Introduction
Breast cancer is a malignant tumor, which severely
affects female health, is life threatening and the incidence of
which has increased gradually over recent years (1,2).
With the prevalence of advanced diagnostic instruments and the
development of standardized systematic therapy, the rate of early
diagnosis in patients with recurrent-metastasis patients has
increased and survival rates have improved, with mortality rates
declining by 1–2% per year in China (1,2). The
present study aimed to investigate the risk factors of breast
cancer, recurrent-metastasis and intervention methods, which are
important to decrease the breast cancer mortality rate. The human
centrosomal ninein-like protein (Nlp) is a novel member of the
γ-tubulin complex binding proteins (GTBPs) and is essential in the
process of mitosis. The primary function of Nlp is to promote
microtubule nucleation, which contributes to centrosomal
maturation, spindle formation and chromosome segregation (3,4). The
centrosome from almost all types of tumor exhibit abnormal
structure, morphology and function. Previous studies have
demonstrated that centrosome activity is important in cell division
and in the transition from G1 phase to S phase (5,6).
Abnormal centrosomes may lead to interruption of the cell cycle,
including the polycaryon phenotype, which causes abnormal cell
transformation, tumorigenesis and the development of malignancy
(7–9). In the present study, the biological
action of Nlp on metastatic capacity of breast cancer was
investigated using advanced transfection technology.
Materials and methods
Cell culture
The MCF-7 breast cancer cell line was provided by
the Basic Center of Shandong Tumor Hospital (Shandong, China), and
the cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM; Hyclone Corporation, Logan, USA), containing 10% fetal
bovine serum (Hyclone Corporation), 100 U/ml penicillin and 100
pg/ml streptomycin (Beyotime Institute of Biotechnology, Haimen,
China) in a humidified incubator at 37°C under 5%
CO2.
Steady transfection with lentivirus
The MCF-7 cells were seeded into 10 cm cell culture
bottles (6×105 cells/ml) containing 10% serum and medium
24 h prior to transfection and were cultured in a humidified
incubator at 37°C under 5% CO2. The cells were not
transfected until the cell density was between 70 and 80%
confluent. The culture medium was replaced with medium without
serum 2 h prior to transfection. The prepared DNA solution,
containing either an enhanced green fluorescent protein (EGFP)-C1
plasmid (40 µl) or a pEGFP-C1-Nlp plasmid (100 µl;
Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China)
was added to the tubes and mixed with Opti-MEM medium to a final
volume of 2.5 ml (Gibco Life Technologies, Carlsbad, CA, USA).
Following addition of the transfection reagent, Attractene (Qiagen,
Hilden, Germany) and agitating lightly, Opti-MEM medium was added
for 20 min at room temperature. The transfection mixture was
transferred into medium containing 5×105 MCF-7 cells and
cultured for 8 h in a humidified incubator at 37°C under 5%
CO2. Following incubation, the medium was replaced with
25 ml fresh 10% serum containing medium for 48 h in a humidified
incubator at 37°C under 5% CO2. The MCF-7 supernatant
was centrifuged at 4,000 x g for 10 min at 4°C and transferred to a
filter cup (Sartorius, Goettingen, Germany). The filter cup was
then inserted into a filtrate collection tube (Sartorius). The
lentivirus was centrifuged at 1,000 x g for 2 min at 4°C and stored
at −80°C.
mRNA expression of Nlp was detected by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
The total RNA was extracted using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA), according to the
manufacturer’s instructions. The purity and concentration of the
RNA samples were determined using an ultraviolet spectrophotometer
(DU 800; Beckman Coulter, Brea, CA, USA) and samples with an
absorbance value >1.7 were assessed by RT-qPCR. A total of 1
µg RNA was used to produce cDNA according to the
manufacturer’s instructions of the TUREscript 1st Strand cDNA
Synthesis kit (Aidlab Biotechnologies Co., Ltd., Beijing, China).
The cDNA reaction system was as follows: 1 µl of Oligo(dt)18
(0.5 µg/µl), 10 µl of 2X RT Reaction mix, 1
µl of TUREscript H-RTase/RI mix and RNase free
H2O at a final volume of 20 µl. The RT-qPCR
assays were performed according to the manufacturer’s instructions
of the 2X SYBR Green qPCR kit (Aidlab Biotechnologies Co., Ltd.,
Beijing, China). PCR was conducted using an Applied Biosystems ABI
Prism 7000 Real-Time PCR System (Applied Biosystems, Foster city,
CA, USA). The cycling conditions were as follows: 94°C for 3 min to
activate the DNA polymerase, followed by 40 cycles of 95°C for 40
sec, 61°C for 60 sec and 72°C for 40 sec, and then extended at 72°C
for 10 min. The specificity of the amplification products were
confirmed by melting curve analysis. The PCR reactions for each
gene were repeated three times and independent experiments were
performed in triplicate. The primer (Shanghai Jingmei
Bioengineering Co., Ltd., Shanghai, China) sequences were as
follows: Nlp, forward 5′-ACCTGGGATTCTGAGGACTTTG-3′ and reverse
5′-ACTTTGCCGTCTCCGTCTTGAT-3′ and GAPDH, forward
5′-CATCAAGAAGGTGGTGAAGC-3′ and reverse 5′-GGAAATTGTGAGGGAGATGC-3′.
