Introduction
Breast cancer is the most commonly diagnosed type of
cancer in females and is the leading cause of cancer-related
mortality in females worldwide (1). In 2013, the American Cancer Society
estimated that 232,340 females would be newly diagnosed with breast
cancer and 39,620 females would succumb to the disease (2). The key objectives of scientists and
clinicians in managing this breast cancer are to prevent the
incidence, detect it early and treat it with effective therapeutic
strategies resulting in long overall survival with minimal side
effects. Therefore, the aim of the current study was to identify
the genes associated with cell growth inhibition that are induced
by Retinoid X receptor (RXR)-selective retinoids (rexinoids), with
an aim to improve prevention and treatment of breast cancer.
Retinoids regulate a variety of biological
functions, including embryogenesis, growth, differentiation, vision
and reproduction (3–5). Retinoids additionally possess
antiproliferative properties, which suggests a chemopreventive and
therapeutic role against cancer (6). In addition, retinoids have been
reported to inhibit normal- or tumor-cell growth through the
regulation of differentiation and/or apoptosis (7–10).
Retinoids exert their effects in target cells via
interaction with retinoic acid receptors (RARs) and RXRs. Each of
these includes three subtypes, termed α, β and γ, which are encoded
by distinct genes. The RARα, RARβ and RARγ genes have been
localized to chromosomes 17q21, 3p24 and 12q13, respectively. The
RXRα, RXRβ and RXRγ genes have been mapped to chromosomes 9q34.3,
6p21.3 and 1q22-23, respectively (11). The RARs bind
all-trans-retinoic acid (ATRA) and 9-cis-retinoic
acid (RA) while RXRs bind 9-cis-RA alone. RXRs are known to
heterodimerize with several steroid hormone receptors, including
RAR, thyroid hormone receptor, vitamin D receptor, peroxisome
proliferator-activated receptor, liver X receptor, pregnane X
receptor and farnesoid X receptor suggesting its involvement in
several signaling pathways (12).
RXRs are also able to homodimerize in transfected cells (13).
In addition to naturally occurring retinoids,
including ATRA, 9-cis-RA and 13-cis-RA, various
synthetic retinoids with varied selectivity have been developed and
are currently available to treat psoriasis, acne, photoaging,
actinic keratosis and certain types of cancer, including acute
promelocytic leukemia, cutaneous T-cell lymphoma and squamous or
basal cell carcinoma (14).
However, the use of RAR-selective retinoids is limited by their
toxicity, which can result in chelitis, hypertriglyceridemia and
hepatosplenomegaly (15).
Rexinoids are important in controlling apoptosis and
can function in a ligand-dependent or ligand-independent manner
(16,17). Notably, rexinoids have been
reported to suppress estrogen receptor (ER)-positive and
ER-negative mammary tumor development with reduced toxicity
compared with RAR-selective retinoids (18–20).
Rexinoids are additionally active in animals with
tamoxifen-resistant breast cancer (17,21)
and in ATRA-resistant breast cancer cells (22). Thus, rexinoids appear to be
promising chemopreventive and therapeutic agents with improved
efficiency as compared with RAR-selective ligands. Among the
rexinoids, LGD1069 (Bexarotene) was confirmed as a safe and
well-tolerated agent in clinical trials of cutaneous T-cell
lymphoma, breast cancer and lung cancer (22,23).
Thus, we focussed on rexinoids and their cognate
receptor, RXR, in breast cells, and aimed to investigate their
regulatory activity on the transcription of genes involved in
growth suppression. In particular, the present study investigated
the RXRα isoform, which has been suggested as a potential
therapeutic target in breast cancer cells, due to the observation
that overexpression of RXRα sensitized breast cancer cells lines to
the antiproliferative effects of RXR-selective ligands (24). In addition, infection with
adenoviral RXRα induced nucleoplasmic overexpression of RXRα and
resulted in apoptosis with treatment with an RXR ligand in
retinoid-resistant MDA-MB-231 cells (25). Thus, in the current study, the
growth-suppressive activity of RXR pan agonists (LGD1069 and
LG100268) and an RXRα specific ligand (Ro25-7386) were investigated
in normal human mammary epithelial cells (HMECs) and four breast
cancer cell lines (MCF-7, T47D, MDA-MB-231 and MDA-MB-435) using an
MTS assay. Subsequently, the genes regulated by rexinoids that may
be involved in their antiproliferative activity were investigated
with an Affymetrix microarray.
Materials and methods
Ligands and antibodies
LGD1069 and LG100268 were provided by Ligand
Pharmaceuticals, Inc. (La Jolla, CA, USA). Ro25-7386 was obtained
from Roche Bioscience (Palo Alto, CA, USA). These compounds were
diluted in dimethyl sulfoxide purchased from Sigma-Aldrich (St.
Louis, MO, USA) to a final concentration of 0.1%. Monoclonal or
polyclonal antibodies (mouse or rabbit) against RXRα (cat. no.
sc-553) B-cell lymphoma 2-associated X protein (Bax; cat. no.
sc-7480), E-cadherin (cat. no. sc-7870), integrin α6 (cat. no.
sc-13542), cell division control protein 42 (CDC42; cat. no.
sc-8401) and actin (cat. no. sc-8432) were purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Cells and culture materials
Human normal mammary epithelial cells (HMECs) were
obtained from Lonza Group (San Diego, CA, USA). Cells between
passages 10 and 11 were used for experiments and the cells were
grown and maintained in mammary epithelial basal medium
supplemented with 13 mg/ml bovine pituitary extract, 0.5% serum, 5
μg/ml insulin, 10 ng/ml human recombinant epidermal growth
factor, 0.5 mg/ml hydrocortisone, 50 μg/ml gentamicin and 50
μg/ml amphotericin-β (all Clonetics, Lonza Group, San Diego,
CA, USA). Cells were maintained in a humidified environment at 37°C
with 5% CO2 in air.
Four different human breast cancer cell lines
(MCF-7, T47D, MDA-MB-231 and MBA-MB-435) purchased from the
American Type Culture Collection (Manassas, VA, USA) were grown and
maintained in appropriate growth media; minimal essential medium
for MCF-7 and RPMI 1640 for T47D, MDA-MB-231 and MBA-MB-435
(Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with
10% heat-inactivated fetal bovine serum (FBS; Welgene, Daegu,
Korea). L-glutamine, penicillin, streptomycin and gentamicin (Life
Technologies Korea, LLC, Seoul, Korea) were used at the usual
concentrations. For all experiments, breast cancer cells were
harvested by trypsinization (0.25% trypsin and 0.02% EDTA; Life
Technologies Korea, LLC), seeded and grown in the appropriate media
containing 10% FBS in a humidified 95% air 5% CO2
atmosphere.
Cell growth rate measurements
The CellTiter 96® AQueous
Non-Radioactive Cell Proliferation Assay (Promega Corporation,
Madison, WI, USA) was used for the measurement of cell growth rate
in breast cancer cells according to the manufacturer’s
instructions. The CellTiter 96® AQueous Assay
is composed of solutions of a novel tetrazolium compound
[3-(4,5-dimethylthiazol-2-yl)-5-(-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium,
inner salt; MTS] and an electron coupling reagent (phenazine
methosulfate; PMS). Briefly, HMECs, MCF-7 and T47D (1,000
cells/well) were plated in 96-well plates. Following a 24 h resting
period, LGD1069, LG100268 and Ro25-7386 were added into the growth
media and cell culture continued for 8–12 days. Each measurement
day (every 2 days), MTS (Promega Corporation) was added to the
cells (20 μl combined MTS/PMS solution per 100 μl
culture medium) and further incubation was conducted for 2 h. MTS
is bioreduced by cells into a formazan product that is soluble in
tissue culture medium. The absorbance of the formazan at 490 nm was
measured directly using an ELISA plate reader (Gemini EM Microplate
reader, Versa Max, Fluorescence readers; Molecular Devices,
Sunnyvale, CA, USA). Each data point was performed in quadruplicate
and the results were presented as the mean absorption (optical
density).
RNA target preparation/Affymetrix
microarray analysis
Total RNA was extracted from different breast cells
treated with rexinoids using the guanidinium isothiocynate method
(TRIzol reagent; Invitrogen Life Technologies) followed by
purification using an RNeasy column (Qiagen, Valencia, CA, USA).
RNA quality was assessed using the 2100 Bioanalyzer Instrument
(Agilent Technologies, Inc., Palo Alto, CA, USA). A total of 10
μg total RNA was processed for use on the microarray using
the Affymetrix GeneChip One-Cycle Target Labeling kit (Affymetrix,
Inc., Santa Clara, CA, USA) according to the manufacturer’s
instructions. The resultant biotinylated cRNA was fragmented and
then hybridized to the Affymetrix U133 Plus 2.0 GeneChip. The
arrays were washed, stained and scanned using the Affymetrix 450
Fluidics Station and GeneChip Scanner 3000 7G (Affymetrix, Inc.)
according to the manufacturer’s recommendations. Expression values
were generated using Microarray Suite software, version 5.0
(Affymetrix, Inc.).
Statistical analysis of microarray
data
Background subtraction and normalization using the
robust multi-array average algorithm method was performed using
GeneSpring GX 11.5 software (Agilent Technologies) for gene
expression. Fold change values for genes were calculated as the
ratio of the signal values of the experimental group compared with
the control group. Alterations in gene expression >2-fold were
considered to be statistically significant. Genes of interest were
selected by referring to the PathArt program which shows
intersection of genes in several signaling pathways.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Cells were cultured to 80–90% confluence. Total RNA
was prepared using the Qiagen RNeasy Mini kit (Qiagen). The RT
reaction was performed using 1 μg total RNA which was
reverse-transcribed into cDNA using a random hexamer primer
(GeneAmp RNA PCR Core kit; Applied Biosystems Life Technologies,
Foster City, CA, USA), according to the manufacturer’s
instructions. cDNA of the 7 selected genes and an internal
reference gene (GAPDH) was produced from each sample and was
quantified using a fluorescence-based real-time detection method
(iCycler; Bio-Rad Laboratories, Inc., Hercules, CA, USA). RT-qPCR
analysis was performed using the standard methods recommended by
the RT-qPCR kit supplier (SYBR® Green Dye-Based Gene
Expression Detection; Applied Biosystems Life Technologies). Primer
sequences used for detection of RXRα-regulated genes are shown in
Table I (Cosmo Genetech, Seoul,
Korea). For the endogenous control, human GAPDH labeled with VIC™
dye provided by Applied Biosystems Life Technologies was used. The
amplification conditions were as follows: 30 sec at 95°C and 3 min
at 95°C, and 30 sec at 95°C and 60 sec at 65°C for 40 cycles,
followed by a final extension for 20 min at 72°C. The ratio between
the values obtained provided the relative gene expression
levels.
 | Table IForward and reverse primers for
amplification of targeted genes with reverse
transcription-quantitative polymerase chain reaction. |
Table I
Forward and reverse primers for
amplification of targeted genes with reverse
transcription-quantitative polymerase chain reaction.
| Target gene | Forward primer | Reverse primer |
|---|
| BAX |
5′-TGGAGCTGCAGAGGATGATTG-3′ |
5′-GAAGTTGCCGTCAGAAAACATG-3′ |
| E-cadherin |
5′-CACTGCCAACTGGCTGGAG-3′ |
5′-GGGTTAGCTCAGCAGTAAAG-3′ |
| FOXO3A |
5′-TCAATCAGAACTTGCTCCACCA-3′ |
5′-GGACTCACTCAAGCCCATGTTG-3′ |
| Integrin α6 | 5-
TTTCCCGTTTCTTTCTTGAGTTGT-3′ |
5′-TGGAAAAGGTAACTTGTGAGCCA-3′ |
| Integrin β4 |
5-TTCCAAATCACAGAGGAGAC-3 |
5-CTTGAGGTTGTCCAGATCAT-3′ |
| PXN |
5′-TGGCTTCGCTGTCGGATTTC-3′ |
5′GTCAAGGGCTGTCACCACTTTATC-3′ |
| PTEN |
5′-AGAGCGTGCAGATAATGACAAG-3′ |
5′-GGATCAGAGTCAGTGGTGTCAG-3′ |
| STAT |
5′-CTGCTGCGGTTCAGTGAGAG-3′ |
5′-CCAAGTGAAAGTGACCCCTCC-3′ |
| Collagen type VI
α3 |
5′-CTGGGCAGACATACCATGTG-3′ |
5′-GCAAGTTCCTTCGTCTTTCG-3′ |
Western blot analysis
Whole cell extracts were prepared using 1X sodium
dodecyl sulfate (SDS) laemmlli lysis buffer (125 mM Tris-HCl, pH
6.8; 1% SDS; 2% β-mercaptoethanol). Total cell lysates with equal
quantities of protein (30 μg) were subjected to 10% SDS-PAGE
and subsequently electrotransferred onto a nitrocellulose membrane
(Bio-Rad Laboratories, Inc.). Membranes were blocked with 5%
skimmed milk in PBST (phosphate-buffered saline containing 0.1%
Tween 20) for 1 h at room temperature, then incubated overnight
with primary antibodies in PBST containing 2.5% bovine serum
albumin (1:1,000 dilution). Subsequent to washing with PBST, the
blot was further incubated for 1 h at room temperature with
peroxidase conjugated anti-rabbit or anti-mouse antibodies (Pierce
Technology Corporation, Holmdel, NJ, USA) in PBST and then
visualized using the enhanced chemiluminescence system (GE
Healthcare Life Sciences, Chalfont, UK). Protein expression was
normalized using β-actin expression.
