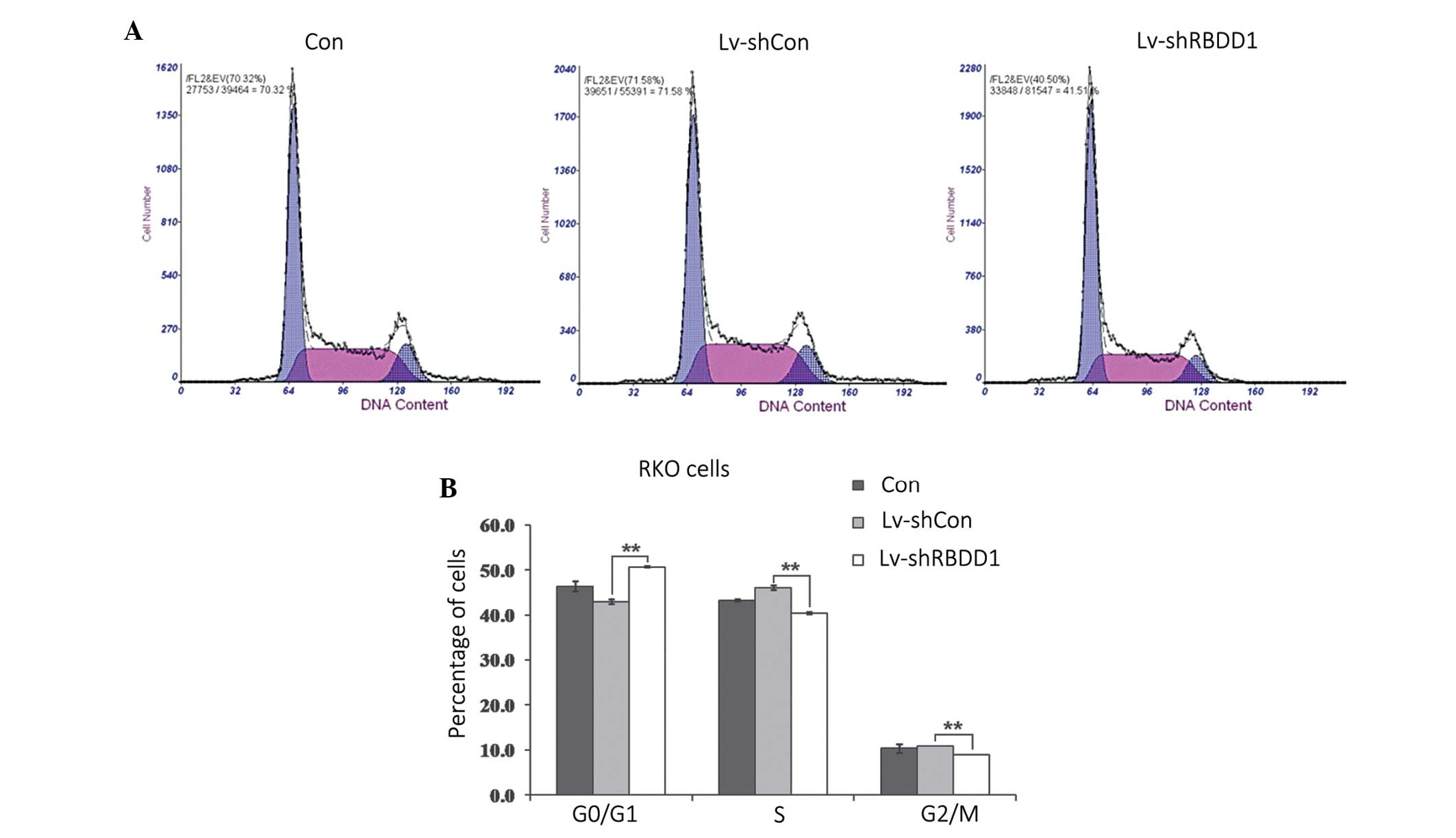
To elucidate whether RHBDD1 knockdown induced cell
growth inhibition by affecting the progression of the cell cycle,
flow cytometric analysis was performed in RKO cells. As shown in
Figure 4, following infection with
Lv-shRHBDD1, an increased number of cells accumulated at the G0/G1
phase and the percentage of cells in the S and G2/M phases were
reduced (P<0.01). These results indicate that RHBDD1 knockdown
may inhibit the growth of RKO cells via cell cycle arrest.
Discussion
Since the initial identification of rhomboid
proteases (18), there have been
indications that they may have a fundamental function within
numerous cell types. However little research has been performed to
determine their role in CRC. Vertebrate rhomboid genes are grouped
into three classes and HBDD1 is classed as an active cellular
rhomboid. Previous research has demonstrated that in Drosophila
that rhomboid proteases regulate EGFR signaling pathways (19,20),
and that rhomboid families may have pivotal roles in the modulation
of EGFR transactivation (21). The
monoclonal antibodies Cetuximab and Panitumumab, which target EGFR,
have been effective against CRC, in clinical practice (22). However, it remains unclear as to
whether the rhomboid protease family is involved in CRC
progression.
