1. Introduction
Apolipoprotein M (ApoM), first identified by Xu Ning
et al (1) in 1999, accounts
for ~5% of high-density lipoprotein (HDL) and <2% of low-density
lipoprotein (LDL). It has a density similar to that of HDL2
(1.063–1.125 g/l) and HDL3 (1.125–1.210 g/l), and is one of the
predominant components of HDL. The ApoM complex also contains HDL
(apoA-I, apoA-II, apoC-I, apoC-II, apoC-III and apoJ) and LDL
(apoB)-associated apolipoprotein (2). Plasma in adults has an ApoM content
of ~0.63–1.13 mmol/l (3). ApoM is
a newly identified protein with potential antioxidant activity and
anti-atherosclerotic effects through cholesterol efflux (2). It has also been associated with
diabetes, however, its specific biological functions and mechanisms
of action remain to be elucidated. In the present review, the roles
of ApoM gene regulation in lipid and HDL metabolism, and its
association with the development of specific diseases are
discussed, based on previous studies.
2. ApoM gene and protein
The ApoM gene is located on human chromosome 6p21.3,
covers a span of 2.3 kbp and is adjacent to the major
histocompatibility complex (MHC) class III region. It is a single
copy gene, containing six exons and five introns. The human ApoM
protein has a molecular weight of ~26 kD and preferentially exists
in HDL, followed by triglyceride-rich lipoprotein and LDL. Its
structure is associated with the lipocalin family (1). Previous expression microarray
analyses in multiple tissues (4)
revealed that the expression of ApoM is highly tissue-specific and
is specifically expressed in the liver and kidneys of humans. The
mouse ApoM gene is located on chromosome 17 and covers a span of
2.8 kbp (1). The sequence homology
of the human and mouse ApoM genes is ~79%, and each contain six
exons. ApoM is expressed in trace quantities in skeletal muscle and
gastrointestinal tissues of human embryos at an early stage (3–5
months), and reaches a peak at 5–9 months (4). At the embryonic stage in mice, the
expression of ApoM is detected at 7.5 days, peaks at 10.5 days and
then begins to decline. Gradually, it becomes expressed,
predominantly in the liver and renal tissues, and in trace
quantities in heart, brain, spleen and testicular tissues of mouse
embryos (5,6). The biological function of ApoM during
the embryonic period remains to be elucidated.
The first 20 amino acid residues of ApoM form a
hydrophobic signal peptide α-helix, which is retained in the mature
protein, due to the lack of cleavage sites on the signal peptide.
This feature is similar to two HDL apolipoproteins, paraoxonase
(PON-1) and haptoglobin-related protein (7–9). The
hydrophobic α-helices of ApoM and PON-1 are anchored to HDL
phospholipid monomolecular layers (9). Following mutation of Glu-22 of ApoM
to Ala, a functional hydrolysis site is produced, leading to the
hydrolysis of the signal peptide. In transgenic mice, mutant ApoM
was no longer associated with lipoprotein and was rapidly filtered
out through the kidney (9–11). In a previous study, in which HEK293
cells were stably transfected with the ApoM gene with a Q-22
mutation (10), culture of cells
with wild-type ApoM in serum-free medium expressed ApoM in trace
quantities compared with the cells with the ApoM mutation. The
addition of serum or purified HDL stimulated the wild-type cell
lines to secrete ApoM, which was further involved in the formation
of HDL. However, in the HepG2 cell line, which synthesized HDL
endogenously when cultured in serum-free medium, ApoM was only
secreted in HDL-like particles, indicating that less ApoM was
secreted without co-expression or exogenous addition of HDL to the
medium. This may be due to the retention by the signal peptide. The
results of this study confirmed that ApoM is an important component
of HDL, and that HDL may promote the secretion of ApoM (10).
The apolipoprotein family, also termed the
calycin-like protein family, has a common conservative structure
consisting of a central core enclosed by an eight-stranded β-barrel
structure, which is usually lined with hydrophobic or aromatic
amino acid residues, to facilitate the association of hydrophobic
molecules, including retinoic acid and 9-cis-retinoic acid
(6,12). Computer simulation imaging of the
three-dimensional structure has indicated that ApoM has the same
conserved structure as the apolipoprotein superfamily (13). The eight anti-parallel β-sheets of
ApoM are connected by seven inserted loops (13,14)
and six cysteines form three disulfide bonds (13). There are two glycosylation sites at
Asn-135 and Asn-148. Asn-135 is located in the periphery of the
sheets and is a potential glycosylation site (13). In the three-dimensional model,
characterized by an eight-stranded β-barrel structure, a fragment
with Asn-135 may have an open or closed conformation (13).
Christoffersen et al (15) analyzed the crystal structure of
amino acid residues 22–188 at the ApoM N-terminus, and demonstrated
that 1-phospho-sphingosine (S1P) was bound to the ‘calyx’-like
ligand domain in the hydrophobic domain of the ApoM molecule. S1P
hydrocarbon chains were observed to be close to the internal
section of the calyx, indicating the interaction between S1P-ApoM
and further involvement of the complex in HDL metabolism. Intrinsic
tryptophan fluorescence analyses has indicated that, on the basis
of structure and function, various subtypes of ApoM bind with
vitamin A, all-trans retinoic acid and 9-cis-retinoic acid
(15), supporting the concept of
ApoM as an apolipoprotein. The above evidence suggests that S1P and
retinoic acids are natural ligands of ApoM.
Plasma ApoM is primarily secreted from the liver,
however, the ApoM expressed in the kidneys may only be metabolized
in the kidneys (1). Studies have
demonstrated that megalin mediates the intake of kidney-derived
ApoM via the proximal tubules (16). Megalin is a membrane receptor,
which is highly expressed in the renal proximal tubule and mediates
the reabsorption of proteins, including lipid transfer proteins.
Kidney-derived ApoM may be a ligand of megalin (16), suggesting that ApoM may be involved
in the reabsorption of small molecular fat-soluble substances by
renal tubules. LDL contains five subtypes of ApoM (17), including three N-glycosylated and
sialyated subtypes, one N-glycosylated, but not sialyated, subtype
and one subtype without N-glycosylation or sialyation. The former
three subtypes account for 80–100% of the total ApoM in LDL. The
latter subtype accounts for ~20% of HDL and also contains five
subtypes of ApoM, which can be separated by hydrophilic interaction
chromatography (18). The
significance of polymorphism of these ApoM subtypes in health and
disease remains to be elucidated.