Statistical analysis
All experiments were performed in triplicate.
Statistical analyses were performed using Microsoft Excel 2007
(Microsoft Corporation, Albuquerque, NM, USA). The data for the MTS
assay and RT-qPCR are expressed as the mean ± standard deviation.
Student’s t-test was used for single variable comparisons, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Anti-proliferative activity of
rexinoids
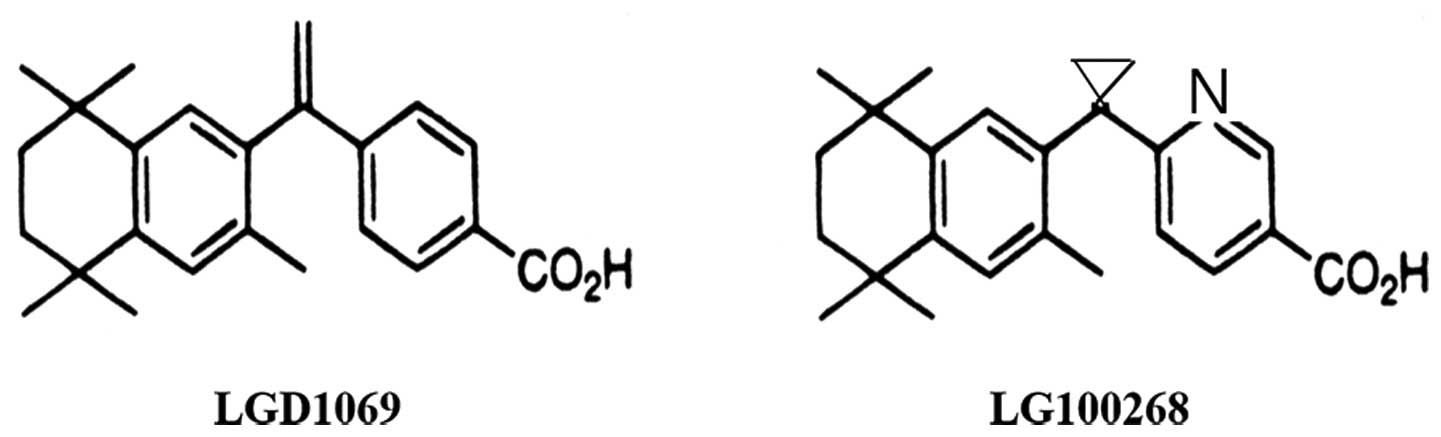
In Fig. 1, the
structures of LGD1069 and LG100268 are presented. The
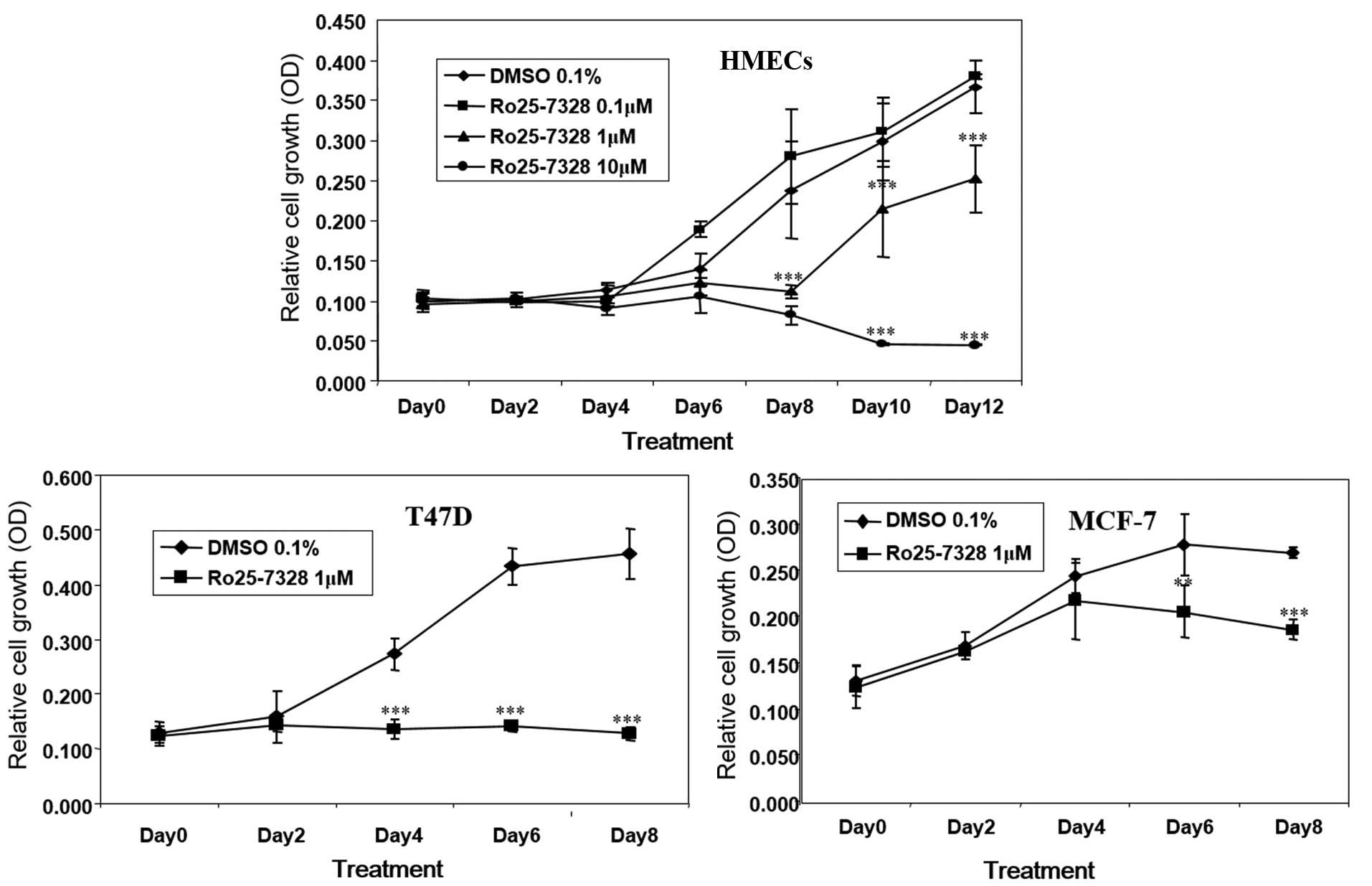
anti-proliferative effects of rexinoids in normal and malignant
breast cells were investigated. It was observed that LGD1069 and
LG100268 significantly suppressed cell growth in HMECs at 10
μM; whereas co-treatment with LGD1069 and LG100268 reduced
cell growth at 1 and 10 μM suggesting that these two
rexinoids possess the capacity to prevent mammary cell growth
(Fig. 2). By contrast, LGD1069
weakly (10 μM, P<0.05) inhibited cell growth in MCF-7
cells while the compound strongly and significantly suppressed cell
growth in a dose-dependent manner in T47D cells (0.1 μM,
P<0.01; 1 and 10 μM, P<0.001) (Fig. 3). Notably, LGD1069 induced mild
inhibition (P<0.05) of cell growth in MDA-MB-231 cells at 10
μM while rexinoids did not affect cell growth in MDA-MB-435
cells (Fig. 3). This result
indicates that LGD1069 is able to inhibit the growth of ER-negative
breast cancer with therapeutic potency.
In addition, Ro25-7386, the RXRα agonist
significantly suppressed cell growth in a dose-dependent manner in
HMECs. Ro25-7386 strongly reduced T47D cell growth at 1 μM
and induced suppression of cell growth in MCF-7 cells at day 8 at 1
μM (Fig. 4). These results
suggest that RXRα is important in the suppression of growth induced
by rexinoids in breast cells.
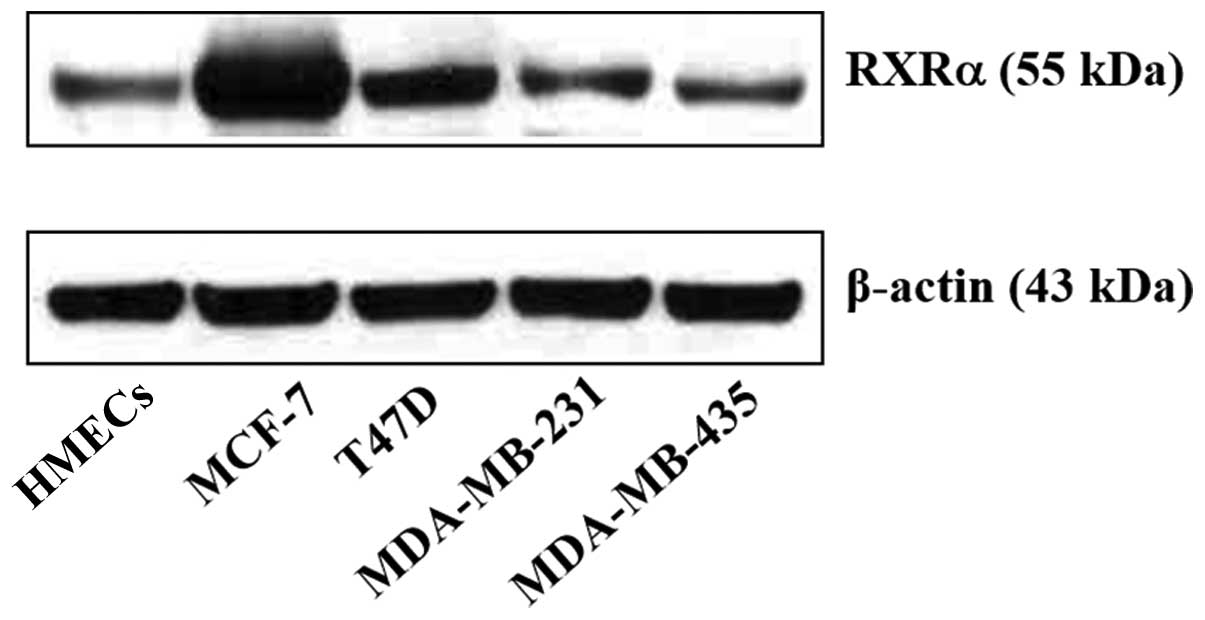
Expression of RXRα in breast cells
The RXRα level in normal and malignant breast cells
was next determined. It was observed that all breast cell lines
express RXRα but with different intensities. MCF-7 and T47D
expressed higher levels of RXRα (Fig.
5). Notably, the ER-negative breast cancer cell lines,
MDA-MB-231 and MDA-MB-435, also expressed RXRα.