GAPDH was used as an internal loading control.
Western blotting to detect the protein
expression of Nlp and CXCR4
The cells were washed with phosphate-buffered saline
(PBS) twice and lysed in radioimmunoprecipitation buffer (50 mM
Tris, pH 7.4; 0.15 M NaCl; 1% Triton X-100; 1% sodium deoxycholate;
0.1% SDS) (Aidlab Biotechnologies Co., Ltd.) for 30 min on ice. The
samples were then centrifuged at 12,000 x g for 3 min at 4°C and
the supernatants were collected and stored at −80°C. Protein
concentrations were determined using a bicinchoninic acid assay
(Aidlab Biotechnologies Co., Ltd.). The total protein (100
µg) was resolved using a 10% SDS-PAGE gel (Beyotime
Institute of Biotechnology), electrotransferred onto polyvinylidene
fluoride membranes (GE Healthcare Life Sciences, Little Chalfont,
UK) and blocked with 5% non-fat dry milk (Shanghai Bright Dairy
Group Co., Ltd., Shanghai, China) for 1 h in Tris-buffered saline
(Boster Biological Technology, Ltd., Wuhan, China). The membranes
were incubated in primary Nlp polyclonal antibody (1:1,000; cat.
no. ab179678; Abcam, Cambridge, CA, USA), rabbit polyclonal C-X-C
chemokine receptor (CXCR)4 (1:2,000; cat. no. ab2074; Abcam) and
rabbit polycloanl actin antibody (1:5,000; cat. no. sc-7210; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The
membranes were washed with PBS, containing 5% non-fat milk and 0.1%
Tween-20, and were subsequently incubated with horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody
(1:2,000; cat. no. SN134; SunShine Bio, Nanjing, China) for 1 h at
room temperature. Finally, the membranes were incubated with
enhanced chemiluminescence reagents (EMD Millipore, Billerica, MA,
USA) for 1 min at room temperature, and developed in a dark
room.
MTT assay to detect cell growth
curves
The cells were seeded into 96-well plates (1,000
cells/well) and six wells were repeated. The cells were incubated
for 24 h and subsequently treated with 20 µl 5 g/l MTT and
incubated at 37°C for 4 h. The supernatants were washed and 150
µl dimethylsulfoxide (Sigma-Aldrich, St. Louis, MO, USA) was
added to each well to terminate the reaction. The optical density
(OD) value was detected at a wavelength of 570 nm on an
enzyme-linked immune detector (Model 680; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The OD value was continuously measured
for 6 days, and the growth curves were produced. The day was set as
abscissa and the absorbance value as the longitudinal
coordinates.
Plate colony formation assay
The cells were collected as a single cell suspension
and were seeded into 6-well plates, each group had three repeated
wells and contained 150–200 cells/well. The single cell suspension
was incubated in a humidified incubator at 37°C under 5%
CO2 for 12 days, until the cell colonies were visible by
eye. The cells were washed twice with PBS and fixed in 100%
methanol (Shenyang Chemical Co., Ltd., Shenyang, China) for 30 min
at room temperature. Following fixation, the cells were stained
with 0.5% crystal violet (Beyotime Institute of Biotechnology) for
5 min, prior to washing and drying. Positive colonies, containing
>50 cells, were observed under a microscope (IX71; Olympus
Corporation, Tokyo, Japan), images were captured and colony numbers
were quantified.
Transwell chamber migration assay
For the cell migration assay, 3×105 cells
in 1 ml medium without fetal calf serum (FCS) were seeded onto a
fibronectin coated polycarbonate membrane insert in a Transwell
apparatus (Corning, Inc., Corning, NY, USA). In the lower chamber,
600 µl DMEM, containing 20% FCS was added as
chemoattractant. The cells were incubated for 24 h at 37°C in a 5%
CO2 atmosphere, the insert was washed with PBS and the
cells on the top surface of the insert were removed using a cotton
swab. The cells adhering to the lower surface were fixed with
methanol for 20 min, stained with 5% Giemsa solution (Merck &
Co., Inc., Rahway, NJ, USA) for 20 min at room temperature, and
quantified under a microscope (IX71; Olympus Corporation) in five
randomly selected fields.