3. Regulation of ApoM
As the human ApoM gene is located at the MHC class
III region (1), with multiple
immune response-associated genes, it may be involved in the immune
response. Xu et al (19)
revealed that platelet-activating factor (PAF) significantly
increased the mRNA expression levels and the secretion of ApoM in
HepG2 cells. PAF or the PAF antagonist, lexipafant, selectively
regulate the expression of ApoM. Subsequently, Xu et al
(20) revealed that transforming
growth factor (TGF)α, epidermal growth factor and hepatocyte growth
factor reduce the mRNA expression levels of ApoM and ApoB in HepG2
cells, however, to a lesser extent compared with TGF-β. These four
cytokines had more significant effects on the mRNA expression
levels of ApoM compared with the mRNA expression levels of ApoB.
Hepatocyte nuclear factor (HNF)-1α is an important transcription
factor in the liver, kidney and pancreas. It is widely involved in
cell growth, differentiation and metabolic processes (21). HNF-1α binds to the promoter of ApoM
and activates the transcriptional activity of the ApoM gene through
conservative sites (−103 to −88), thereby directly regulating the
protein expression levels of ApoM (22). The expression of ApoM in the liver
or renal tissues in the HNF-1α gene knockout (HNF-1α−/−)
mice was minimal, whereas the levels of ApoM in the HNF-1α
transgenic (HNF-1α+/+) mice were 25 times higher.
Disorders of bile acid transport in HNF-1α−/− mice may
cause increased bile acid and cholesterol, therefore, affecting HDL
metabolism (23). Mosialou et
al (24) demonstrated that the
pro-inflammatory substance, phorbol ester, can induce the AP-1
transcription factor at the ApoM promoter region (−53/−47), to
recruit c-Jun and JunB protein kinase C associated proteins at
−214/−14, and competitively bind to the HNF-1 binding site of the
ApoM promoter, thereby leading to the downregulation of ApoM.
Liver receptor homolog-1 (LRH-1) is important in the
control of the early stage inflammatory reaction in hepatitis,
reabsorption of intestinal crypt cells, bile acid biosynthesis,
reverse cholesterol transport and other processes. Venteclef et
al (25) demonstrated that
ApoM was one of the target genes of LRH-1. LRH-1 binds to the LRH-1
response element in the proximal promoter region of ApoM (−83 to
−67) and directly regulates the expression of ApoM in humans and
mice. Liver X receptor (LXR) belongs to a liver receptor
superfamily and is a ligand-activated transcription factor involved
in regulation of inflammation and lipid metabolism (26). Zhang et al (27) revealed that LXR and its agonist,
T0901317, can reduce the expression of ApoM in vivo and
in vitro. Calayir et al (28) observed a decrease in the levels of
ApoM in liver treated with T0901317, however, the expression levels
of ApoM in the intestinal tissues and the adenosine
triphosphate-binding cassette transporter (ABCA1) were increased.
ABCA1 mediates cholesterol efflux to lipid-poor apoA-I, and ApoM
can promote the formation of pre-β-HDL via ABCA1, which is a basic
step for the formation of HDL (29). ABCA1 can regulate the expression of
ApoM in HepG2 cells via the retinoid X receptor (RXR)/LXR pathway
(30). Zhang et al
(31) demonstrated that the
expression of ApoM was reduced in HepG2 cells treated with
9-cis-retinoic acid and in combination with T0901317, can reduce
the mRNA expression levels of ApoM in HepG2 cells by 65%. However,
several studies have suggested that LXR has multiple effects on
ApoM. ApoM has high affinity for vitamin A and its derivatives,
all-trans retinoic acid and 9-cis-retinoic acid (15). Mosialou et al (32) demonstrated that the levels of ApoM
were significantly increased in HepG2 cells treated with
9-cis-retinoic acid. RXRα, HNF-4 and HNF-1, regulated by RXR, were
all increased within 2–6 h following treatment, however, these
returned to baseline levels within 8–24 h. HNF-4α can bind to
hormone response elements (HREs) at the proximal promoter (−33 to
−21) of ApoM. Following mutation in HRE, the primary activity of
the ApoM promoter declines to 10% of the control group, and
HNF-4α-mediated transcription of the ApoM promoter is completely
prevented. In addition to HNF-4α, a variety of receptor ligands of
HREs can significantly induce the gene transcription of human ApoM
and the activity of the ApoM promoter in HepG2 cells, and mutations
in proximal HREs can lead to loss of function (32). Shih et al (23) demonstrated that HNF-1 regulates the
expression of fragile X mental retardation syndrome-related protein
1 (FXR-1) through nuclear receptor subfamily 1, group H, member 4
(Nr1h4) gene, encoding FXR-1, and thereby regulates the activities
of Src homology region 2 domain-containing phosphatase-1 (SHP-1)
and cholesterol 7 α-hydroxylase (CYP7A1). These results suggested
that in bile acid and cholesterol metabolism, HNF-1 and FXR-1 may
regulate ApoM through interactions with a number of factors.
Venteclef et al (25)
revealed that bile acids suppress the expression of ApoM in an
SHP-dependent manner in vitro and in vivo, by
inhibiting the transcriptional activity of LRH-1 on ApoM. It has
also been confirmed (27) that, in
HepG2 cells treated with HNF-1α small interfering RNA, the levels
of ApoM are significantly reduced, as are apoA-I, CYP7A1, FXR and
SHP-1 (27). This suggests that,
in addition to directly regulating ApoM, HNF-1α may interact with
other regulatory factors. Estrogen and androgen are also involved
in the regulation of ApoM (33).