Identification of target genes regulated
by rexinoids in normal and malignant breast cells by Affymetrix
microarray
Finally, the genes regulated by rexinoids in normal
(HMECs) and malignant (MCF-7, T47D and MDA-MB-231) breast cells
were identified. Gene expression profiles were established using
the Affymetrix microarray (human genome U133A 2.0). Among them,
several genes involved in cell death, cell growth/maintenance,
signal transduction and response to stimulus were identified.
In HMECs, 638 genes upregulated and 347 genes
downregulated by Ro25-7386 with alterations in fold induction
>2-fold were identified. A total of 22 genes were strongly
upregulated (>10-fold) and 5 genes were strongly downregulated
(>4-fold) in expression levels by Ro25-7386 (Table IIA and B). Among them, several
genes were notable, including integrin β4, E-cadherin (CDH1),
C-terminal binding protein 1 (CtBP1), integrin α6, paxillin (PAX),
BAX, forkhead box O3A (FOXO3A) and signal transducer and activator
of transcription 3 (STAT3) (upregulated genes), and collagen type
VI α3 and cell division cycle 42 (CDC42) (downregulated genes).
 | Table IIGenes up- and downregulated by
Ro25-7386 in human mammary epithelial cells. |
Table II
Genes up- and downregulated by
Ro25-7386 in human mammary epithelial cells.
A, Genes upregulated
by Ro25-7386
|
|---|
| Probe set | Gene | Fold change |
|---|
| 213872_at |
gb:BE465032/DB_XREF=gi:9510807/DB_XREF=hv76g09.×1/CLONE=
IMAGE:3179392/FEA=EST/CNT=34/TID=Hs.173685.1/TIER=Stack/STK=15/UG=Hs.
173685/LL= 81688/UG_GENE=FLJ12619/UG_TITLE=hypothetical protein
FLJ12619 | 27.55 |
| 204989_s_at | Integrin, β4 | 26.60 |
| 210317_s_at | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein,
epsilon polypeptide | 22.14 |
| 200935_at | Calreticulin | 21.26 |
| 201130_s_at | Cadherin 1, type 1,
E-cadherin (epithelial) | 20.66 |
| 201123_s_at | Eukaryotic
translation initiation factor 5A | 19.04 |
| 200751_s_at | Heterogeneous nuclear
ribonucleoprotein C (C1/C2) | 18.97 |
| 214007_s_at | PTK9 protein tyrosine
kinase 9 | 17.37 |
| 203392_s_at | C-terminal binding
protein 1 | 16.35 |
| 204427_s_at | Coated vesicle
membrane protein | 16.00 |
| 216971_s_at | Plectin 1,
intermediate filament binding protein 500 kDa | 15.17 |
| 217211_at | Consensus includes
gb:D50604/DEF=Human β-cytoplasmic actin (ACTBP9)
pseudogene/FEA=CDS/DB_XREF=gi:2094759/UG=Hs.248007 Human
β-cytoplasmic actin (ACTBP9) pseudogene | 14.35 |
| 215780_s_at | SET translocation
(myeloid leukemia-associated) | 12.51 |
| 201971_s_at | ATPase,
H+ transporting, lysosomal 70 kDa, V1 subunit A | 11.75 |
| 204426_at | Coated vesicle
membrane protein | 11.74 |
| 220494_s_at |
gb:NM_018678.1/DEF=Homo sapiens
lipopolysaccharide specific response-68 protein (LSR68),
mRNA./FEA=mRNA/GEN=LSR68/PROD=lipopolysaccharide specific
response-68 protein/DB_XREF=gi:8923914/UG=Hs.103189
lipopolysaccharide specific response-68 protein/ | 11.18 |
| 215177_s_at | Integrin, α6 | 10.97 |
| 215434_x_at | AG1 | 10.35 |
| 214693_x_at | Hypothetical
protein MGC8902///AG1///hypothetical protein
DJ328E19.C1.1///hypothetical protein LOC200030///hypothetical
protein LOC348482 | 10.34 |
| 211905_s_at | Integrin, β4 | 10.34 |
| 201048_x_at | RAB6A, member RAS
oncogene family | 10.03 |
| 214701_s_at | Fibronectin 1 | 10.01 |
| 210092_at | Mago-nashi homolog,
proliferation-associated (Drosophila) |
9.74 |
| 212107_s_at | DEAH
(Asp-Glu-Ala-His) box polypeptide 9 |
9.68 |
| 202118_s_at | Copine III |
9.48 |
| 217234_s_at | Villin 2
(ezrin) |
9.09 |
| 208853_s_at | Calnexin |
7.59 |
| 201742_x_at | Splicing factor,
arginine/serine-rich 1 (splicing factor 2, alternate splicing
factor) |
7.44 |
| 208750_s_at | ADP-ribosylation
factor 1 |
7.31 |
| 203803_at | Prenylcysteine
oxidase 1 |
7.31 |
| 211162_x_at | Stearoyl-CoA
desaturase (δ-9-desaturase) |
7.30 |
| 202856_s_at | Solute carrier
family 16 (monocarboxylic acid transporters), member 3 |
7.26 |
| 200796_s_at | Myeloid cell
leukemia sequence 1 (BCL2-related) |
7.25 |
| 213606_s_at | Rho GDP
dissociation inhibitor (GDI) α |
7.25 |
| 201373_at | Plectin 1,
intermediate filament binding protein 500kDa |
7.19 |
| 208057_s_at | GLI-Kruppel family
member GLI2 |
7.04 |
| 217294_s_at | Enolase 1, (α) |
6.99 |
| 213875_x_at | Chromosome 6 open
reading frame 62 |
6.93 |
| 91816_f_at | Ring finger and KH
domain containing 1 |
6.90 |
| 200806_s_at | Heat shock 60 kDa
protein 1 (chaperonin) |
6.69 |
| 214845_s_at | Calumenin |
6.66 |
| 211823_s_at | Paxillin |
5.75 |
| 206665_s_at | BCL2-like 1 |
5.40 |
| 208637_x_at | Actinin, α1 |
5.11 |
| 208677_s_at | Basigin (OK blood
group) |
4.66 |
| 221499_s_at | Syntaxin 16 |
4.16 |
| 209226_s_at | Transportin 1 |
3.90 |
| 201752_s_at | Adducin 3 (γ) |
3.90 |
| 200766_at | Cathepsin D
(lysosomal aspartyl protease) |
3.90 |
| 203085_s_at | Transforming growth
factor, β1 (Camurati-Engelmann disease) |
3.75 |
| 211833_s_at | BCL2-associated X
protein |
3.65 |
| 208852_s_at | Calnexin |
3.49 |
| 210655_s_at | Forkhead box
O3A |
3.33 |
|
B, Genes
downregulated by Ro25-7386
|
| Probe set | Gene | Fold change |
|
| 203991_s_at | Ubiquitously
transcribed tetratricopeptide repeat, X chromosome | −5.32 |
| 220568_at |
gb:NM_018582.1/DEF=Homo sapiens
hypothetical protein PRO1483 (PRO1483),
mRNA./FEA=mRNA/GEN=PRO1483/PROD=hypothetical protein
PRO1483/DB_XREF=gi:8924047/UG=Hs.279694 hypothetical protein
PRO1483/FL=gb:AF116635.1
gb:NM_018582.1 | −4.72 |
| 213705_at | Methionine
adenosyltransferase II, α |
− −4.64 |
| 201438_at | Collagen, type VI,
α3 |
− −4.59 |
| 217665_at | Consensus includes
gb:AA420614/FEA=EST/DB_XREF=gi:2094586/DB_XREF= est:
nc62g02.r1/CLONE=IMAGE:745874/UG=Hs.188826 ESTs, Moderately similar
to G02654 ribosomal protein L39 H. sapiens | −4.17 |
| 209459_s_at | 4-aminobutyrate
aminotransferase | −3.99 |
| 220992_s_at | Chromosome 1 open
reading frame 25///chromosome 1 open reading frame 25 | −3.81 |
| 222294_s_at | Eukaryotic
translation initiation factor 2C, 2 | −3.78 |
| 221995_s_at | Consensus includes
gb:BF195165/FEA=EST/DB_XREF=gi:11081754/DB_XREF= est:
7n16b01.×1/CLONE=IMAGE:3564624/UG=Hs.182695 hypothetical protein
MGC3243 | −3.71 |
| 215095_at | Esterase
D/formylglutathione hydrolase | −3.68 |
| 212675_s_at | KIAA0582 | −3.66 |
| 210187_at | FK506 binding
protein 1A, 12 kDa | −3.65 |
| 204634_at | NIMA (never in
mitosis gene a)-related kinase 4 | −3.59 |
| 203791_at | Dmx-like 1 | −3.53 |
| 205583_s_at | Chromosome X open
reading frame 45 | −3.53 |
| 218352_at | Regulator of
chromosome condensation (RCC1) and BTB (POZ) domain containing
protein 1 | −3.52 |
| 209788_s_at | Type 1 tumor
necrosis factor receptor shedding aminopeptidase regulator | −3.48 |
| 212959_s_at | MGC4170
protein | −3.47 |
| 205802_at | Transient receptor
potential cation channel, subfamily C, member 1 | −3.43 |
| 202732_at | Protein kinase
(cAMP-dependent, catalytic) inhibitor γ | − −3.40 |
| 202149_at | Neural precursor
cell expressed, developmentally downregulated 9 | −3.39 |
| 213225_at | Protein phosphatase
1B (formerly 2C), magnesium-dependent, β isoform | −3.39 |
| 213624_at | Sphingomyelin
phosphodiesterase, acid-like 3A | −3.39 |
| 207855_s_at | Mid-1-related
chloride channel 1 | −3.37 |
| 204415_at | Interferon,
α-inducible protein (clone IFI-6-16) | −3.29 |
| 210017_at | Mucosa associated
lymphoid tissue lymphoma translocation gene 1 | −3.12 |
| 205420_at | Peroxisomal
biogenesis factor 7 | −3.05 |
| 219317_at | Polymerase (DNA
directed) iota | −3.01 |
| 204176_at | Kelch-like ECT2
interacting protein | −3.00 |
| 203741_s_at | Adenylate cyclase
7 | −2.95 |
| 205034_at | Cyclin E2 | −2.94 |
| 204078_at | Synaptonemal
complex protein SC65 | −2.90 |
| 203881_s_at | Dystrophin
(muscular dystrophy, Duchenne and Becker types) | −2.88 |
| 209717_at | Ecotropic viral
integration site 5 | −2.87 |
| 213473_at | BRCA1 associated
protein | −2.86 |
| 215949_x_at | Immunoglobulin
heavy constant μ | −2.83 |
| 205668_at | Lymphocyte antigen
75 | −2.83 |
| 219688_at | Bardet-Biedl
syndrome 7 | −2.82 |
| 207845_s_at | Anaphase promoting
complex subunit 10 | −2.80 |
| 208920_at | Sorcin | −2.79 |
| 218002_s_at | Chemokine (C-X-C
motif) ligand 14 | −2.53 |
| 208727_s_at | Cell division cycle
42 (GTP binding protein, 25 kDa) | −2.25 |
In MCF-7 cells, 83 genes were upregulated and 98
genes were downregulated by Ro25-7328 with alterations in fold
induction >2-fold were identified (Table III). Among them, several genes
were recognized including transforming growth factor β2,
immunoglobulin heavy constant γ1, protein kinase Cδ binding
protein, interleukin 6 receptor and neurophilin 2 (upregulated
genes), and cathepsin S, zinc finger protein 36, integrin β4,
transforming growth factor β1, PAX and CtBP1 (downregulated
genes).
 | Table IIIGenes up- and downregulated by
Ro25-7386 in MCF-7 cells. |
Table III
Genes up- and downregulated by
Ro25-7386 in MCF-7 cells.
A, Genes
upregulated by Ro25-7386
|
|---|
| Probe set | Gene | Fold change |
|---|
| 209909_s_at | Transforming growth
factor, β2 | 4.94 |
| 211430_s_at | Immunoglobulin
heavy constant γ 1 (G1m marker) | 3.82 |
| 213010_at | Protein kinase C, δ
binding protein | 3.76 |
| 63825_at | Abhydrolase domain
containing 2 | 3.40 |
| 208993_s_at | Peptidyl-prolyl
isomerase G (cyclophilin G) | 3.39 |
| 204681_s_at | Rap guanine
nucleotide exchange factor (GEF) 5 | 3.26 |
| 213536_s_at |
gb:AA910614/DB_XREF=gi:3049904/DB_XREF=ok61b04.s1/CLONE=IMAGE:
1518415/FEA=EST/CNT=42/TID=Hs.84285.2/TIER=Stack/STK=12/UG=Hs.