Statistical analysis
Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA) and all data are expressed
as the mean ± standard error of the mean. A least significant
difference-t test was used for two sample comparisons from two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Overexpression of Nlp is established in
the MCF-7 breast cancer cell line
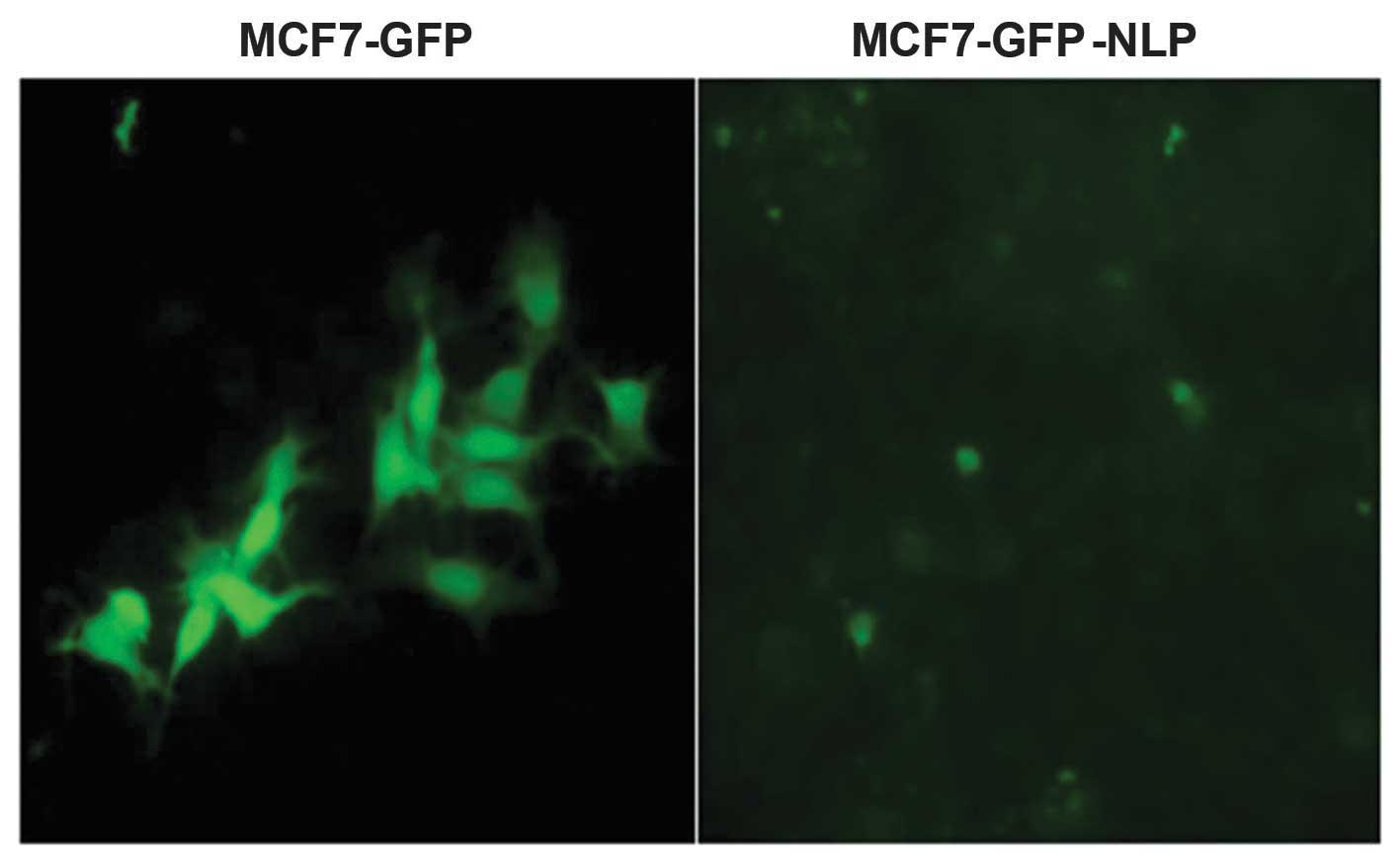
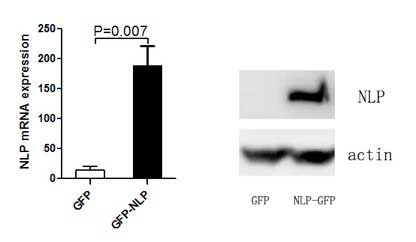
The pEGFP-C1-Nlp plasmid or the pEGFP-C1 plasmid
were transfected into MCF-7 cells to establish MCF7-GFP-NLP and
MCF7-GFP cells. The mRNA and protein expression levels of Nlp were
detected by RT-qPCR and western blotting (Figs. 1 and 2).