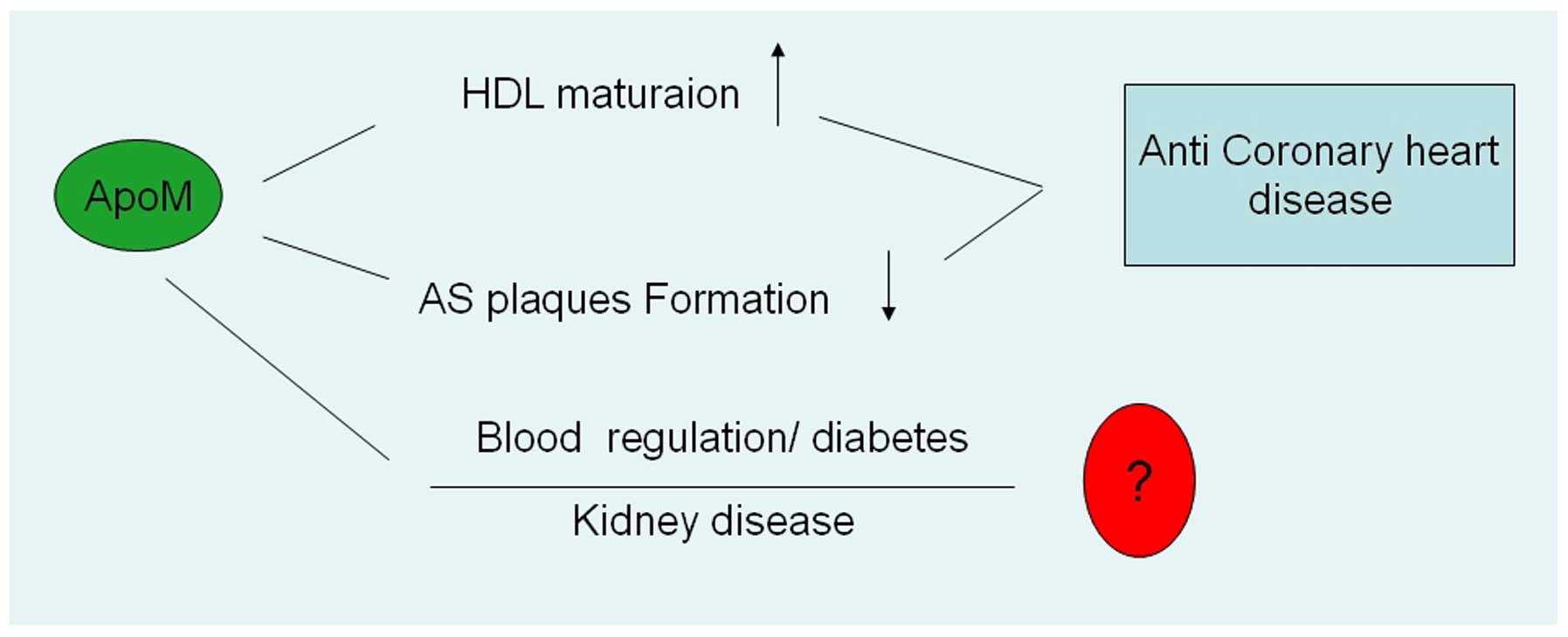
4. Association with diseases
Although the ApoM is predominant in HDL, the
biological functions and mechanism of ApoM in HDL and disease
remain to be elucidated. ApoM gene regulation has been reported in
lipid and HDL metabolism. ApoM also participate in the formation of
atherosclerotic plaques. ApoM is associated with diabetes and the
development of kidney disease and other diseases (Fig 1).
Association with coronary heart disease
(CHD)
HDL particles have anti-atherosclerotic effects,
predominantly by enabling the reversal of cholesterol transport to
the liver, to be excreted along with bile (34–36).
Mature HDL originates from nascent HDL, which is either produced by
the liver or from the lipolysis of very low density lipoprotein
(VLDL) and chylomicrons, along with fat-free ApoA-I or poor fat
preβ-HDL (29). Intracellular
cholesterol, transferred by preβ-HDL, is esterified by lecithin
cholesterol acyltransferase (LCAT) from its mature form, which
further promotes the maturation of preβ-HDL and increases the
particle size (37,38). Esterified cholesterol in mature HDL
particles can be phagocytized by the liver cells via pinocytosis,
and the triglycerides and cholesterol esters can be indirectly
exchanged to LDL (39). The
particle density of ApoM is close to that of HDL2 and HDL3. In
vitro experiments have confirmed that HDL (ApoM+) is more
effective as an antioxidant compared with HDL (ApoM-) (2), and promotes cholesterol efflux from
THP-1 cells (2,39,40).
Furthermore, LDL (ApoM+) is more resistant to oxidation compared
with LDL (ApoM-) (2). ApoM can
affect HDL particle conversion through LCAT (41). It has been confirmed that ApoM
enables preβ-HDL to be transformed into mature HDL particles, which
are involved in cholesterol efflux and may have
anti-atherosclerotic effects (42). In LDL receptor-deficient mice fed a
high cholesterol diet, overexpression of the ApoM gene increases
plasma concentrations of ApoM and delays the development of early
atherosclerosis (42). In a
previous study involving the treatment of C57BL/6 mice with ApoM
siRNA, the expression of ApoM was reduced by 90% and HDL-c was
reduced by 25%. In addition, preβ-HDL was almost absent in
TCF1−/− mice and mice treated with TCF siRNA. In
transgenic mice injected with HDL2 particles, large particle HDL1,
rather than small particle HDL, was detected, suggesting that
ApoM-HDL2 particles were transformed into preβ-HDL and promoted HDL
maturation (37–39). In LDLR−/− mice injected
with adenovirus-ApoM, the plasma levels of HDL were increased by
40% compared with control mice, the average incidence of
atherosclerosis of the aortic root was reduced to 28% of the
control group, and lesion areas, assessed using positive oil red O
staining, of the large artery and aortic root, were reduced by 70%.
This suggested that ApoM protected the hypercholesterolemia
LDLR−/− mice from atherosclerotic plaque formation
(42). Although in vivo and
in vitro investigations have indicated that ApoM exhibits
anti-atherosclerotic effects, Ahnström suggested, through case
control investigations, that ApoM levels in patients with CHD were
unchanged compared with healthy control subjects (43). The effects of ApoM on human
patients with CHD requires clarification in future experiments.
Christoffersen reported that ApoM is not associated with CHD as it
is, in part, associated with plasma LDL (40). In LDLR−/− mice,
overexpression of ApoM led to increased levels of VLDL/LDL-c, and
ApoM gene silencing led to decreased levels of VLDL/LDL-c. However,
ApoM did not affect the levels of VLDL/LDL-c in mice with complete
LDLR. In mice with an absence of functional LDLR, the ApoM-VLDL/LDL
rich particles were removed and were at lower levels compared with
the control VLDL/LDL particles, suggesting that ApoM decreased
VLDL/LDL catabolism without depending on LDLR. ApoM reduced the
atherosclerotic area in ApoE−/− mice and wild-type mice
fed a high fat/high cholesterol diet. However, in
LDLR−/− mice, the anti-atherosclerotic effect of ApoM
was reduced, due to increased levels of VLDL/LDL. Loss of LDLR
function led to increased levels of ApoM and VLDL/LDL, which
reflected another aspect of the anti-atherosclerotic effects of
ApoM. Christoffersen (40) also
demonstrated a reciprocal interchange of ApoM between various
lipoproteins in vivo. In wild-type mice, plasma ApoM was
primarily associated with HDL particles, however, in transgenic
mice, it was also observed in other abnormally elevated
lipoproteins. In LDLR−/− mice, ApoM was enriched in LDL
particles. In apoE−/− mice fed with normal diets, ApoM
was predominantly present in HDL and was also detected in LDL and
VLDL. In mice fed with a high fat/high cholesterol diet, ApoM was
significantly increased in the VLDL of the intestine, but was only
detected in trace quantities in HDL. The levels of HDL and ApoM
levels in apoA-I−/− mice were significantly lower
compared with wild type mice. However, ApoM remained primarily in
the HDL particles (17,40,44,45).