A previous study in hepatoma cells suggested that
RHBDD1 may be a positive regulator for HCC cell growth and
apoptosis using recombinant lentivirus-mediated silencing of RHBDD1
in HepG2 cells (17). It is
therefore conceivable that RHBDD1 has a similar, essential role in
CRC tumorigenesis. In the present study, we noted that RHBDD1 was
widely expressed in numerous human CRC cell lines. To illuminate
the functional role of RHBDD1 in the CRC cells, the present study
used lentivirus-mediated siRNA to silence the expression of RHBDD1
in the RKO colon cancer cells. Knock down of RHBDD1 expression was
found to markedly suppress cell proliferation and colony formation
in the RKO cells. Furthermore, flow cytometric analysis revealed
that depletion of RHBDD1 in the RKO cells led to cell cycle arrest
in the G0/G1 phase.
A previous study from a murine model indicated that
RHBDD1 has anti-apoptotic potential and that spermatogonia GC-1
cells, a mouse derived spermatogonia line, were more sensitive to
apoptotic stimuli following knock down of RHBDD1 expression
(23). Further investigation is
required to elucidate the modulation of RHBDD1 on CRC cell
apoptosis and its regulatory mechanism in CRC development and
progression.
In conclusion, the findings of the present study
demonstrated that RHBDD1 could promote CRC cell growth via cell
cycle control. RHBDD1 may serve as a potential therapeutic target
in human CRC.
Acknowledgments
The authors of the present study are grateful for
the financial support received from the Academic Leaders Training
Program of Pudong Health Bureau of Shanghai (no. PEWd2010-05).
References
|
1
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Merika E, Saif MW, Katz A, Syrigos K and
Morse M: Review. Colon cancer vaccines: an update. In Vivo.
24:607–628. 2010.PubMed/NCBI
|
|
3
|
Cunningham D, Atkin W, Lenz HJ, et al:
Colorectal cancer. Lancet. 375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
5
|
Lemberg MK: Intramembrane proteolysis in
regulated protein trafficking. Traffic. 12:1109–1118. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brown MS, Ye J, Rawson RB and Goldstein
JL: Regulated intramembrane proteolysis: a control mechanism
conserved from bacteria to humans. Cell. 100:391–398. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kroos L and Yu YT: Regulation of sigma
factor activity during Bacillus subtilis development. Curr Opin
Microbiol. 3:553–560. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weihofen A and Martoglio B:
Intramembrane-cleaving proteases: controlled liberation of proteins
and bioactive peptides. Trends Cell Biol. 13:71–78. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freeman M: Rhomboid proteases and their
biological functions. Annu Rev Genet. 42:191–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wolfe MS and Kopan R: Intramembrane
proteolysis: theme and variations. Science. 305:1119–1123. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Urban S: Rhomboid proteins: conserved
membrane proteases with divergent biological functions. Genes Dev.
20:3054–3068. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koonin EV, Makarova KS, Rogozin IB,
Davidovic L, Letellier MC and Pellegrini L: The rhomboids: a nearly
ubiquitous family of intramembrane serine proteases that probably
evolved by multiple ancient horizontal gene transfers. Genome Biol.
4:R192003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wasserman JD, Urban S and Freeman M: A
family of rhomboid-like genes: Drosophila rhomboid-1 and
roughoid/rhomboid-3 cooperate to activate EGF receptor signaling.
Genes Dev. 14:1651–1663. 2000.PubMed/NCBI
|
|
14
|
McQuibban GA, Saurya S and Freeman M:
Mitochondrial membrane remodelling regulated by a conserved
rhomboid protease. Nature. 423:537–541. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Guan X, Fok KL, et al: A novel
member of the Rhomboid family, RHBDD1, regulates BIK-mediated
apoptosis. Cell Mol Life Sci. 65:3822–3829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin YN, Gui FM, Shen H, et al: Expression
of RHBDD1 gene in patients with chronic myeloid leukemia and its
clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
21:12–15. 2013.Chinese. PubMed/NCBI
|
|
17
|
Liu XN, Tang ZH, Zhang Y, et al:
Lentivirus-mediated silencing of rhomboid domain containing 1
suppresses tumor growth and induces apoptosis in hepatoma HepG2
cells. Asian Pac J Cancer Prev. 14:5–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Urban S and Dickey SW: The rhomboid
protease family: a decade of progress on function and mechanism.
Genome Biol. 12:2312011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JR, Urban S, Garvey CF and Freeman M:
Regulated intracellular ligand transport and proteolysis control
EGF signal activation in Drosophila. Cell. 107:161–171. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Urban S, Lee JR and Freeman M: Drosophila
rhomboid-1 defines a family of putative intramembrane serine
proteases. Cell. 107:173–182. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zou H, Thomas SM, Yan ZW, Grandis JR, Vogt
A and Li LY: Human rhomboid family-1 gene RHBDF1 participates in
GPCR-mediated transactivation of EGFR growth signals in head and
neck squamous cancer cells. FASEB J. 23:425–432. 2009. View Article : Google Scholar :
|
|
22
|
Normanno N, Tejpar S, Morgillo F, De Luca
A, Van Cutsem E and Ciardiello F: Implications for KRAS status and
EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol.
6:519–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Song W, Li S, et al: GC-1 mRHBDD1
knockdown spermatogonia cells lose their spermatogenic capacity in
mouse seminiferous tubules. BMC Cell Biol. 10:252009. View Article : Google Scholar : PubMed/NCBI
|