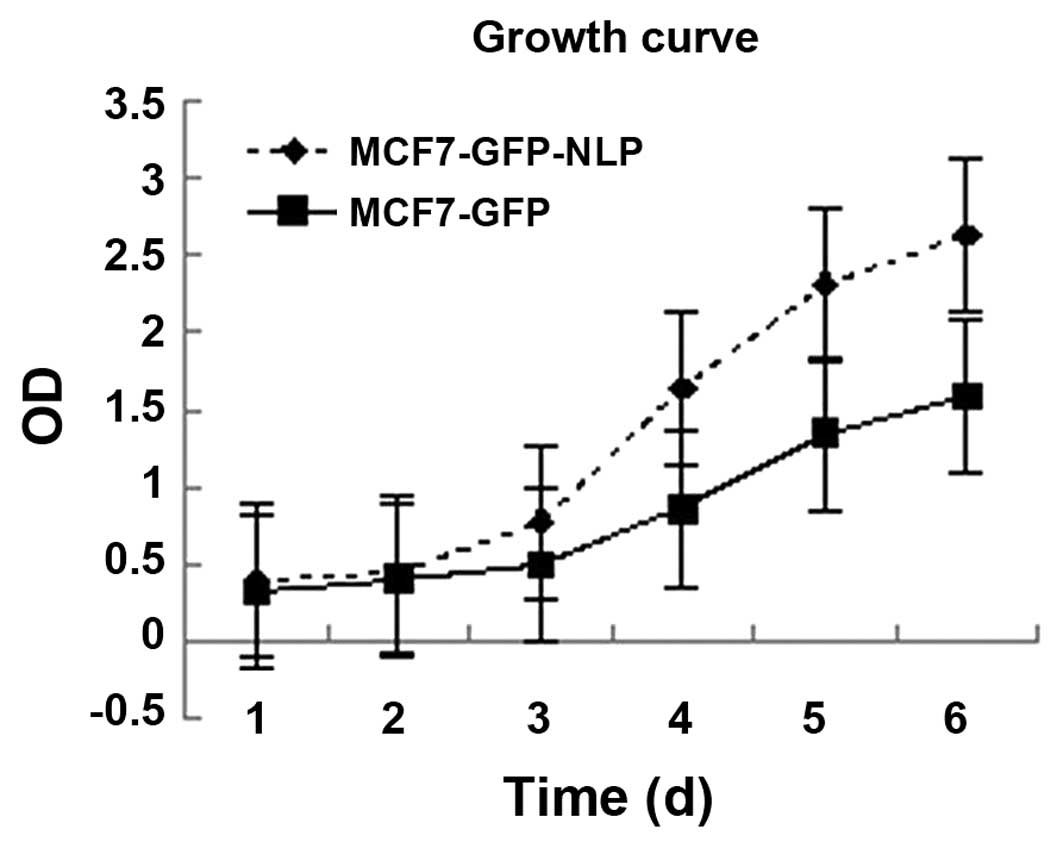
Effect of high expression of Nlp on the
growth of MCF-7 cells detected using an MTT assay
Under the same growth conditions, the growth rate
was more rapid in the MCF7-GFP-NLP cells (P<0.05) compared with
the MCF7-GFP cells, which indicated that Nlp promoted the growth of
MCF-7 cells (Fig. 3).
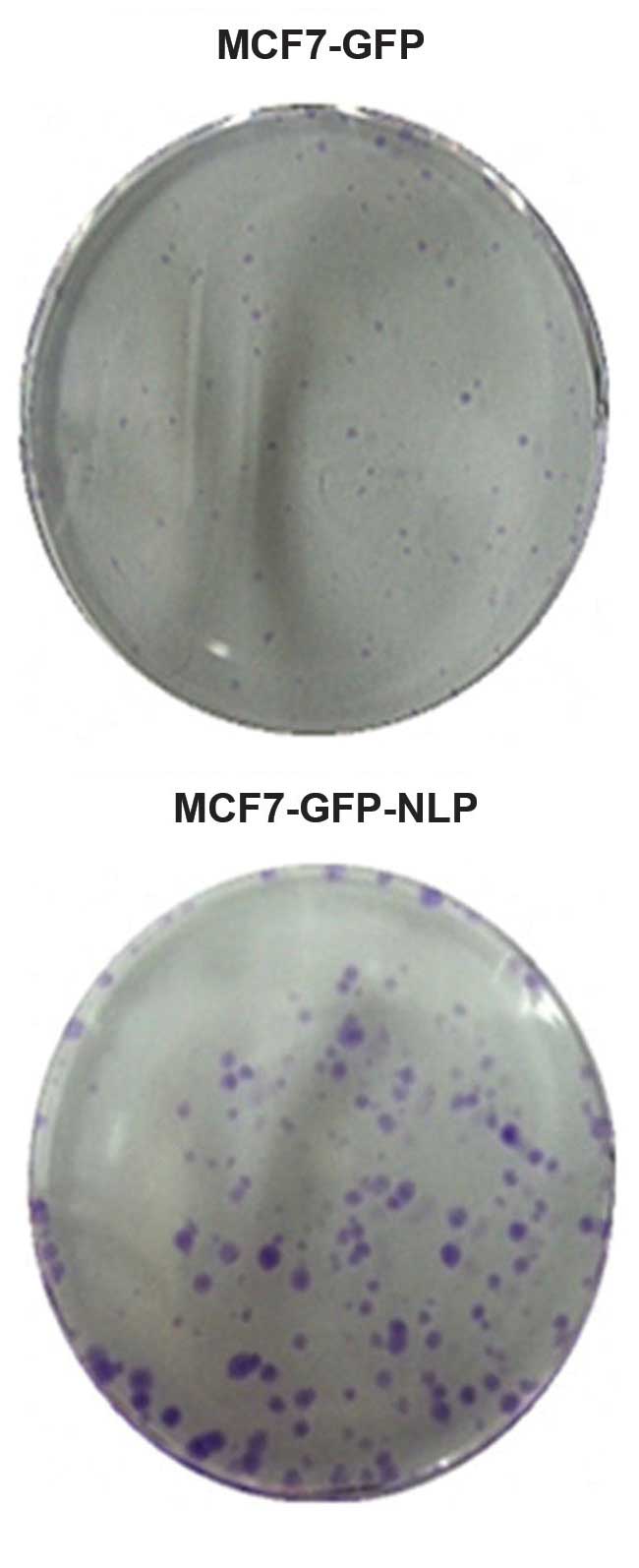
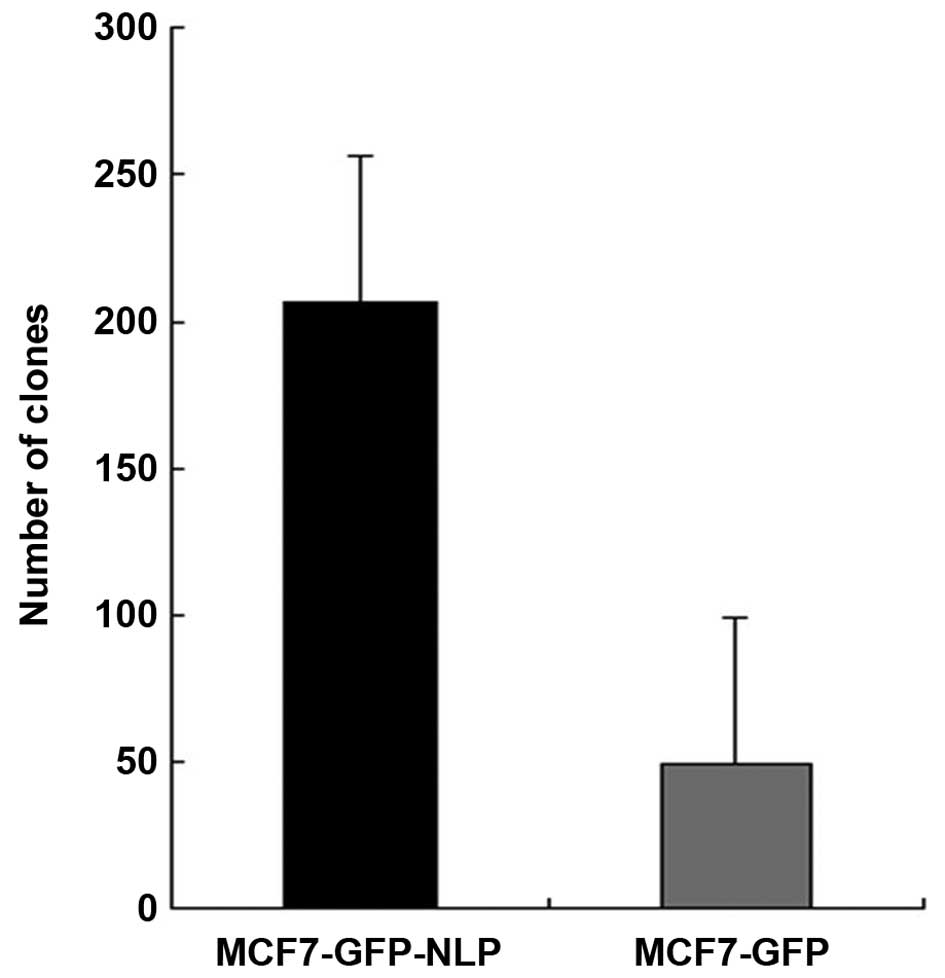
Colony formation assay to detect cell
proliferation ability
The results demonstrated that colony numbers in
MCF-GFP cells and MCF7-GFP-NLP cells were 49±3.45 and 206±14.35,
respectively, under identical conditions. The colony formation rate
was markedly increased in the MCF7-GFP-NLP cells (P<0.05)
compared with the MCF7-GFP cells, which indicated that Nlp promoted
MCF7 cell proliferation (Figs. 4
and 5).