These findings suggest that additional investigations into the
cross metabolic conversion of various lipoproteins are required to
further improve understanding of the function of ApoM in lipid
metabolism.
Elsøe et al (46,47)
observed cholesterol metabolism in peripheral cells in
ApoM-knockout mice (ApoM−/−) and ApoM-transgenic
(ApoM−Tg) mice, and demonstrated that ApoM-HDL particles
increased the efflux of 3H-labeled cholesterol from
macrophages in the ApoM−Tg mice by >24±3% compared
with the wild-type mice (P<0.05). Following intra-peritoneal
injection of foam cells with 3H-labeled cholesterols
into ApoM−/− and ApoM−Tg mice, increased
plasma ApoM did not significantly promote the transport of
intracellular cholesterol to the plasma. However, following
continuous intravenous infusion of 13C-2-labeled
cholesterol/fat emulsion into ApoM−/−,
ApoM−Tg and wild-type mice for 5 h, the enrichment rate
of free cholesterols and 13C-2-labeled cholesterol in
the blood of the ApoM−Tg mice were lower compared with
the ApoM−/− and wild-type mice. This suggested that
levels of plasma ApoM promotes the mobilization of intracellular
cholesterol transport in plasma. However, ApoM had no significant
effect on the excretion of cholesterol into the feces.
The anti-inflammatory effects of ApoM may also be
associated with its anti-atherosclerotic effects (48). In patients with sepsis with or
without shock, and sepsis and infection with or without systemic
inflammatory response syndrome, the average plasma levels of ApoM
are all reduced, suggesting that ApoM can be used as a novel
biomarker of sepsis (49,50). The majority of S1P is bound to HDL
in plasma, which increases the protective effects of HDL on
endothelial cells (51). S1P-ApoM
can activate the S1P-1 receptor and affect the functions of
endothelial cells. S1P-ApoM is important in lipid metabolism,
lipoprotein metabolism and the pathogenesis of atherosclerosis
(52). These studies suggest novel
directions for the investigation of ApoM. Through the
overexpression and intraperitoneal injection of 17C-S1P,
Makoto (53) demonstrated that
17C-S1P only existed in the liver cells and plasma, and
was not present in the peripheral blood cells. Following the
overexpression of ApoM in HepG2 cells and the mouse liver, the
levels of S1P in the medium/plasma and peripheral blood
cells/hepatocytes increased. Following adenoviral mediated
overexpression of ApoM in hepatocytes, the levels of S1P in the
plasma were increased, but no increase was observed in the
peripheral blood cells. Further results indicated that S1P can
associate with ApoM and affect the metabolism of S1P. Analysis of a
single nucleotide of the proximal promoter region of the ApoM gene
revealed that a single nucleotide C in ApoM T-778C, T-855C of the
CHD group increased the risk of CHD, and the plasma total
cholesterol levels in populations with CC and CT genotypes were
significantly higher than a population with the TT genotype. These
results suggested that polymorphisms of the ApoM gene promoter were
risk factors for CHD (54–55). A previous meta-analysis (56) indicated that the ApoM T-778C
polymorphism was associated with plasma LDL-c, and the levels of
total cholesterol and LDL-c in CT+CC groups were higher compared
with those in the TT group. The CT+CC genotypes exhibited a higher
risk of CHD.
Association with diabetes
An analysis of ~2,500 single nucleotide
polymorphisms (SNPs), located in the MHC region, demonstrated that
ApoM may be associated with the development of type 1 diabetes.
ApoM was also revealed to interact with Kruppel-like factor 6, a
tumor suppressor of prostate cancer, located on 10p15, and
C8orf30A, a brain protein on 8q24 of unknown function, which were
not associated with type 1 diabetes. These results require further
clarification in further investigations (57). Wu et al (58) revealed that an SNP of the T-778C
polymorphic locus in the proximal promoter of ApoM was
significantly associated with type 1 diabetes, and Zhou et
al (59) demonstrated that
T-778C was associated with cholesterol levels and susceptibility
for type 2 diabetes, suggesting that T-778C of the ApoM gene
promoter may be involved in the co-pathogenesis of type 1 and type
2 diabetes. The investigation of 17 SNPs of ApoM revealed that ApoM
polymorphisms were associated with the susceptibility for type 2
diabetes in a population in Hong Kong, however, in patients who had
suffered from the disease for >10 years, the rs805297C allele
was significantly associated with type 2 diabetes and the rs707922
TT genotype significantly increased the levels of TC and LDL-C in
patients with type 2 diabetes (59).
Leptin is a hormone-like protein, primarily secreted
by fat cells, and ob is the protein product of the gene. Leptin is
important in regulating food intake and energy metabolism in the
body. It affects obesity, diabetes, cardiovascular disease,
ischemia-reperfusion injury and other cellular metabolic pathways
(60). Xu et al (61,62)
performed a case control study on obese females and females with
normal body weight, and demonstrated that ApoM was positively
correlated with levels of leptin and negatively correlated with
levels of cholesterol. Further results (63) have demonstrated that the in
vivo expression levels of ApoM in ob/ob mice with leptin
deficiency and db/db mice with leptin receptor deficiency, were
significantly lower compared with control groups. However,
treatment with leptin significantly increased the expression levels
of ApoM in ob/ob mice. Luo et al (64) demonstrated that in vitro
treatment with recombinant leptin, at doses higher than the
physiological dose (>100 ng/ml), inhibits the expression of ApoM
in HepG2 cells in a dose-dependent manner, and leptin at
physiological doses (10 ng/ml) can downregulate the expression of
ApoM. ApoM may therefore be a significant independent predictor in
the assessment of HDL apoA-I and apoA-II catabolism in overweight
and obese male patients with insulin resistance (65). ApoM is positively correlated with
PCSK9 of the LDLR pathway in populations with a lean body mass,
however, it is not correlated in overweight or obese patients
(66).