84285/LL=7329/UG_GENE=UBE2I/UG_TITLE=ubiquitin-conjugating enzyme
E2I (homologous to yeast UBC9) | 3.20 |
| 213087_s_at | Eukaryotic
translation elongation factor 1 δ (guanine nucleotide exchange
protein) | 3.09 |
| 217489_s_at | Interleukin 6
receptor | 3.07 |
| 205443_at | Small nuclear RNA
activating complex, polypeptide 1, 43 kDa | 3.04 |
| 213747_at | Consensus includes
gb:AA047234/FEA=EST/DB_XREF=gi:1525134/DB_XREF=est:zf50b09.s1/CLONE=IMAGE:380345/UG=Hs.223014
antizyme inhibitor | 2.99 |
| 221815_at | Abhydrolase domain
containing 2 | 2.95 |
| 212451_at | KIAA0256 gene
product | 2.93 |
| 205363_at | Butyrobetaine (γ),
2-oxoglutarate dioxygenase (γ-butyrobetaine hydroxylase) 1 | 2.92 |
| 212952_at | Consensus includes
gb:AA910371/FEA=EST/DB_XREF=gi:3049661/DB_XREF=est:
ok83h10.s1/CLONE=IMAGE:1520611/UG=Hs.16488 calreticulin | 2.90 |
| 210136_at | Myelin basic
protein | 2.88 |
| 214255_at | ATPase, class V,
type 10A | 2.87 |
| 213789_at | Consensus includes
gb:N58493/FEA=EST/DB_XREF=gi:1202383/DB_XREF=est:
yv72d01.s1/CLONE=IMAGE:248257/UG=Hs.75105 emopamil-binding protein
(sterol isomerase) | 2.86 |
| 217464_at | Consensus includes
gb:L48784/DEF=050 Homo sapiens
cDNA/FEA=mRNA/DB_XREF=gi:1066715/UG=Hs.182426 ribosomal protein
S2 | 2.83 |
| 210841_s_at | Neuropilin 2 | 2.82 |
| 204378_at | Breast carcinoma
amplified sequence 1 | 2.80 |
| 208859_s_at | α
thalassemia/mental retardation syndrome X-linked (RAD54 homolog,
S. cerevisiae) | 2.76 |
| 221018_s_at | Tudor domain
containing 1///tudor domain containing 1 | 2.76 |
| 218876_at | Brain specific
protein///brain specific protein | 2.73 |
| 215081_at | KIAA1024
protein | 2.71 |
| 201510_at | E74-like factor 3
(ets domain transcription factor, epithelial-specific) | 2.69 |
| 210089_s_at | Laminin, α4 | 2.68 |
| 218859_s_at | Chromosome 20 open
reading frame 6 | 2.65 |
| 211626_x_at | v-ets
erythroblastosis virus E26 oncogene like (avian)///v-ets
erythroblastosis virus E26 oncogene like (avian) | 2.64 |
| 214316_x_at |
gb:AI378706/DB_XREF=gi:4188559/DB_XREF=tb91f09.×1/CLONE=IMAGE:2061737/FEA=EST/CNT=13/TID=Hs.16488.3/TIER=Stack/STK=13/UG=Hs.16488/LL=811/UG_GENE=CALR/UG_TITLE=calreticulin | 2.64 |
| 220657_at | Kelch-like 11
(Drosophila) | 2.61 |
| 206490_at | Discs, large
(Drosophila) homolog-associated protein 1 | 2.60 |
| 208383_s_at | Phosphoenolpyruvate
carboxykinase 1 (soluble) | 2.59 |
| 214884_at |
gb:AL033403/DB_XREF=gi:3859054/FEA=mRNA/CNT=15/TID=Hs.89543.1/TIER=ConsEnd/STK=0/UG=Hs.89543/LL=4168/UG_GENE=MCF2/UG_TITLE=MCF.2
cell line derived transforming sequence/DEF=Human DNA sequence from
clone 88D7 on chromosome Xq25-26.3 Contains F9 (coagulation factor
IX (plasma thromboplastic component, Christmas disease, haemophilia
B)), dbl oncogene. EST, STS, GSS | 2.59 |
| 201506_at | Transforming growth
factor, β-induced, 68 kDa | 2.18 |
| 213979_s_at | Consensus includes
gb:BF984434/FEA=EST/DB_XREF=gi:12387246/DB_XREF=est:
602307971F1/CLONE=IMAGE:4399313/UG=Hs.239737 C-terminal binding
protein 1 | 2.50 |
| 211253_x_at | Peptide YY | 2.38 |
| 206879_s_at | Neuregulin 2 | 2.33 |
| 208835_s_at | Cisplatin
resistance-associated overexpressed protein | 2.33 |
| 201506_at | Transforming growth
factor, β-induced, 68 kDa | 2.18 |
|
B, Genes
downregulated by Ro25-7386
|
| Probe set | Gene | Fold change |
|
| 202901_x_at | Cathepsin S | −64.37 |
| 201367_s_at | Zinc finger protein
36, C3H type-like 2 | −5.67 |
| 213606_s_at | Rho GDP
dissociation inhibitor (GDI) α | −−5.01 |
| 211136_s_at | Cleft lip and
palate associated transmembrane protein 1 | −4.59 |
| 204989_s_at | Integrin, β4 | −4.51 |
| 213042_s_at | ATPase, Ca++
transporting, ubiquitous | −4.42 |
| 216971_s_at | Plectin 1,
intermediate filament binding protein 500 kDa | −4.37 |
| 201167_x_at | Rho GDP
dissociation inhibitor (GDI) α | −4.14 |
| 219529_at | Chloride
intracellular channel 3 | −3.97 |
| 218813_s_at | SH3-domain
GRB2-like endophilin B2 | −3.93 |
| 211905_s_at | Integrin, β4 | −3.87 |
| 211672_s_at | Actin related
protein 2/3 complex, subunit 4, 20 kDa///actin related protein 2/3
complex, subunit 4, 20kDa | −3.70 |
| 207521_s_at | ATPase, Ca++
transporting, ubiquitous | −3.44 |
| 213986_s_at | Chromosome 19 open
reading frame 6 | −3.43 |
| 207824_s_at | MYC-associated zinc
finger protein (purine-binding transcription factor) | −3.42 |
| 203085_s_at | Transforming growth
factor, β1 (Camurati-Engelmann disease) | −3.34 |
| 203953_s_at | Claudin 3 | −3.26 |
| 211019_s_at | Lanosterol synthase
(2,3-oxidosqualene-lanosterol cyclase) | −3.22 |
| 209872_s_at | Plakophilin 3 | −3.20 |
| 214326_x_at | Jun D
proto-oncogene | −3.14 |
| 208677_s_at | Basigin (OK blood
group) | −3.12 |
| 201245_s_at | OTU domain,
ubiquitin aldehyde binding 1 | −3.08 |
| 203751_x_at | Jun D
proto-oncogene | −3.08 |
| 203370_s_at | PDZ and LIM domain
7 (enigma) | −3.05 |
| 203028_s_at | Cytochrome b-245, α
polypeptide | −3.02 |
| 210954_s_at | KIAA0669 gene
product | −2.99 |
| 211823_s_at | Paxillin | −2.97 |
| 200968_s_at | Peptidylprolyl
isomerase B (cyclophilin B) | −2.93 |
| 205463_s_at | Platelet-derived
growth factor α polypeptide | −2.87 |
| 210317_s_at | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein, ε
polypeptide | −2.87 |
| 211300_s_at | Tumor protein p53
(Li-Fraumeni syndrome) | −2.84 |
| 214251_s_at | Nuclear mitotic
apparatus protein 1 | −2.81 |
| 207722_s_at | BTB (POZ) domain
containing 2 | −2.80 |
| 216969_s_at | Kinesin family
member 22 | −2.79 |
| 203809_s_at | v-akt murine
thymoma viral oncogene homolog 2 | −2.76 |
| 218848_at | Hypothetical
protein MGC2655 | −2.73 |
| 212090_at | Glutamate receptor,
ionotropic, N-methyl D-asparate-associated protein 1 (glutamate
binding) | −2.69 |
| 201373_at | Plectin 1,
intermediate filament binding protein 500 kDa | −2.68 |
| 218302_at | Presenilin enhancer
2 | −2.68 |
| 213887_s_at | Polymerase (RNA) II
(DNA directed) polypeptide E, 25 kDa | −2.67 |
| 201369_s_at | Zinc finger protein
36, C3H type-like 2 | |
| 203392_s_at | C-terminal binding
protein 1 | −2.50 |
| 200796_s_at | Myeloid cell
leukemia sequence 1 (BCL2-related) | −2.43 |
| 206665_s_at | BCL2-like 1 | −2.35 |
In T47D cells, 16 upregulated genes and 3
downregulated genes modulated by LGD1069 were observed (Table IV), whereas 3 upregulated genes
and 5 downregulated genes were identified to be modulated by
LG100268 (Table V) with
alterations in fold induction >2-fold. According to the data,
several notable genes induced by LGD1069 and LG100268 in T47D cells
were identified, including cytochrome P450, dehydrogenase/reductase
member 3, metallothionein, neuro-oncological ventral antigen 1 and
regulator of G-protein signaling 1 (for LGD1069), and chemokine,
glutamate receptor, colon carcinoma-related protein and
insulin-like growth factor binding protein 7 (for LG100268). In
addition, 3 upregulated genes and 5 downregulated genes by
Ro25-7386 were identified with alterations in fold induction
>2-fold in T47D cells. Among them, chemokine (upregulated
genes), and glutamate receptor, ionotropic kainite 2, colon
carcinoma-related protein, insulin-like growth factor binding
protein 7 and growth differentiation factor 8 were identified
(Table VI).
 | Table IVGenes up- and downregulated by
LGD1069 in T47D cells. |
Table IV
Genes up- and downregulated by
LGD1069 in T47D cells.
A, Genes
upregulated by LGD1069
|
|---|
| Probe set | Gene | Fold change |
|---|
| 215653_at | Consensus includes
gb:AF339805.1/DEF=Homo sapiens clone IMAGE:248602, mRNA
sequence./FEA=mRNA/DB_XREF=gi:13507343/UG=Hs.326719 Homo sapiens
clone IMAGE:248602, mRNA sequence | 4.74 |
| 206424_at | Cytochrome P450,
family 26, subfamily A, polypeptide 1 | 2.98 |
| 202481_at |
Dehydrogenase/reductase (SDR family)
member 3 | 2.79 |
| 211689_s_at | Transmembrane
protease, serine 2///transmembrane protease, serine 2 | 2.61 |
| 213629_x_at | Metallothionein 1F
(functional) | 2.32 |
| 215924_at | Consensus includes
gb:AK022102.1/DEF=Homo sapiens cDNA FLJ12040 fis, clone
HEMBB1001944./FEA=mRNA/DB_XREF=gi:10433423/UG=Hs.296687 Homo
sapiens cDNA FLJ12040 fis, clone HEMBB1001944 | 2.32 |
| 208581_x_at | Metallothionein
1X | 2.31 |
| 210827_s_at | E74-like factor 3
(ets domain transcription factor, epithelial-specific) | 2.28 |
| 217165_x_at | Metallothionein 1F
(functional) | 2.23 |
| 204326_x_at | Metallothionein
1X | 2.19 |
| 206461_x_at | Metallothionein
1H | 2.15 |
| 204470_at | Chemokine (C-X-C
motif) ligand 1 (melanoma growth stimulating activity, α) | 2.14 |
| 204745_x_at | Metallothionein
1G | 2.13 |
| 217028_at | Chemokine (C-X-C
motif) receptor 4 | 2.04 |
| 212185_x_at | Consensus includes
gb:NM_005953.1/DEF=Homo sapiens metallothionein 2A (MT2A),
mRNA./FEA=CDS/GEN=MT2A/PROD=metallothionein
2A/DB_XREF=gi:5174763/UG=Hs.118786 metallothionein
2A/FL=gb:NM_005953.1 | 2.02 |
| 211456_x_at |
gb:AF333388.1/DB_XREF=gi:13310411/FEA=FLmRNA/CNT=1/TID=Hs.326774.0/TIER=FL/STK=0/UG=Hs.326774/DEF=Homo
sapiens metallothionein 1H-like protein mRNA, complete
cds./PROD=metallothionein 1H-like protein/FL=gb:AF333388.1 | 2.01 |
|
B, Genes
downregulated by LGD1069
|
| Probe set | Gene | Fold change |
|
| 207437_at | Neuro-oncological
ventral antigen 1 | −3.04 |
| 210806_at | KIAA0998 | −2.33 |
| 202989_at | Regulator of
G-protein signaling 1 | −2.13 |
 | Table VGenes up- and downregulated by
LG100268 in T47D cells. |
Table V
Genes up- and downregulated by
LG100268 in T47D cells.