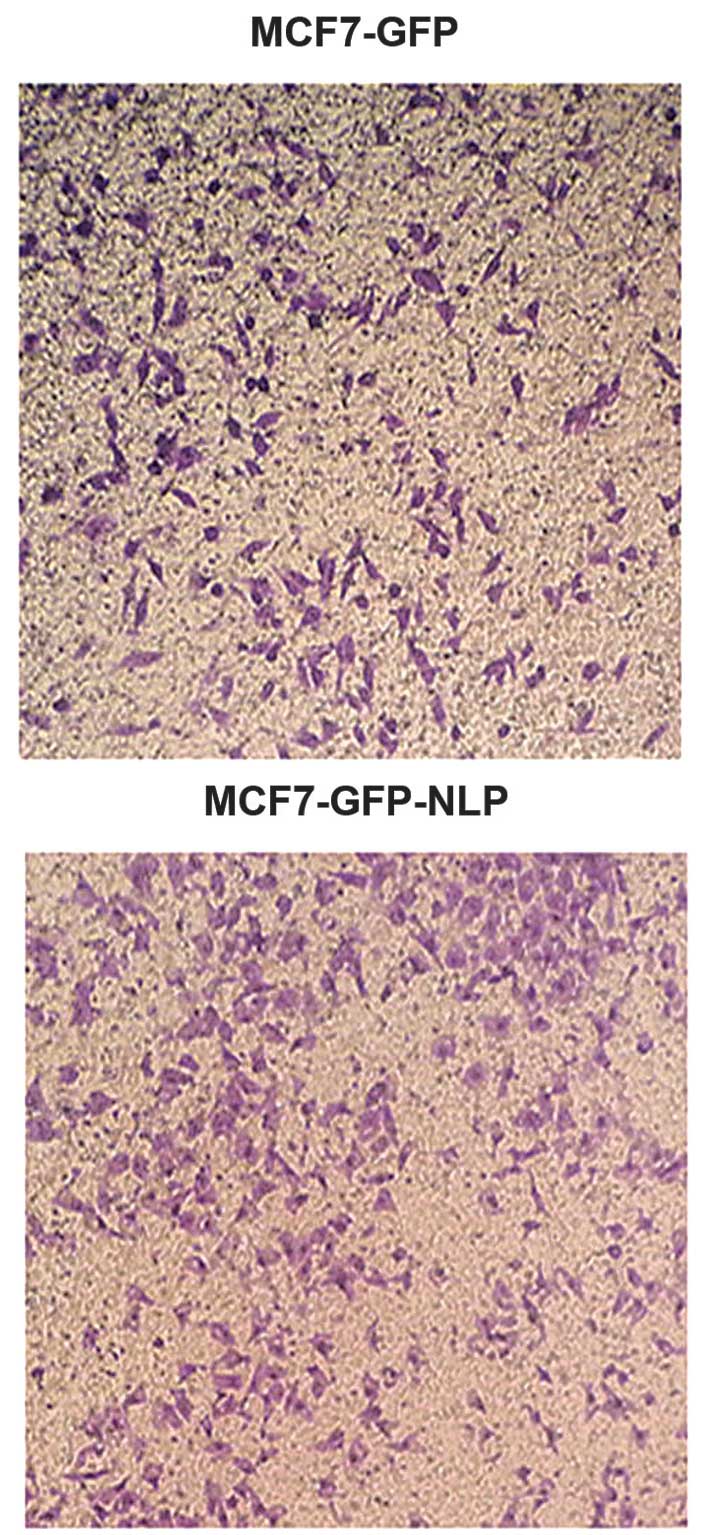
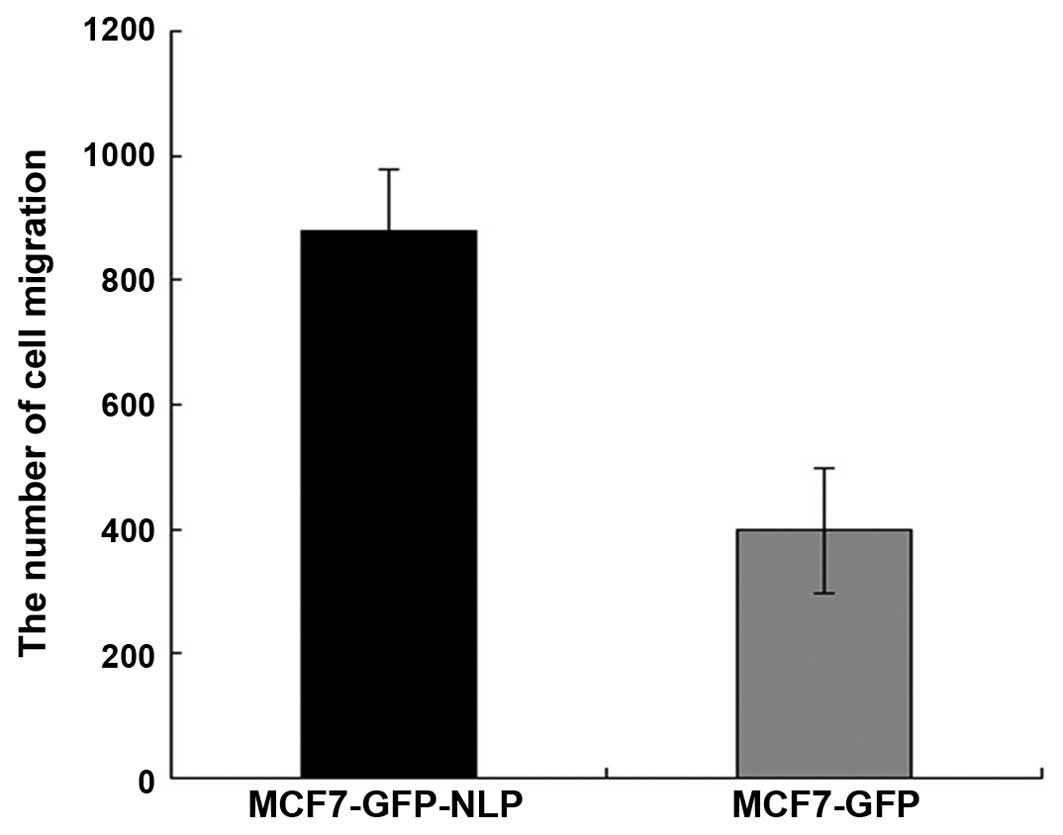
Effect of high expression of Nlp on the
migration ability in vitro
Under the same conditions, quantification of the
cells migrated under the membrane in MCF7-GFP-Nlp cells and
MCF7-GFP cells were 878±18.22 and 398±8.02, respectively. The
migration ability was increased in the MCF7-GFP-Nlp cells compared
with the MCF7-GFP cells and a significant difference was observed
between two groups (P<0.05; Figs.
6 and 7).
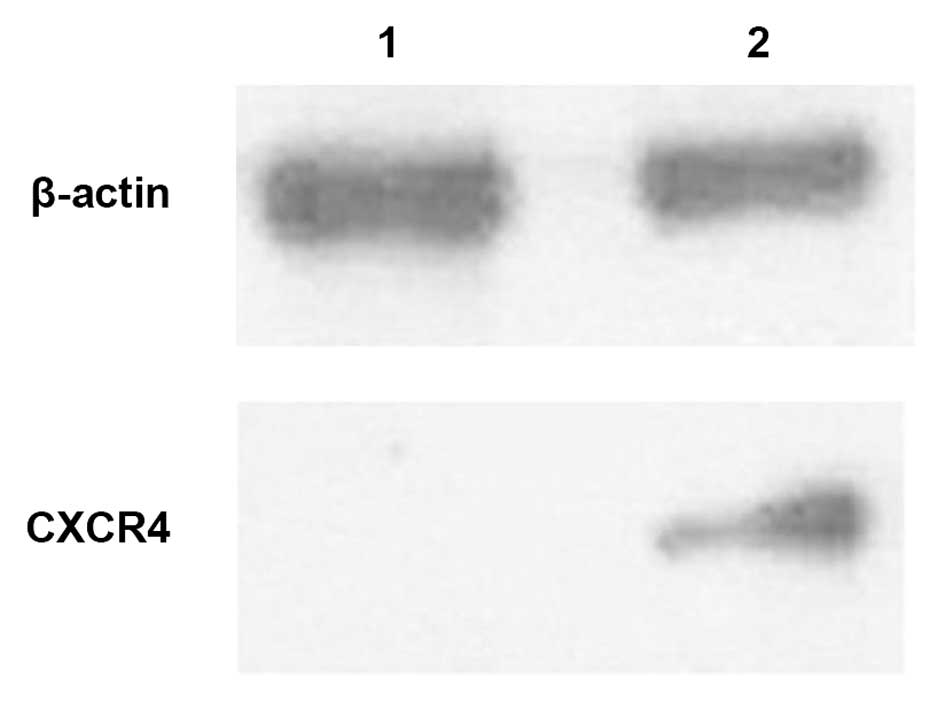
Expression of CXCR4 was detected by
western blotting in the MCF7-GFP-Nlp cells and MCF7-GFP cells
The results of the western blotting revealed that
the protein expression levels of CXCR4 were higher in the
MCF7-GFP-Nlp cells compared with the MCF7-GFP cells (Fig. 8).
Discussion
Nlp is overexpressed in breast, lung and ovarian
cancer, and head and neck squamous cell carcinoma. Notably,
centrosomal Nlp causes spontaneous tumorigenesis in transgenic mice
overexpressing Nlp (10–12). Previous studies have reported that
centrosomal abnormalities occur in certain low-grade tumors and
exhibit an demonstrate an increased trend in invasive tumors. In
ovarian cancer tissues, the higher the pathological classification,
the higher the number of centrosome abnormalities that are present.
Notably, the presence of centrosome abnormalities are higher in
malignant ovarian cancer (13–16).