The pathogenesis of maturity-onset diabetes of the
young (MODY) is primarily due to heterozygous mutations in the
HNF-1α gene, leading to a deficiency of insulin secretion in
pancreatic β cells of the islet. Richter (22) demonstrated that plasma
concentrations of ApoM in patients with MODY3 were 36% lower than
in the normal control group (0.65, vs. 1.0; P=0.02), and that ApoM
was deficient in HNF-1α−/− mice (22). In a case control study, Cervin
(67) suggested that the levels of
ApoM in females carrying the HNF-1α P291fsinsC mutation were 10%
lower than those in the control group, and that ApoM was not useful
as a biomarker of the HNF-1α mutation (67). However, Mughal (68) reported that the levels of ApoM in
HNF1A-MODY patients were significantly lower compared with patients
with type 1 diabetes. Detection of the levels of ApoM can
differentiate HNF1α-MODY and type 1 diabetes, and thereby provide
an experimental basis for further diagnosis and treatment. In a
mouse model of alloxan-induced diabetes, the plasma levels of
insulin and ApoM were significantly lower compared with mice
injected with saline, and insulin treatment increased the
expression of ApoM in the liver and kidney tissues (69). High blood sugar levels can
significantly inhibit the in vivo and in vitro
expression of ApoM (70). Plasma
levels of ApoM in mice with alloxan-induced diabetes were lower
compared with normal mice, however, insulin levels were elevated
following short-term infusion of high glucose, and the levels of
ApoM in mice with high blood sugar were significantly decreased
(69,70). In HepG2 cells, glucose and insulin
reduced the levels of ApoM, suggesting that insulin had indirect
regulatory effects on ApoM in vivo (70). In HepG2 cells, low doses of insulin
and insulin-like growth factor 1 suppressed the levels of ApoM in
the cells in a time- and dose-dependent manner. The insulin-induced
downregulation of ApoM can be inhibited by the insulin-specific
receptor inhibitor, AG1024, and the phosphatidylinositol 3-kinase
(PI3-K) inhibitor, LY294002, and peroxisome proliferator-activated
receptors β/δ agonists (GW501516) suppress the expression of ApoM
in HepG2 cells. This suggests that ApoM can be regulated through
the PI3-K pathway (71), and the
above-mentioned findings suggest that insulin may regulate ApoM
through the PI3-K pathway in vivo and in vitro.
PI3-K/Akt is an important signaling pathway for insulin metabolism.
Wolfrum et al (42)
demonstrated that insulin activates phosphorylated PI3-K/Akt and
induces the phosphorylation of Ser-156 at conservative sites of
hepatocyte nuclear factor Foxa2, which interacts with Akt in the
PI3-K pathway and inhibits the Foxa2-dependent transcription
pathway. A Foxa2 Thr-156 mutant can resist the Akt-mediated
phosphorylation and Foxa2-dependent transcriptional inactivation.
Foxa2 is known to bind to the −474 to −462 region of the ApoM
promoter to directly regulate ApoM. The functions of ApoM and its
role in the development of diabetes require further investigation
(72).
Association with other diseases
Human ApoM is specifically expressed in the liver
and kidney (4). However, Luo et
al (73) revealed that ApoM is
expressed in human intestinal tissues in trace quantities and may
be associated with lymph node metastasis of colorectal cancer. Gu
et al (74) compared the
serum levels of ApoM in 126 patients with hepatitis B virus (HBV)
and 118 healthy controls, and demonstrated that the serum levels of
ApoM in patients were 27% higher compared with the control group.
HBV increased the promoter activity of ApoM and increased the
translation and expression of the gene. Plasma levels of ApoM in
patients with hepatocellular carcinoma (HCC) are significantly
higher compared with normal subjects, however, the mRNA expression
levels of ApoM in cancer tissues are lower compared with adjacent
tissues (75). In a mouse model of
hepatic ischemia-reperfusion, a gradual increase in the mRNA
expression levels of ApoM was observed within 0.5–3 h, and the
levels of ApoM declined to baseline levels within 6–24 h, however,
the function of ApoM in ischemia remains to be elucidated (76). The human ApoM gene is adjacent to
the MHC class III region, which is closely associated with the
pathogenesis of rheumatoid arthritis (RA). Single gene analysis of
patients with RA and healthy controls revealed that allele A was
associated with blood lipid disorders in patients with RA and ApoM
C-1065A, and may increase the risk of atherosclerosis in patients
with RA (77). The ApoM T-778C and
C-1065A SNPs have been associated with high risk of stroke in the
Chinese population (78), and the
ApoM gene polymorphism has also been associated with emphysema and
altered lung function (79).
Obstructive sleep apnea (OSA) may be a risk factor
for CHD. Proteomic analysis has revealed that the protein
expression levels of ApoM in patients with OSA are abnormal, and
multivariate regression analysis indicated that ApoM was an
independent risk factor for OSA (80). In addition, patients with metabolic
syndrome exhibit lower plasma levels of ApoM, however, this cannot
predict vascular media thickness (IMT). Plasma levels of ApoM in
patients with MetS was reported to be ~15% lower than in
individuals without MetS (P=0.036), and multivariate regression
analysis suggested that IMT was only associated with MetS (P=0.05)
(81). Ahnström et al
(82) compared 343 patients with
abdominal aortic aneurysm (AAA) with 214 healthy volunteers, and
demonstrated that the levels of ApoA-I, apoB and ApoM in patients
with AAA were significantly lower than the healthy controls. The
levels of ApoA-I were 1.62, vs. 2.08 g/l, levels of apoB were 0.91,
vs. 1.04 g/l and the levels of ApoM were 0.72, vs. 0.91 mmol/l (all
P<0.0001). However, multivariate analysis demonstrated that only
apoA-I and apoB were associated with AAA. Ahnström et al
(83) also demonstrated that the
levels of Apo in patients with critical limb ischemia, a relatively
severe type of atherosclerosis, were lower compared with those in
the healthy control group. The levels of ApoA-I were 1.23, vs. 2.08
g/l, levels of apoB were 0.93 vs. 1.04 g/l and levels of ApoM
levels were 0.75, vs. 0.91 mmol/l (all P<0.0001). However,
multivariate regression analysis revealed that only apoA-I was an
independent risk factor for critical limb ischemia. In the kidneys
of 11 Sprague Dawley rats, Xu et al (76) induced 45 min ischemia, with
reperfusion 24 h after the ischemia, and demonstrated that plasma
levels of urea nitrogen and creatine were significantly increased
between 6 and 24 h. The plasma levels of ApoM were increased
significantly 45 min after ischemia and 2 h after reperfusion, and
urine levels of ApoM were also significantly increased between 2
and 6 h after reperfusion. These results suggested that ApoM
functions as an acute inflammatory protein and can serve as a
marker of acute kidney injury.