A, Genes
upregulated by LG100268
|
|---|
| Probe set | Gene | Fold change |
|---|
| 215653_at | Consensus includes
gb:AF339805.1/DEF=Homo sapiens clone IMAGE:248602, mRNA
sequence./FEA=mRNA/DB_XREF=gi:13507343/UG=Hs.326719 Homo sapiens
clone IMAGE:248602, mRNA sequence | 4.74 |
| 215924_at | Consensus includes
gb:AK022102.1/DEF=Homo sapiens cDNA FLJ12040 fis, clone
HEMBB1001944./FEA=mRNA/DB_XREF=gi:10433423/UG=Hs.296687 Homo
sapiens cDNA FLJ12040 fis, clone HEMBB1001944 | 2.87 |
| 204470_at | Chemokine (C-X-C
motif) ligand 1 (melanoma growth stimulating activity, α) | 2.26 |
|
B, Genes
downregulated by LG100268
|
| Probe set | Gene | Fold change |
| 215655_at | Glutamate receptor,
ionotropic, kainate 2 | −3.29 |
| 220327_at | Colon
carcinoma-related protein | −2.94 |
| 213910_at | Insulin-like growth
factor binding protein 7 | −2.66 |
| 207145_at | Growth
differentiation factor 8 | −2.49 |
| 210806_at | KIAA0998 | −2.30 |
 | Table VIGenes up- and downregulated by
Ro25-7386 in T47D cells. |
Table VI
Genes up- and downregulated by
Ro25-7386 in T47D cells.
A, Genes
upregulated by Ro25-7386
|
|---|
| Probe set | Gene | Fold change |
|---|
| 215653_at | Consensus includes
gb:AF339805.1/DEF=Homo sapiens clone IMAGE:248602, mRNA
sequence./FEA=mRNA/DB_XREF=gi:13507343/UG=Hs.326719 Homo sapiens
clone IMAGE:248602, mRNA sequence | 4.74 |
| 215924_at | Consensus includes
gb:AK022102.1/DEF=Homo sapiens cDNA FLJ12040 fis, clone
HEMBB1001944./FEA=mRNA/DB_XREF=gi:10433423/UG=Hs.296687 Homo
sapiens cDNA FLJ12040 fis, clone HEMBB1001944 | 2.87 |
| 204470_at | Chemokine (C-X-C
motif) ligand 1 (melanoma growth stimulating activity, α) | 2.26 |
|
B, Genes
downregulated by Ro25-7386
|
| Probe set | Gene | Fold change |
|
| 215655_at | Glutamate receptor,
ionotropic, kainate 2 | −3.29 |
| 220327_at | Colon
carcinoma-related protein | −2.94 |
| 213910_at | Insulin-like growth
factor binding protein 7 | −2.66 |
| 207145_a | Growth
differentiation factor 8 | −2.49 |
| 210806_at | KIAA0998 | −2.30 |
In MDA-MB-231 cells, a total of 335 upregulated
genes and 320 downregulated genes modulated by LGD1069 were
observed (Table VII); whereas
118 upregulated genes and 432 downregulated genes were modulated by
LGD100268 (Table VIII) with
alterations in fold induction >2-fold. According to the data,
several notable genes were identified, including several types of
hypothetical protein, zinc finger homeobox 1b, recombination
activating gene 2 and tumor protein D52 (for LGD1069), and zinc
finger protein 21, Mdm2, and gonadotropin-releasing hormone 1 (for
LG100268).
 | Table VIIGenes up- and downregulated by
LGD1069 in MDA-MB-231. |
Table VII
Genes up- and downregulated by
LGD1069 in MDA-MB-231.
A, Genes
upregulated by LGD1069
|
|---|
| Probe set | Gene | Fold change |
|---|
| 219948_x_at | Hypothetical
protein FLJ21934 | 232.43 |
| 209672_s_at | Hypothetical
protein FLJ20323 | 69.61 |
| 207750_at |
gb:NM_018510.1/DEF=Homo sapiens
hypothetical protein PRO1866 (PRO1866), mRNA.
/FEA=mRNA/GEN=PRO1866/PROD=hypothetical protein
PRO1866/DB_XREF=gi:
8924091/UG=Hs.283031 hypothetical protein PRO1866/FL=gb:AF119858.1
gb:NM_018510.1 | 30.50 |
| 203603_s_at | Zinc finger
homeobox 1b | 10.18 |
| 217698_at | Consensus includes
gb:AV651668/FEA=EST/DB_XREF=gi:9872682/DB_XREF=est:AV651668/CLONE=GLCCSC04/UG=Hs.282480
ESTs | 10.11 |
| AFFX-r2- | E.
coli/GEN=bioB/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 9.76 |
| Ec-bioB- | 2393-2682 of
gb:J04423.1/DEF=E.coli 7,8-diamino-pelargonic acid (bioA), biotin
synthetase | |
| M_at | (bioB),
7-keto-8-amino-pelargonic acid synthetase (bioF), bioC protein, and
dethiobiot | |
| 205386_s_at | Mdm2, transformed
3T3 cell double minute 2, p53 binding protein (mouse) | 9.65 |
| 216119_s_at | Chromosome 20 open
reading frame 28 | 9.42 |
| AFFX- | E.
coli/GEN=bioB/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 9.32 |
| BioB-M_at | 2482-2739 of
gb:J04423.1/DEF=E.coli 7,8-diamino-pelargonic acid (bioA), biotin
synthetase (bioB), 7-keto-8-amino-pelargonic acid synthetase
(bioF), bioC protein, and dethiobiot | |
| 209613_s_at | Alcohol
dehydrogenase IB (class I), β polypeptide | 8.85 |
| AFFX-r2- | E.
coli/GEN=bioB/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 8.78 |
| Ec-bioB-3_at | 2772-3004 of
gb:J04423.1/DEF=E.coli 7,8-diamino-pelargonic acid (bioA), biotin
synthetase (bioB), 7-keto-8-amino-pelargonic acid synthetase
(bioF), bioC protein, and dethiobiot | |
| 217194_at | Consensus includes
gb:AB007970.1/DEF=Homo sapiens mRNA, chromosome 1 specific
transcript KIAA0501./FEA=mRNA/DB_XREF=gi:3413945/UG=Hs.223020 Homo
sapiens mRNA, chromosome 1 specific transcript KIAA0501 | 7.08 |
| 205524_s_at | Hyaluronan and
proteoglycan link protein 1 | 7.06 |
| 215514_at | Consensus includes
gb:AL080072.1/DEF=Homo sapiens mRNA; cDNA DKFZp564M0616 (from clone
DKFZp564M0616)./FEA=mRNA/DB_XREF=gi:5262482/UG=Hs.21195 Homo
sapiens mRNA; cDNA DKFZp564M0616 (from clone DKFZp564M0616) | 6.85 |
| 214774_x_at | Trinucleotide
repeat containing 9 | 6.70 |
| 215526_at | Consensus includes
gb:AL050145.1/DEF=Homo sapiens mRNA; cDNA DKFZp586C2020 (from clone
DKFZp586C2020)./FEA=mRNA/DB_XREF=gi:4884356/UG=Hs.225986 Homo
sapiens mRNA; cDNA DKFZp586C2020 (from clone DKFZp586C2020) | 6.22 |
| 211091_s_at | Neurofibromin 2
(bilateral acoustic neuroma) | 6.21 |
| 221959_at | Hypothetical
protein MGC39325 | 6.11 |
| 206863_x_at |
gb:U76376.1/DB_XREF=gi:1923234/GEN=HRK/FEA=FLmRNA/CNT=9/TID=Hs.87247.0/TIER=ConsEnd/STK=0/UG=Hs.87247/LL=8739/DEF=Homo
sapiens activator of apoptosis Hrk (HRK) mRNA, complete
cds./PROD=activator of apoptosis Hrk/FL=gb:NM_003806.1
gb:U76376.1 | 6.09 |
| 206202_at | Mesenchyme homeo
box 2 (growth arrest-specific homeo box) | 5.75 |
| 205288_at | CDC14 cell division
cycle 14 homolog A (S. cerevisiae) | 5.62 |
| 220931_at | Hypothetical
protein MGC5590 | 5.40 |
| 216795_at | CDNA: FLJ23194 fis,
clone REC00490 | 5.29 |
| 206410_at | Nuclear receptor
subfamily 0, group B, member 2 | 5.23 |
| 207647_at | Chromodomain
protein, Y-linked, 1///chromodomain protein, Y-linked, 1B | 5.19 |
| 215112_x_at | MCF.2 cell line
derived transforming sequence-like 2 | 5.11 |
| 216775_at | Ubiquitin specific
protease 53 | 4.90 |
| 220109_at | Transferrin | 4.88 |
| 217132_at | Clone 24587 mRNA
sequence | 4.86 |
| 216737_at | CDNA: FLJ20872 fis,
clone ADKA02604 | 4.84 |
| 220036_s_at |
Lipocalin-interacting membrane
receptor | 4.70 |
| AFFX-r2- | E.
coli/GEN=bioD/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 4.66 |
| Ec-bioD-3_at | 5312-5559 of
gb:J04423.1, not 100% identical/DEF=E.coli 7,8-diamino-pelargonic
acid (bioA), biotin synthetase (bioB), 7-keto-8-amino-pelargonic
acid synthetase (bioF), bioC pro | |
| 220564_at | Chromosome 10 open
reading frame 59 | 4.64 |
| 211611_s_at | Tenascin
XB///tenascin XB///cAMP responsive element binding protein-like
1///cAMP responsive element binding protein-like 1 | 4.61 |
| AFFX- | E.
coli/GEN=bioD/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | |
| BioDn-3_at | 5286-5570 of
gb:J04423.1, not 100% identical/DEF=E.coli 7,8-diamino-pelargonic
acid (bioA), biotin synthetase (bioB), 7-keto-8-amino-pelargonic
acid synthetase (bioF), bioC pro | 4.49 |
| 207272_at | Zinc finger protein
80 (pT17) | 4.49 |
| 210690_at | Killer cell
lectin-like receptor subfamily C, member 4 | 4.47 |
| 216625_at | Consensus includes
gb:AL050032.1/DEF=Homo sapiens mRNA; cDNA DKFZp566F1224 (from clone
DKFZp566F1224)./FEA=mRNA/DB_XREF=gi:4884272/UG=Hs.306307 Homo
sapiens mRNA; cDNA DKFZp566F1224 (from clone DKFZp566F1224) | 4.37 |
| 207245_at | UDP
glycosyltransferase 2 family, polypeptide B17 | 4.35 |
| 208014_x_at | Neuronal thread
protein AD7c-NTP | 4.32 |
| 214767_s_at | Heat shock protein,
α-crystallin-related, B6 | 4.31 |
| 216697_at | Triple functional
domain (PTPRF interacting) | 4.28 |
| 222341_x_at | Consensus includes
gb:AW973235/FEA=EST/DB_XREF=gi:8163081/DB_XREF=est:
EST385333/UG=Hs.293697 ESTs | 4.27 |
| 207262_at | Apolipoprotein
F | 4.25 |
| 222320_at | Consensus includes
gb:AW970584/FEA=EST/DB_XREF=gi:8160429/DB_XREF=est:
EST382665/UG=Hs.291033 ESTs | 4.14 |
| 206201_s_at | Mesenchyme homeo
box 2 (growth arrest-specific homeo box) | 4.06 |
| 208019_at | Zinc finger protein
157 (HZF22) | 4.01 |
| 204991_s_at | Neurofibromin 2
(bilateral acoustic neuroma) | 3.97 |
| 207607_at | Achaete-scute
complex-like 2 (Drosophila) | 3.88 |
| AFFX-r2- | E.