5. Prospects
ApoM is important in the development of CHD. It can
promote HDL maturation and reverse the formation of atheromatous
plaques. Therefore, it can serve as a novel marker for the
diagnosis and target for the treatment of CHD. In the pathogenesis
of diabetes, the complex mechanisms of insulin and blood sugar
regulation, and their association with the development of diabetes,
remain to be elucidated. ApoM is highly expressed in kidney
tissues, is a ligand for renal proteins and can prevent the loss of
lipid molecules in kidneys. However, its role in kidney disease
also remains to be elucidated and its physiological and
pathological functions in renal tissues require further
investigation.
Acknowledgments
The authors would like to thank the participants for
their dedication of this study. This study was supported by grants
from the Science and technology Project in Wuhu (no. 2013cxy04),
the National Natural Science Foundation of China (no. 81200632) and
Anhui Provincial Natural Science Foundation (to Yao Zhang, no.
1508085MH149).
References
|
1
|
Xu N and Dahlbäck B: A novel human
apolipoprotein (apoM). J Biol Chem. 274:31286–31290. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Christoffersen C, Nielsen LB, Axler O,
Andersson A, Johnsen AH and Dahlbäck B: Isolation and
characterization of human apolipoprotein M-containing lipoproteins.
J Lipid Res. 47:1833–1843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Axler O, Ahnström J and Dahlbäck B: An
ELISA for apolipoprotein M reveals a strong correlation to total
cholesterol in human plasma. J Lipid Res. 48:1772–1780. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang XY, Dong X, Zheng L, et al: Specific
tissue expression and cellular localization of human apolipoprotein
M as determined by in situ hybridization. Acta Histochem.
105:67–72. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang XY, Jiao GQ, Hurtig M, et al:
Expression pattern of apolipoprotein M during mouse and human
embryogenesis. Acta Histochem. 106:123–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Faber K, Axler O, Dahlbäck B and Nielsen
LB: Characterization of apoM in normal and genetically modified
mice. J Lipid Res. 45:1272–1278. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Flower DR, North AC and Attwood TK:
Structure and sequence relationships in the lipocalins and related
proteins. Protein Sci. 2:753–761. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sorenson RC, Bisgaier CL, Aviram M, Hsu C,
Billecke S and La Du BN: Human serum Paraoxonase/Arylesterase’s
retained hydrophobic N-terminal leader sequence associates with
HDLs by binding phospholipids: apolipoprotein A-I stabilizes
activity. Arterioscler Thromb Vasc Biol. 19:2214–2225. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Axler O, Ahnström J and Dahlbäck B:
Apolipoprotein M associates to lipoproteins through its retained
signal peptide. FEBS Lett. 582:826–828. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Duan J, Dahlbäck B and Villoutreix BO:
Proposed lipocalin fold for apolipoprotein M based on
bioinformatics and site-directed mutagenesis. FEBS Lett.
499:127–132. 2001. View Article : Google Scholar
|
|
11
|
Christoffersen C, Ahnström J, Axler O,
Christensen EI, Dahlbäck B and Nielsen LB: The signal peptide
anchors apolipoprotein M in plasma lipoproteins and prevents rapid
clearance of apolipoprotein M from plasma. J Biol Chem.
283:18765–18772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flower DR, North AC and Sansom CE: The
lipocalin protein family: structural and sequence overview. Biochim
Biophys Acta. 1482:9–24. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sevvana M, Ahnström J, Egerer-Sieber C,
Lange HA, Dahlbäck B and Muller YA: Serendipitous fatty acid
binding reveals the structural determinants for ligand recognition
in apolipoprotein M. J Mol Biol. 393:920–936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahnström J, Faber K, Axler O and Dahlbäck
B: Hydrophobic ligand binding properties of the human lipocalin
apolipoprotein M. J Lipid Res. 48:1754–1762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Christoffersen C, Obinata H, Kumaraswamy
SB, et al: Endothelium-protective sphingosine-1-phosphate provided
by HDL-associated apolipoprotein M. Proc Natl Acad Sci USA.
108:9613–9618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Faber K, Hvidberg V, Moestrup SK, Dahlbäck
B and Nielsen LB: Megalin is a receptor for apolipoprotein M and
kidney-specific megalin-deficiency confers urinary excretion of
apolipoprotein M. Mol Endocrinol. 20:212–218. 2006. View Article : Google Scholar
|
|
17
|
Karlsson H, Lindqvist H, Tagesson C and
Lindahl M: Characterization of apolipoprotein M isoforms in
low-density lipoprotein. J Proteome Res. 5:2685–2690. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tetaz T, Detzner S, Friedlein A, Molitor B
and Mary JL: Hydrophilic interaction chromatography of intact,
soluble proteins. J Chromatogr A. 1218:5892–5896. 2011. View Article : Google Scholar
|
|
19
|
Xu N, Zhang XY, Dong X, Ekström U, Ye Q
and Nilsson-Ehle P: Effects of platelet-activating factor, tumor
necrosis factor and interleukin-1alpha on the expression of
apolipoprotein M in HepG2 cells. Biochem Biophys Res Commun.
292:944–950. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu N, Hurtig M, Zhang XY, Ye Q and
Nilsson-Ehle P: Transforming growth factor-beta down-regulates
apolipoprotein M in HepG2 cells. Biochim Biophys Acta. 1683:33–37.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Swenson ES, Guest I, Ilic Z, et al:
Hepatocyte nuclear factor-1 as marker of epithelial phenotype
reveals marrow-derived hepatocytes, but not duct cells, after liver
injury in mice. Stem Cells. 26:1768–1777. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Richter S, Shih DQ, Pearson ER, et al:
Regulation of apolipoprotein M gene expression by MODY3 gene
hepatocyte nuclear factor-1alpha: haploinsufficiency is associated
with reduced serum apolipoprotein M levels. Diabetes. 52:2989–2995.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shih DQ, Bussen M, Sehayek E, et al:
Hepatocyte nuclear factor-1alpha is an essential regulator of bile
acid and plasma cholesterol metabolism. Nat Genet. 27:375–382.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mosialou I, Krasagakis K and Kardassis D:
Opposite regulation of the human apolipoprotein M gene by
hepatocyte nuclear factor 1 and Jun transcription factors. J Biol
Chem. 286:17259–17269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venteclef N, Haroniti A, Tousaint JJ,
Talianidis I and Delerive P: Regulation of anti-atherogenic
apolipoprotein M gene expression by the orphan nuclear receptor
LRH-1. J Biol Chem. 283:3694–3701. 2008. View Article : Google Scholar
|
|
26
|
Zhang Y and Mangelsdorf DJ: LuXuRies of
lipid homeostasis: the unity of nuclear hormone receptors,
transcription regulation, and cholesterol sensing. Mol Interv.
2:78–87. 2002. View Article : Google Scholar
|
|
27
|
Zhang Y, Chen CJ, Yang QL, Cheng LQ, Wang
H and Huang LZ: Effect of interfering hepatocyte nuclear factor-1
alfa in HepG2 on the expressions of apoM, apoA-I and the
correlative key enzyme of cholesterol metabolism. Zhonghua Gan Zang
Bing Za Zhi. 19:121–126. 2011.In Chinese. PubMed/NCBI
|
|
28
|
Calayir E, Becker TM, Kratzer A, et al:
LXR-agonists regulate ApoM expression differentially in liver and
intestine. Curr Pharm Biotechnol. 9:516–521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fielding CJ and Fielding PE: Cellular
cholesterol efflux. Biochim Biophys Acta. 1533:175–189. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Di D, Wang Z, Liu Y, et al: ABCA1
upregulating apolipoproein M expression mediates via the RXR/lXR
pathway in HepG2 cells. Biochem Biophys Res Commun. 421:152–156.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Zhu Z, Luo G, Zheng L,
Nilsson-Ehle P and Xu N: Liver X receptor agonist downregulates
hepatic apoM expression in vivo and in vitro. Biochem Biophys Res
Commun. 371:114–117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mosialou I, Zannis VI and Kardassis D:
Regulation of human apolipoprotein m gene expression by orphan and
ligand-dependent nuclear receptors. J Biol Chem. 285:30719–30730.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yi-Zhou Y, Guo Qing J, Jian Z, et al:
Dihydrotestosterone regulating apolipoprotein M expression mediates
via protein kinase C in HepG2 cells. Lipids Health Dis. 11:1682012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Silver DL, Jiang XC, Arai T, Bruce C and
Tall AR: Receptors and lipid transfer proteins in HDL metabolism.
Ann NY Acad Sci. 902:103–111. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stein O and Stein Y: Atheroprotective
mechanisms of HDL. Atherosclerosis. 144:285–301. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Assmann G and Nofer JR: Atheroprotective
effects of high-density lipoproteins. Annu Rev Med. 54:321–341.
2003. View Article : Google Scholar
|
|
37
|
Phillips MC, Gillotte KL, Haynes MP,
Johnson WJ, Lund-Katz S and Rothblat GH: Mechanisms of high density
lipoprotein-mediated efflux of cholesterol from cell plasma
membranes. Atherosclerosis. 137(Suppl): 13–17. 1998. View Article : Google Scholar
|
|
38
|
Santamarina-Fojo S, Lambert G, Hoeg JM and
Brewer HB Jr: Lecithin-cholesterol acyltransferase: role in
lipoprotein metabolism, reverse cholesterol transport and
atherosclerosis. Curr Opin Lipidol. 11:267–275. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sparks DL and Pritchard PH: Transfer of
cholesteryl ester into high density lipoprotein by cholesteryl
ester transfer protein: effect of HDL lipid and apoprotein content.
J Lipid Res. 30:1491–1498. 1989.PubMed/NCBI
|
|
40
|
Christoffersen C, Pedersen TX, Gordts PL,
Roebroek AJ, Dahlbäck B and Nielsen LB: Opposing effects of
apolipoprotein m on catabolism of apolipoprotein B-containing
lipoproteins and atherosclerosis. Circ Res. 106:1624–1634. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Christoffersen C, Jauhiainen M, Moser M,
et al: Effect of apolipoprotein M on high density lipoprotein
metabolism and atherosclerosis in low density lipoprotein receptor
knock-out mice. J Biol Chem. 283:1839–1847. 2008. View Article : Google Scholar
|
|
42
|
Wolfrum C, Poy MN and Stoffel M:
Apolipoprotein M is required for prebeta-HDL formation and
cholesterol efflux to HDL and protects against atherosclerosis. Nat
Med. 11:418–422. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ahnström J, Axler O, Jauhiainen M, et al:
Levels of apolipoprotein M are not associated with the risk of
coronary heart disease in two independent case-control studies. J
Lipid Res. 49:1912–1917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rye KA, Duong M, Psaltis MK, et al:
Evidence that phospholipids play a key role in pre-beta apoA-I
formation and high-density lipoprotein remodeling. Biochemistry.
41:12538–12545. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Christoffersen C, Jauhiainen M, Moser M,
et al: Effect of apolipoprotein M on high density lipoprotein
metabolism and atherosclerosis in low density lipoprotein receptor
knock-out mice. J Biol Chem. 283:1839–1847. 2008. View Article : Google Scholar
|
|
46
|
Elsøe S, Ahnström J, Christoffersen C, et
al: Apolipoprotein M binds oxidized phospholipids and increases the
antioxidant effect of HDL. Atherosclerosis. 221:91–97. 2012.
View Article : Google Scholar
|
|
47
|
Elsøe S, Christoffersen C, Luchoomun J,
Turner S and Nielsen LB: Apolipoprotein M promotes mobilization of
cellular cholesterol in vivo. Biochim Biophys Acta. 1831:1287–1292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang XS, Zhao SP, Hu M and Luo YP:
Apolipoprotein M likely extends its anti-atherogenesis via
anti-inflammation. Med Hypotheses. 69:136–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Christoffersen C and Nielsen LB:
Apolipoprotein M-a new biomarker in sepsis. Crit Care. 16:1262012.
View Article : Google Scholar
|
|
50
|
Kumaraswamy SB, Linder A, Akesson P and
Dahlbäck B: Decreased plasma concentrations of apolipoprotein M in
sepsis and systemic inflammatory response syndromes. Crit Care.
16:R602012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sattler KJ, Elbasan S, Keul P, et al:
Sphingosine 1-phosphate levels in plasma and HDL are altered in
coronary artery disease. Basic Res Cardiol. 105:821–832. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Arkensteijn BW, Berbée JF, Rensen PC,
Nielsen LB and Christoffersen C: The apolipoprotein
m-sphin-gosine-1-phosphate axis: biological relevance in
lipoprotein metabolism, lipid disorders and atherosclerosis. Int J
Mol Sci. 14:4419–4431. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kurano M, Tsukamoto K, Ohkawa R, et al:
Liver involvement in sphingosine 1-phosphate dynamism revealed by
adenoviral hepatic overexpression of apolipoprotein M.
Atherosclerosis. 229:102–109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu WW, Zhang Y, Tang YB, et al: A genetic
variant of apolipoprotein M increases susceptibility to coronary
artery disease in a Chinese population. Clin Exp Pharmacol Physiol.