coli/GEN=bioD/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 3.83 |
| Ec-bioD-5_at | 5024-5244 of
gb:J04423.1/DEF=E.coli 7,8-diamino-pelargonic acid (bioA), biotin
synthetase (bioB), 7-keto-8-amino-pelargonic acid synthetase
(bioF), bioC protein, and dethiobiot | |
| 211315_s_at | Calcium channel,
voltage-dependent, α 1G subunit | 3.78 |
| 205953_at | Leucine-rich
repeats and immunoglobulin-like domains 2 | 3.75 |
| 207781_s_at | Zinc finger protein
6 (CMPX1) | 3.74 |
| 216068_at | Sodium- and
chloride-activated ATP-sensitive potassium channel | 3.69 |
| 214899_at | Hypothetical
BC331191_1 | 3.59 |
| 208212_s_at | Anaplastic lymphoma
kinase (Ki-1) | 3.58 |
|
B, Genes
downregulated by LGD1069
|
| Probe set | Gene | Fold change |
|
| 215117_at | Recombination
activating gene 2 | −60.45 |
| 217535_at | Consensus includes
gb:AV720514/FEA=EST/DB_XREF=gi:10817666/DB_XREF=est: -
AV720514/CLONE=GLCGSB09/UG=Hs.282721 ESTs, Weakly similar to
ALU7_HUMAN ALU SUBFAMILY SQ SEQUENCE CONTAMINATION WARNING ENTRY
H.sapiens | 16.22 |
| 201691_s_at | Tumor protein
D52 | −16.09 |
| 207674_at | Fc fragment of IgA,
receptor for | −6.54 |
| 215172_at | DKFZP566K0524
protein | −5.85 |
| 218541_s_at | Chromosome 8 open
reading frame 4 | −5.79 |
| 215350_at | Spectrin repeat
containing, nuclear envelope 1 | −5.69 |
|
AFFX-HUMRGE/M10098_5_at | H. sapiens/GEN=18S
rRNA/DB_XREF=gb:M10098.1/NOTE=SIF corresponding to nucleotides
115-595 of gb:M10098.1/DEF=Human 18S rRNA gene, complete. | −5.59 |
| 213652_at | Proprotein
convertase subtilisin/kexin type 5 | −5.57 |
| 216050_at | Transcribed locus,
moderately similar to NP_803425.1 DNA segment, Chr 19, brigham
& women’s genetics 1357 expressed [Mus musculus] | −5.43 |
| 222342_at | Consensus includes
gb:AW979196/FEA=EST/DB_XREF=gi:8170484/DB_XREF=est: -
EST391306/UG=Hs.292713 ESTs, Moderately similar to ALU1_HUMAN ALU
SUBFAMILY J SEQUENCE CONTAMINATION WARNING ENTRY H.sapiens | 5.41 |
| 205638_at | Brain-specific
angiogenesis inhibitor 3 | −5.04 |
| 217464_at | Consensus includes
gb:L48784/DEF=050 Homo sapiens
cDNA/FEA=mRNA/DB_XREF=gi:1066715/UG=Hs.182426 ribosomal protein
S2 | −4.97 |
| 205848_at | Growth
arrest-specific 2 | −4.86 |
| 206588_at | Deleted in
azoospermia-like | −4.75 |
| 213826_s_at | Consensus includes
gb:AA292281/FEA=EST/DB_XREF=gi:1940261/DB_XREF=
est:zt51b03.s1/CLONE=IMAGE:725837/UG=Hs.181307 H3 histone, family
3A | −4.74 |
| 220432_s_at | Cytochrome P450,
family 39, subfamily A, polypeptide 1 | −4.48 |
| 209227_at | Tumor suppressor
candidate 3 | −4.41 |
| 211712_s_at | Annexin
A9///annexin A9 | −4.31 |
|
AFFX-HUMRGE/M10098_M_at | H. sapiens/GEN=18S
rRNA/DB_XREF=gb:M10098.1/NOTE=SIF corresponding to nucleotides
688-1219 of gb:M10098.1/DEF=Human 18S rRNA gene, complete. | −4.28 |
|
AFFX-HUMRGE/M10098_3_at | Signal recognition
particle 68 kDa | −4.20 |
| 202648_at |
gb:BC000023.1/DB_XREF=gi:12652562/FEA=FLmRNA/CNT=966/TID=Hs.298262.
- 0/TIER=ConsEnd/STK=0/UG=Hs.298262/LL=6223/UG_GENE=RPS19/DEF= Homo
sapiens, ribosomal protein S19, clone MGC:1630, mRNA, complete
cds./PROD= ribosomal protein S19/FL=gb:M81757.1 g | 4.15 |
| 207815_at | Platelet factor 4
variant 1 | −4.15 |
| 205363_at | Butyrobetaine (γ),
2-oxoglutarate dioxygenase (γ-butyrobetaine hydroxylase) 1 | −4.14 |
| 213856_at | CD47 antigen
(Rh-related antigen, integrin-associated signal transducer) | −4.11 |
| 216087_at | MRNA full length
insert cDNA clone EUROIMAGE 117929 | −4.11 |
| 211264_at | Glutamate
decarboxylase 2 (pancreatic islets and brain, 65 kDa) | −4.03 |
| 220771_at | Melanoma
antigen | −3.83 |
| 220474_at | Solute carrier
family 25 (mitochondrial oxodicarboxylate carrier), member 21 | −3.81 |
| 220281_at | Solute carrier
family 12 (sodium/potassium/chloride transporters), member 1 | −3.80 |
| 217524_x_at | Consensus includes
gb:AA018923/FEA=EST/DB_XREF=gi:1482314/DB_XREF= -
est:ze58d03.s1/CLONE=IMAGE:363173/UG=Hs.261204 ESTs | 3.72 |
| 211776_s_at | Erythrocyte
membrane protein band 4.1-like 3///erythrocyte membrane protein
band 4.1-like 3 | −3.69 |
| 212681_at | Erythrocyte
membrane protein band 4.1-like 3 | −3.69 |
| 217333_at | Consensus includes
gb:AL031903/DEF=Human DNA sequence from clone 1032F13 on chromosome
Xq25-26.3. Contains a pseudogene similar to Keratin 18 (KRT18,
Cytokeratin 18) and ESTs/FEA=CDS/DB_XREF=gi:3766260/UG=Hs.247763
Human DNA sequence from clone 1032F1 | −3.69 |
| 210721_s_at |
p21(CDKN1A)-activated kinase 7 | −3.63 |
| 210327_s_at | Alanine-glyoxylate
aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria;
serine-pyruvate aminotransferase) | −3.57 |
| 206265_s_at |
Glycosylphosphatidylinositol specific
phospholipase D1 | −3.54 |
| 205847_at | Protease, serine,
22 | −3.52 |
| 202901_x_at | Cathepsin S | −3.42 |
| 204681_s_at | Rap guanine
nucleotide exchange factor (GEF) 5 | −3.35 |
| 222227_at | Zinc finger protein
236 | −3.35 |
| 207465_at | PRO0628
protein | −3.34 |
 | Table VIIIGenes upregulated and downregulated
by LG100268 in MDA-MB-231 cells. |
Table VIII
Genes upregulated and downregulated
by LG100268 in MDA-MB-231 cells.
A, Genes
upregulated by LG100268 in MDA-MB-231
|
|---|
| Probe set | Gene | Fold change |
|---|
| 219948_x_at | Hypothetical
protein FLJ21934 | 88.95 |
| 207750_at |
gb:NM_018510.1/DEF=Homo sapiens
hypothetical protein PRO1866 (PRO1866),
mRNA./FEA=mRNA/GEN=PRO1866/PROD=hypothetical protein
PRO1866/DB_XREF=gi:8924091/UG =Hs.283031 hypothetical protein
PRO1866/FL=gb:AF119858.1 gb:NM_018510.1 | 26.42 |
| 209672_s_at | Hypothetical
protein FLJ20323 | 14.63 |
| 215514_at | Consensus includes
gb:AL080072.1/DEF=Homo sapiens mRNA; cDNA DKFZp564M0616 (from clone
DKFZp564M0616)./FEA=mRNA/DB_XREF=gi:5262482/UG=Hs.21195 Homo
sapiens mRNA; cDNA DKFZp564M0616 (from clone DKFZp564M0616) | 9.11 |
| 215309_at | Transcribed locus,
weakly similar to XP_092995.4 zinc finger protein 21 (KOX 14)
[Homo sapiens] | 8.12 |
| 214774_x_at | Trinucleotide
repeat containing 9 | 7.58 |
| 203603_s_at | Zinc finger
homeobox 1b | 5.77 |
| 205386_s_at | Mdm2, transformed
3T3 cell double minute 2, p53 binding protein (mouse) | 5.20 |
| 205419_at | Epstein-Barr virus
induced gene 2 (lymphocyte-specific G protein-coupled
receptor) | 4.18 |
| 216978_x_at | Consensus includes
gb:U50277.1/DEF=Human breast cancer suppressor element Ishmael
Upper CP1 mRNA, partial cds./FEA=mRNA/PROD=suppressor element
Ishmael Upper CP1/DB_XREF=gi:1224126/UG=Hs.121485 Human breast
cancer suppressor element Ishmael Upper CP | 3.93 |
| 220931_at | Hypothetical
protein MGC5590 | 3.81 |
| 219995_s_at | Hypothetical
protein FLJ13841 | 3.77 |
| 208076_at | Histone 1, H4d | 3.6 |
| 214255_at | ATPase, Class V,
type 10A | 3.55 |
| 207987_s_at |
Gonadotropin-releasing hormone 1
(luteinizing-releasing hormone) | 3.52 |
| 205651_x_at | Rap guanine
nucleotide exchange factor (GEF) 4 | 3.46 |
| 220401_at | Hypothetical
protein FLJ21369 | 3.39 |
| 207241_at | Chromosome 4 open
reading frame 6 | 3.35 |
| 215623_x_at | SMC4 structural
maintenance of chromosomes 4-like 1 (yeast) | 3.17 |
| 216119_s_at | Chromosome 20 open
reading frame 28 | 3.13 |
| 217194_at | Consensus includes
gb:AB007970.1/DEF=Homo sapiens mRNA, chromosome 1 specific
transcript KIAA0501./FEA=mRNA/DB_XREF=gi:3413945/UG=Hs.223020 Homo
sapiens mRNA, chromosome 1 specific transcript KIAA0501 | 3.10 |
| 206381_at | Sodium channel,
voltage-gated, type II, α 2 | 3.09 |
| 212182_at | Nudix (nucleoside
diphosphate linked moiety X)-type motif 4 | 2.98 |
| 215112_x_at | MCF.2 cell line
derived transforming sequence-like 2 | 2.94 |
| 213747_at | Consensus includes
gb:AA047234/FEA=EST/DB_XREF=gi:1525134/DB_XREF=
est:zf50b09.s1/CLONE=IMAGE:380345/UG=Hs.223014 antizyme
inhibitor | 2.84 |
| 221683_s_at | Centrosome protein
cep290 | 2.80 |
| 211611_s_at | Tenascin
XB///tenascin XB///cAMP responsive element binding protein-like
1///cAMP responsive element binding protein-like 1 | 2.74 |
| 205421_at | Solute carrier
family 22 (extraneuronal monoamine transporter), member 3 | 2.66 |
| 213764_s_at | Microfibrillar
associated protein 5 | 2.62 |
| 217505_at | Hypothetical
protein MGC22679 | 2.61 |
| 222320_at | Consensus includes
gb:AW970584/FEA=EST/DB_XREF=gi:8160429/DB_XREF=est:
EST382665/UG=Hs.291033 ESTs | 2.61 |
| 216466_at | Neuron navigator
3 | 2.59 |
| AFFX-r2- | E.
coli/GEN=bioB/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 2.55 |
| Ec-bio | 2393-2682 of
gb:J04423.1/DEF=E.coli 7,8-diamino-pelargonic acid (bioA), biotin
synthetase | |
| B-M_at | (bioB),
7-keto-8-amino-pelargonic acid synthetase (bioF), bioC protein, and
dethiobiot | |
| 216775_at | Ubiquitin specific
protease 53 | 2.54 |
| 206201_s_at | Mesenchyme homeo
box 2 (growth arrest-specific homeo box) | 2.53 |
| AFFX- | E.
coli/GEN=bioD/DB_XREF=gb:J04423.1/NOTE=SIF corresponding to
nucleotides | 2.48 |
| BioDn-5_at | 4980-5256 of
gb:J04423.1, not 100% identical/DEF=E.coli 7,8-diamino-pelargonic
acid (bioA), biotin synthetase (bioB), 7-keto-8-amino-pelargonic
acid synthetase (bioF), bioC pro | |
| 216894_x_at | Cyclin-dependent
kinase inhibitor 1C (p57, Kip2) | 2.46 |
| 208019_at | Zinc finger protein
157 (HZF22) | 2.45 |
| 215803_at | Hypothetical
protein FLJ10178 | 2.44 |
| 222320_at | CDNA: FLJ23194 fis,
clone REC00490 | 2.44 |
|
B, Genes
downregulated by LG100268
|
| Probe set | Gene | Fold change |
|
| 217237_at | Zinc finger protein
423 | −78.6 |
| 215014_at | Consensus includes
gb:AL512727.1/DEF=Homo sapiens mRNA; cDNA DKFZp547P042 (from clone
DKFZp547P042)./FEA=mRNA/DB_XREF=gi:12224870/UG=Hs.232127 Homo
sapiens mRNA; cDNA DKFZp547P042 (from clone DKFZp547P042) | −17.74 |
| 213753_x_at | Eukaryotic
translation initiation factor 5A | −7.65 |
| 212382_at | Transcription
factor 4 | −5.74 |
|
AFFX-HUMRGE/M10098_5_at | H. sapiens/GEN=18S
rRNA/DB_XREF=gb:M10098.1/NOTE=SIF corresponding to nucleotides
115-595 of gb:M10098.1/DEF=Human 18S rRNA gene, complete | −5.58 |
| 211712_s_at | Annexin
A9///annexin A9 | −5.49 |
| 209227_at | Tumor suppressor
candidate 3 | −5.11 |
| 216917_s_at | Synaptonemal
complex protein 1 | −4.82 |
|
AFFX-HUMRGE/M10098_M_at | H. sapiens/GEN=18S
rRNA/DB_XREF=gb:M10098.1/NOTE=SIF corresponding to nucleotides
688-1219 of gb:M10098.1/DEF=Human 18S rRNA gene, complete | −4.31 |
| 210697_at | Zinc finger protein
257 | −4.11 |
| 215013_s_at | Ubiquitin specific
protease 34 | −3.97 |
| 209657_s_at | Heat shock
transcription factor 2 | −3.96 |
| 221009_s_at | Angiopoietin-like
4 | −3.90 |
| 205612_at | Multimerin 1 | −3.79 |
| 207613_s_at |
Calcium/calmodulin-dependent protein
kinase (CaM kinase) II α | −3.55 |
| 37232_at | KIAA0586 | −3.38 |
|
AFFX-HUMRGE/M10098_3_at | Signal recognition
particle 68 kDa | −3.37 |
| 204422_s_at | Fibroblast growth
factor 2 (basic) | −3.33 |
| 220638_s_at | Cas-Br-M (murine)
ecotropic retroviral transforming sequence c | −3.32 |
| 208098_at | Olfactory receptor,
family 12, subfamily D, member 3///olfactory receptor, family 12,
subfamily D, member 3///olfactory receptor, family 5, subfamily V,
member 1///olfactory receptor, family 5, subfamily V, member 1 | −3.29 |
| 213826_s_at | Consensus includes
gb:AA292281/FEA=EST/DB_XREF=gi:1940261/DB_XREF -
=est:zt51b03.s1/CLONE=IMAGE:725837/UG=Hs.181307 H3 histone, family
3A | 3.25 |
| 208453_s_at | X-prolyl
aminopeptidase (aminopeptidase P) 1, soluble | −3.20 |
| 207485_x_at | Butyrophilin,
subfamily 3, member A1 | −3.18 |
| 211032_at | COBL-like
1///COBL-like 1 | −3.11 |
| 220619_at | Chromodomain
helicase DNA binding protein 7 | −3.04 |
| 209318_x_at | Pleiomorphic
adenoma gene-like 1 | −3.00 |
| 201547_at | Jumonji, AT rich
interactive domain 1B (RBP2-like) | −2.99 |
| 206996_x_at | Calcium channel,
voltage-dependent, β1 subunit | −2.98 |
| 220114_s_at | Stabilin 2 | −2.95 |
| 216709_at | Hypothetical gene
supported by BC013370; BC034583 | −2.93 |
| 203555_at | Protein tyrosine
phosphatase, non-receptor type 18 (brain-derived) | −2.92 |
| 13267_at | KIAA1117 | −2.91 |
| 201122_x_at | Eukaryotic
translation initiation factor 5A | −2.89 |
| 213495_s_at |
gb:AW166067/DB_XREF=gi:6397592/DB_XREF=xf44g10.×1/CLONE=IMAGE:
-
2620962/FEA=EST/CNT=75/TID=Hs.98614.2/TIER=Stack/STK=51/UG=Hs.98614/LL=6238/UG_GENE=RRBP1/UG_TITLE=ribosome
binding protein 1 (dog 180kD homolog) | 2.89 |
| 220301_at | Chromosome 18 open
reading frame 14 | −2.88 |
| 214837_at | Albumin | −2.85 |
| 209700_x_at | Phosphodiesterase
4D interacting protein (myomegalin) | −2.84 |
| 216805_at | Transcribed locus,
moderately similar to XP_375099.1 hypothetical protein LOC283585
[Homo sapiens] | −2.84 |
| 221671_x_at | Immunoglobulin κ
constant | −2.79 |
| 214001_x_at |
gb:AW302047/DB_XREF=gi:6711724/DB_XREF=xr52f08.×1/CLONE=IMAGE:2763783/
-
FEA=EST/CNT=24/TID=Hs.76230.2/TIER=Stack/STK=20/UG=Hs.76230/LL=6204/UG_GENE=RPS10/UG_TITLE=ribosomal
protein S10 | 2.72 |
| 210047_at | Solute carrier
family 11 (proton-coupled divalent metal ion transporters), member
2 | −2.69 |
| 208367_x_at | Cytochrome P450,
family 3, subfamily A, polypeptide 4 | −2.66 |
| 219252_s_at | Family with
sequence similarity 51, member A1 | −2.65 |
| 205827_at |
Cholecystokinin | −2.63 |
Confirmation of the alterations of
modulation of RXRα target genes of HMECs by RT-qPCR and western
blot analysis
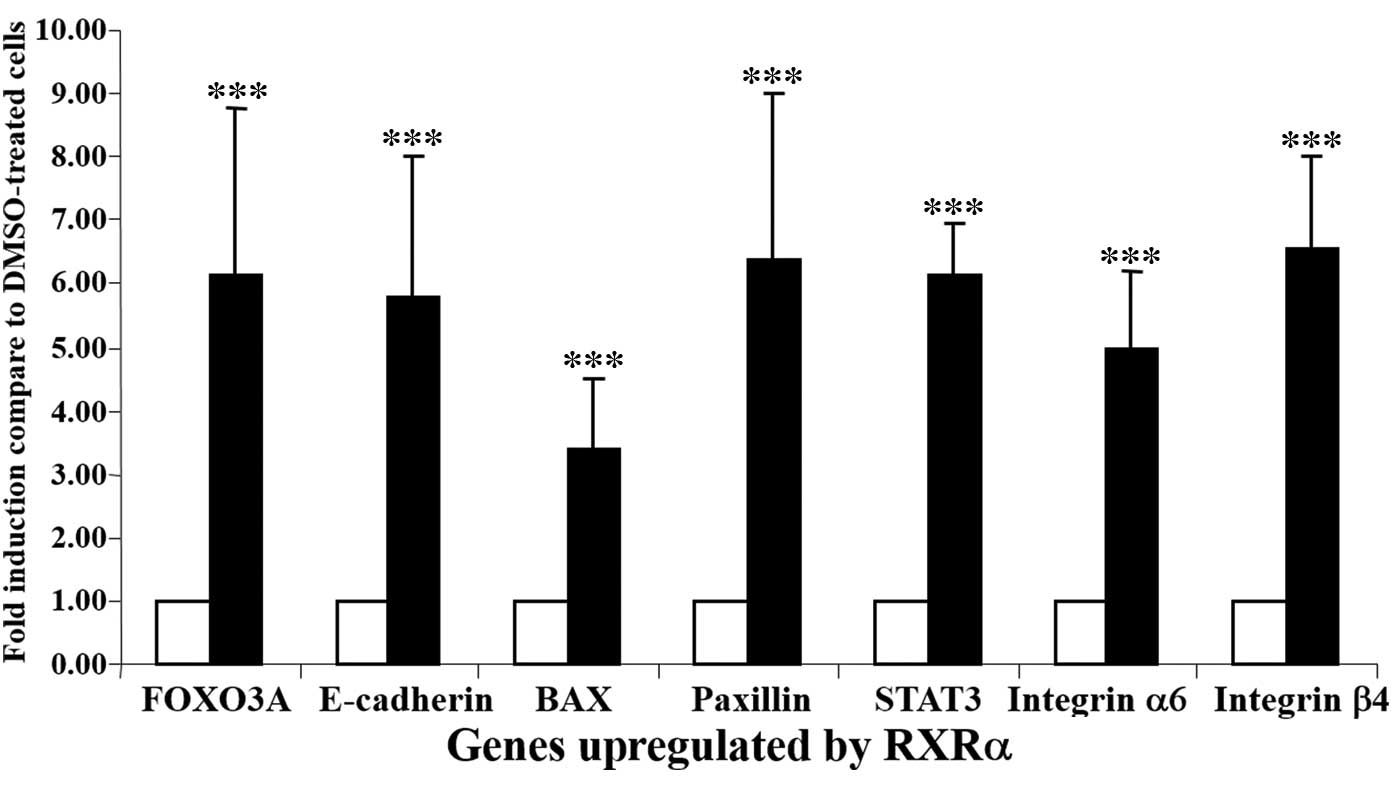
The induction of a total of 7 genes by rexinoid
(mRNA levels) was confirmed by RT-qPCR assays. These 7 genes are as
follows: Integrin β4, integrin α6, CDH1, PAX, BAX, FOXO3A and
STAT3; and upregulation of these genes by Ro25-7386 was confirmed
as demonstrated in Fig. 6. The
alterations in fold induction of protein levels of certain genes
were confirmed by western blot analysis; thus upregulation of BAX,
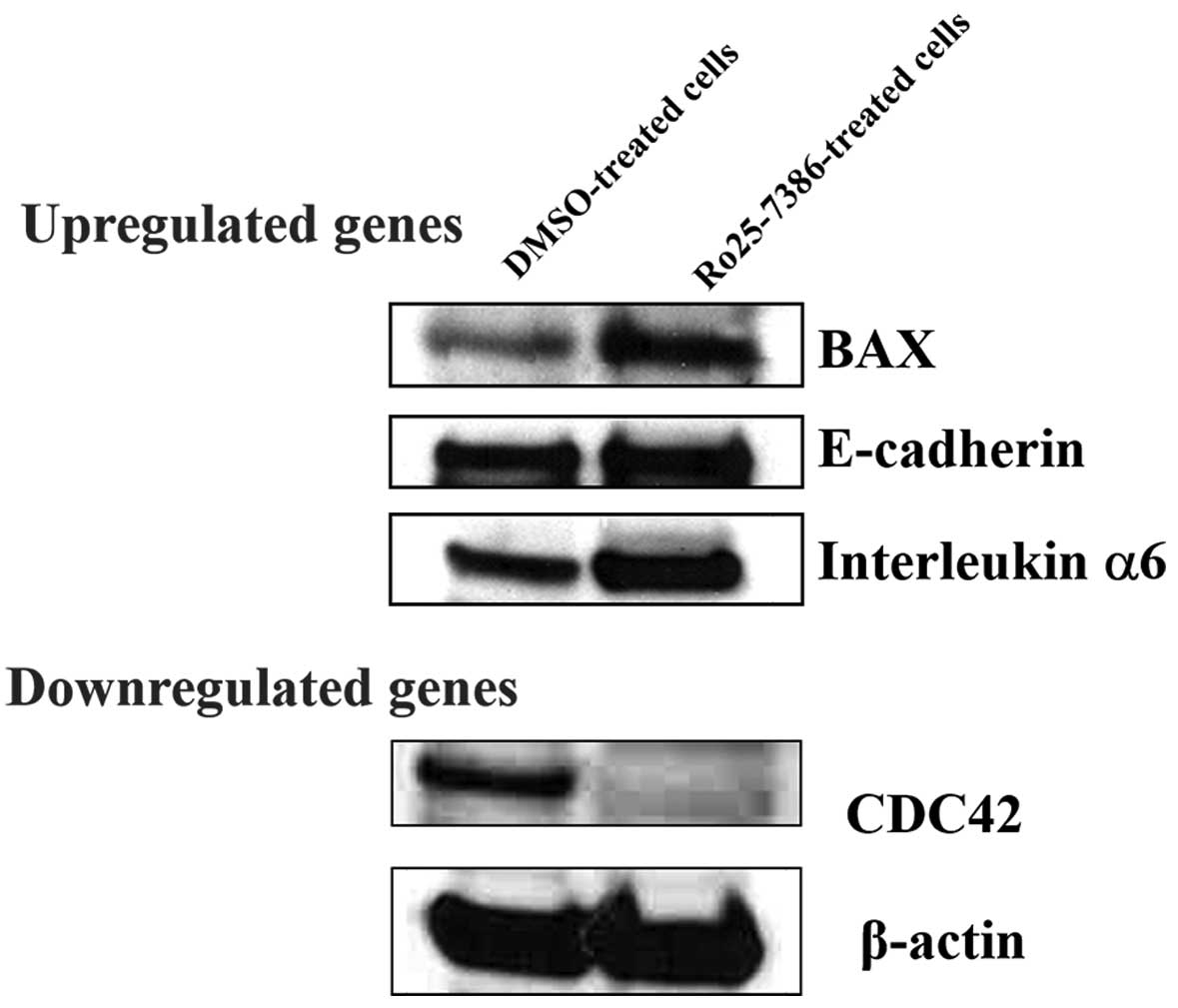
CDH1, interleukin α6 and the downregulation of CDC42 is shown in
Fig. 7.