35:546–551. 2008. View Article : Google Scholar
|
|
55
|
Jiao GQ, Yuan ZX, Xue YS, et al: A
prospective evaluation of apolipoprotein M gene T-778C polymorphism
in relation to coronary artery disease in Han Chinese. Clin
Biochem. 40:1108–1112. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Z, Chu G and Yin RX: Apolipoprotein
M T-778C polymorphism is associated with serum lipid levels and the
risk of coronary artery disease in the Chinese population: a
meta-analysis. Lipids Health Dis. 12:1352013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Brorsson C, Hansen NT, Lage K, Bergholdt
R, Brunak S and Pociot F; Diabetes Genetics Consortium:
Identification of T1D susceptibility genes within the MHC region by
combining protein interaction networks and SNP genotyping data.
Diabetes Obes Metab. 11(Suppl 1): 60–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu X, Niu N, Brismar K, et al:
Apolipoprotein M promoter polymorphisms alter promoter activity and
confer the susceptibility to the development of type 1 diabetes.
Clin Biochem. 42:17–21. 2009. View Article : Google Scholar
|
|
59
|
Zhou JW, Tsui SK, Ng MC, et al:
Apolipoprotein M gene (APOM) polymorphism modifies metabolic and
disease traits in type 2 diabetes. PLoS One. 6:e173242011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang YY, Proenca R, Margherita M, et al:
Positional cloning of the mouse obese gene and its human homologue.
Nature. 372:425–432. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ahima RS and Flier JS: Leptin. Annu Rev
Physiol. 62:413–437. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu N, Nilsson-Ehle P and Ahrén B:
Correlation of apolipoprotein M with leptin and cholesterol in
normal and obese subjects. J Nutr Biochem. 15:579–582. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xu N, Nilsson-Ehle P, Hurtig M and Ahrén
B: Both leptin and leptin-receptor are essential for apolipoprotein
M expression in vivo. Biochem Biophys Res Commun. 321:916–921.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Luo G, Hurtig M, Zhang X, Nilsson-Ehle P
and Xu N: Leptin inhibits apolipoprotein M transcription and
secretion in human hepatoma cell line, HepG2 cells. Biochim Biophys
Acta. 1734:198–202. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ooi EM, Watts GF, Chan DC, et al:
Association of apolipoprotein M with high-density lipoprotein
kinetics in overweight-obese men. Atherosclerosis. 210:326–330.
2010. View Article : Google Scholar
|
|
66
|
Kappelle PJ, Lambert G, Dahlbäck B,
Nielsen LB and Dullaart RP: Relationship of plasma apolipoprotein M
with proprotein convertase subtilisin-kexin type 9 levels in
non-diabetic subjects. Atherosclerosis. 214:492–494. 2011.
View Article : Google Scholar
|
|
67
|
Cervin C, Axler O, Holmkvist J, et al: An
investigation of serum concentration of apoM as a potential MODY3
marker using a novel ELISA. J Intern Med. 267:316–321. 2010.
View Article : Google Scholar
|
|
68
|
Mughal SA, Park R, Nowak N, et al:
Apolipoprotein M can discriminate HNF1A-MODY from Type 1 diabetes.
Diabet Med. 30:246–250. 2013. View Article : Google Scholar
|
|
69
|
Xu N, Nilsson-Ehle P and Ahrén B:
Suppression of apolipoprotein M expression and secretion in
alloxan-diabetic mouse: Partial reversal by insulin. Biochem
Biophys Res Commun. 342:1174–1177. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang X, Jiang B, Luo G, Nilsson-Ehle P
and Xu N: Hyperglycemia down-regulates apolipoprotein M expression
in vivo and in vitro. Biochim Biophys Acta. 1771:879–882. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu N, Ahrén B, Jiang J and Nilsson-Ehle P:
Down-regulation of apolipoprotein M expression is mediated by
phosphatidylinositol 3-kinase in HepG2 cells. Biochim Biophys Acta.
1761:256–260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wolfrum C, Howell JJ, Ndungo E and Stoffel
M: Foxa2 activity increases plasma high density lipoprotein levels
by regulating apolipoprotein M. J Biol Chem. 283:16940–16949. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Luo G, Zhang X, Mu Q, et al: Expression
and localization of apolipoprotein M in human colorectal tissues.
Lipids Health Dis. 9:1022010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gu JG, Zhu CL, Cheng DZ, Xie Y, Liu F and
Zhou X: Enchanced levels of apolipoprotein M during HBV infection
feedback suppresses HBV replication. Lipids Health Dis. 10:1542011.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jiang J, Wu C, Luo G, et al: Expression of
apolipoprotein M in human hepatocellular carcinoma tissues. Acta
Histochem. 113:53–57. 2011. View Article : Google Scholar
|
|
76
|
Xu X, Ye Q, Xu N, et al: Effects of
ischemia-reperfusion injury on apolipoprotein M expression in the
liver. Transplant Proc. 38:2769–2773. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Park YJ, Yoo SA, Lee JH, Chung YJ, Cho CS
and Kim W: The APOM polymorphism as a novel risk factor for
dyslipidaemia in rheumatoid arthritis: a possible shared link
between disease susceptibility and dyslipidaemia. Clin Exp
Rheumatol. 31:180–188. 2013.
|
|
78
|
Zhao DX, He ZY, Qin X, et al: Association
of apolipoprotein M gene polymorphisms with ischemic stroke in a
Han Chinese population. J Mol Neurosci. 43:370–375. 2011.
View Article : Google Scholar
|
|
79
|
Burkart KM, Manichaikul A, Wilk JB, et al:
APOM and high-density lipoprotein cholesterol are associated with
lung function and per cent emphysema. Eur Respir J. 43:1003–1017.
2014. View Article : Google Scholar :
|
|
80
|
Kim J, Lee S, In K, et al: Increase in
serum haptoglobin and apolipoprotein M in patients with obstructive
sleep apnoea. J Sleep Res. 18:313–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dullaart RP, Plomgaard P, de Vries R,
Dahlbäck B and Nielsen LB: Plasma apolipoprotein M is reduced in
metabolic syndrome but does not predict intima media thickness.
Clin Chim Acta. 406:129–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ahnström J, Gottsäter A, Lindblad B and
Dahlbäck B: Plasma concentrations of apolipoproteins A-I, B and M
in patients with abdominal aortic aneurysms. Clin Biochem.
43:407–410. 2010. View Article : Google Scholar
|
|
83
|
Ahnström J, Gottsater A, Lindblad B and
Dahlbäck B: Plasma concentrations of apolipoproteins A-I, B and M
in patients with critical limb ischemia. Clin Biochem. 43:599–603.
2010. View Article : Google Scholar
|