Thorough investigation of the notable genes-CDH1,
FOXO3A, BAX (HMEC-Ro25-7386), insulin-like growth factor binding
protein 7 and growth differentiation factor 8 (T47D-Ro25-7386) and
cathepsin S, TGFβ2, basigin, MCL-1 and BCL2L1 (MCF-7-Ro25-7386),
may aid in the clarification of how RXRα agonists function to
inhibit breast cell growth. Such notable genes are implicated in
breast cancer management and are important for the treatment of
breast cancer. The current study may aid in the elucidation of
novel preventive/therapeutic targets for breast cancer, and may
contribute to the development of novel molecules, which may be able
to inhibit breast cancer development.
Discussion
In order to investigate the molecular mechanism by
which retinoids suppress breast cancer development, the current
study focused upon RXR-specific ligands (rexinoids). These have
been reported to suppress breast cancer development with minimal
toxicity compared with RAR-specific ligands (21), and it was the RXRα isoform that was
specifically focused upon in the present study that serves an
important role in tumor suppression.
The human RXRα gene spans over 40 kilobases in size
and consists of a minimum of 10 exons separated by introns ranging
in size from 700 base pairs (intron 3) to >7.8 kb (intron 4)
(26). It was observed that all of
the cell lines examined expressed RXRα. Notably, ER-negative breast
cancer cells, which do not respond to retinoid treatment, such as
MDA-MB-231 and MDA-MB-435 also expressed RXRα. This suggests that
RXRα is non-functional, losing DNA binding activity or failing to
recruit essential co-activators required for the activation of the
gene in ER-negative cells. Different and inappropriate
sub-localization of the receptor may also explain the
unresponsiveness of the cells to retinoid treatment.
LGD1069, LG100268 and Ro25-7386 were observed to
suppress the growth of breast cells, including the normal HMECs and
ER-positive breast cancer cells (MCF-7 and T47D). LGD1069 was
observed to induce a mild inhibition of MDA-MB-231 cell growth at a
dose of 10 μM. LG100268 did not affect the cell growth as
compared with LGD1069 in all four breast cancer cell lines
suggesting its weaker activity. This result indicates that LGD1069
may possess the ability to inhibit the growth of ER-negative breast
cancer.
The genes of interest were selected by referring to
the PathArt program, which demonstrated the association between
genes of several signaling pathways (data not shown). The
alterations in gene expression were then analyzed using the
Affymetrix microarray (human genome U133A 2.0) to determine which
genes are associated with the inhibition of cell growth induced by
the rexinoids. Among them, several genes were identified that are
involved in cell death, cell growth/maintenance, signal
transduction and response to stimulus, including E-cadherin, CtBP1,
integrin β4, integrin α6, PAX, BAX, FOXO3A, STAT3, collagen type VI
α3 and CDC42. It was additionally confirmed that Ro25-7386
upregulates the mRNA expression levels of FOXO3A, E-cadherin, BAX,
PAX, STAT3, integrin α6 and integrin β4. In addition, Ro25-7386 was
observed to increase the levels of BAX, E-cadherin and integrin α6
but reduce the level of CDC42. These results suggest that RXRa may
have a role in the prevention and treatment of breast cancer
development.
Further investigation regarding the functions of
selected genes may aid in the elucidation of novel
preventive/therapeutic targets for breast cancer, and may
additionally contribute to the development of novel molecules,
which may inhibit breast cancer progression.
Acknowledgments
The current study was supported by the Department of
Defense Breast Cancer Research Program Grants (grant no.
W81XWH04-1-0505). The present study was also supported in part by
the Basic Science Research Program through the National Research
Foundation of Korea funded by the Ministry of Education, Science
and Technology (grant no. NRF-2012R1A1A3004797) and in part by a
grant from the Traditional Korean Medicine Research and Development
Project, Ministry of Health & Welfare, Republic of Korea (grant
no. B120014).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar
|
|
3
|
Abu J, Batuwangala M, Herbert K and
Symonds P: Retinoic acid and retinoid receptors: potential
chemopreventive and therapeutic role in cervical cancer. Lancet
Oncol. 6:712–720. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Luca LM, Darwiche N, Celli G, Kosa K,
Jones C, Ross S and Chen LC: Vitamin A in epithelial
differentiation and skin carcinogenesis. Nutr Rev. 52:S45–S52.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gudas LJ, Sporn MB and Roberts AB:
Cellular biology and biochemistry of the retinoids. The Retinoids:
Biology, Chemistry and Medicine. Sporn MB, Roberts AB and Goodman
DS: Raven Press, Ltd; New York: pp. 443–520. 1994
|
|
6
|
Niles RM: Recent advances in the use of
vitamin A (retinoids) in the prevention and treatment of cancer.
Nutrition. 16:1084–1089. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simeone AM and Tari AM: How retinoids
regulate breast cancer cell proliferation and apoptosis. Cell Mol
Life Sci. 61:1475–1484. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Love JM and Gudas LJ: Vitamin A,
differentiation and cancer. Curr Opin Cell Biol. 6:825–831. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vivat-Hannah V, You D, Rizzo C, Daris JP,
Lapointe P, Zusi FC, Marinier A, Lorenzi MV and Gottardis MM:
Synergistic cytotoxicity exhibited by combination treatment of
selective retinoid ligands with taxol (Paclitaxel). Cancer Res.
61:8703–8711. 2001.PubMed/NCBI
|
|
10
|
Altucci L, Rossin A, Raffelsberger W,
Reitmair A, Chomienne C and Gronemeyer H: Retinoic acid-induced
apoptosis in leukemia cells is mediated by paracrine action of
tumor-selective death ligand TRAIL. Nat Med. 7:680–686. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chambon P: A decade of molecular biology
of retinoic acid receptors. FASEB J. 10:940–954. 1996.PubMed/NCBI
|
|
12
|
Giguère V: Retinoic acid receptors and
cellular retinoid binding proteins: Complex interplay in retinoid
signaling. Endocr Rev. 15:61–79. 1994.PubMed/NCBI
|
|
13
|
Zusi FC, Lorenzi MV and Vivat-Hannah V:
Selective retinoids and rexinoids in cancer therapy and
chemoprevention. Drug Discov Today. 7:1165–1174. 2002. View Article : Google Scholar
|
|
14
|
Thacher SM, Vasudevan J and Chandraratna
RA: Therapeutic applications for ligands of retinoid receptors.
Curr Pharm Des. 6:25–58. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagpal S and Chandraratna RA: Recent
developments in receptor-selective retinoids. Curr Pharm Des.
6:919–931. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong G, Kim HT, Wu K, et al: The retinoid
X receptor-selective retinoid, LGD1069, downregulates
cyclooxygenase-2 expression in human breast cells through
transcription factor crosstalk: implications for molecular-based
chemoprevention. Cancer Res. 65:3462–3469. 2005.PubMed/NCBI
|
|
17
|
Nagy L, Thomazy VA, Heyman RA and Davies
PJ: Retinoid-induced apoptosis in normal and neoplastic tissues.
Cell Death Differ. 5:11–19. 1998. View Article : Google Scholar
|
|
18
|
Lippman SM and Lotan R: Advances in the
development of retinoids as chemopreventive agents. J Nutr.
130(Suppl 2): 479S–482S. 2000.PubMed/NCBI
|
|
19
|
Gottardis MM, Bischoff ED, Shirley MA,
Wagoner MA, Lamph WW and Heyman RA: Chemoprevention of mammary
carcinoma by LGD1069 (Targretin): an RXR-selective ligand. Cancer
Res. 56:5566–5570. 1996.PubMed/NCBI
|
|
20
|
Wu K, Zhang Y, Xu XC, et al: The retinoid
X receptor-selective retinoid, LGD1069, prevents the development of
estrogen receptor-negative mammary tumors in transgenic mice.
Cancer Res. 62:6376–6380. 2002.PubMed/NCBI
|
|
21
|
Wu K, Kim HT, Rodriquez JL, et al:
Suppression of mammary tumorigenesis in transgenic mice by the
RXR-selective retinoid, LGD1069. Cancer Epidemiol Biomarkers Prev.
11:467–474. 2002.PubMed/NCBI
|
|
22
|
Crowe DL and Chandraratna RA: A retinoid X
receptor (RXR)-selective retinoid reveals that RXR-alpha is
potentially a therapeutic target in breast cancer cell lines and
that it potentiates antiproliferative and apoptotic responses to
peroxisome proliferator-activated receptor ligands. Breast Cancer
Res. 6:R546–R555. 2004. View
Article : Google Scholar
|
|
23
|
Farol LT and Hymes KB: Bexarotene: a
clinical review. Expert Rev Anticancer Ther. 4:180–188. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rigas JR and Dragnev KH: Emerging role of
rexinoids in non-small cell lung cancer: focus on bexarotene.
Oncologist. 10:22–33. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma Y, Koza-Taylor PH, DiMattia DA, Hames
L, Fu H, Dragnev KH, Turi T, Beebe JS, Freemantle SJ and Dmitrovsky
E: Microarray analysis uncovers retinoid targets in human bronchial
epithelial cells. Oncogene. 22:4924–4932. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li G, Walch E, Yang X, Lippman SM and
Clifford JL: Cloning and characterization of the human retinoid X
receptor alpha gene: conservation of structure with the mouse
homolog. Biochem Biophys Res Commun. 69:54–57. 2000. View Article : Google Scholar
|