A previous study demonstrated that the overexpression of Nlp is
observed in head and neck squamous cell carcinoma, which is
associated with the clinicopathological characteristics (17). In addition, it has also been
confirmed that the expression of Nlp significantly correlates with
the tumor grade, and that the overexpression of Nlp is marginally
associated with a decrease in overall survival rates (18). Furthermore, Nlp induces tumor
development by interfering with the cell cycle, mitosis and cell
apoptosis (19,20). However, the effect of Nlp on breast
tumor metastasis remains to be elucidated.
The MCF-7 breast cancer cell line retains several
characteristics of differentiated mammary epithelium, including the
ability to process estradiol via cytoplasmic estrogen receptors and
the capability of forming domes. The present study established
MCF7-GFP-Nlp and MCF7-GFP cells and observed, through growth curves
that the MCF7-GFP-Nlp cells grew more rapidly compared with
theMCF7-GFP cells. In addition, plate colony forming assays
demonstrated that the MCF7-GFP-Nlp cells exhibited a increased
colony formation capacity compared with the MCF7-GFP cells. These
results indicated that Nlp promoted MCF-7 cell proliferation.
Transwell chambers are considered as a permeability support, and
usually, a Transwell chamber is put into culture plates and medium
is added to the top and bottom chambers, with the cells were seeded
into the top chamber. Since the membrane is permeable, cells can
migrate to the lower chamber (21,22).
The results of the present study revealed that the overexpression
of Nlp promoted cell migration in the Transwell model in
vitro.
Changes in tumor cell migration capacity is an
important step affecting tumor invasion and metastasis. CXCR4 is
encoded by 352 amino acids and is a seven-transmembrane G-protein
chemokine receptor. In 1996, Feng et al identified that
CXCR4 is a coreceptor for human immunodeficiency virus-1 entry,
following which several studies have investigated CXCR4 (23). It has been demonstrated that CXCR4
is involved in the invasion and metastasis of several types of
cancer, including breast carcinoma (24). Hiller and Chu (25,26)
demonstrated that CXCR4 is important in several types of cancer,
including breast cancer, and revealed that CXCR4 was highly
expressed in areas common for breast cancer metastasis, including
the axillary lymph nodes. Hernandez et al confirmed that
CXCL12-CXCR4 is important in the process of breast tumor cell
growth, angiogenesis, invasion and metastasis (27,28).
A meta-analysis investigation based on thirteen eligible studies,
consisting of 3,865 patients with breast cancer, demonstrated that
the overexpression of CXCR4 was significantly associated with lymph
node status and distant metastasis. In addition, the overexpression
of CXCR4 indicated a poor overall and disease-free survival rates
(29). The present study
demonstrated that the expression of CXCR4 was higher in the
MCF7-GFP-NLP cells compared with the MCF7-GFP control cells, which
implied that Nlp improved the migration capacity of breast cancer
cell lines through activated CXCL12 and CXCR4.
In conclusion, the results of the present study
indicated that an increase in the expression of Nlp resulted in a
malignant phenotype, which induced tumor cell proliferation and
invasion. Furthermore, the results confirmed that Nlp exhibited
certain biological characteristics, including promoting breast
tumorigenesis and development, to provide a novel molecular index
for breast cancer diagnosis. Therefore, Nlp may be an effective
target of antitumor drugs for therapy against specific types of
tumor.
Acknowledgments
This study was supported by the Science-Technology
Development Funds of Shandong Province (no. 2011GSF11823).
References
|
1
|
Andre F, Slimane K, Bachelot T, et al:
Breast cancer with synchronous metastases: trends in survival
during a 14-year period. J Clin Oncol. 22:3302–3308. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giordano SH, Buzdar AU, Smith TL, et al:
Is breast cancer survival improving? Cancer. 100:44–52. 2004.
View Article : Google Scholar
|
|
3
|
Li J and Zhan Q: The role of centrosomal
Nlp in the control of mitotic progression and tumourigenesis. Br J
Cancer. 104:1523–1528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bettencourt-Dias M and Glover DM:
Centrosome biogenesis and function: Centrosomics brings new
understanding. Nat Rev Mol Cell Biol. 8:451–463. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Doxsey SJ: Centrosomes as command centres
for cellular control. Nat Cell Biol. 3:E105–108. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raff JW: Centrosomes: Central no more?
Curr Biol. 11:R159–R161. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lingle WL, Lutz WH, Ingle JN, et al:
Centrosome hypertrophy in human breast tumors: Implications for
genomic stability and cell polarity. Proc Natl Acad Sci USA.
95:2950–2955. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krämer A, Neben K and Ho AD: Centrosome
aberrations in hematological malignancies. Cell Biol Int.
29:375–383. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nigg EA: Origins and consequences of
centrsome aberrations in human cancers. Int J Cancer.
119:2717–2723. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao S, Liu R, Wang Y, et al: Centrosomal
Nlp is an oncogenic protein that is gene-amplified in human tumors
and causes spontaneous tumorigenesis in transgenic mice. J Clin
Invest. 120:498–507. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu D, Qu H, Fu M, et al: Increased
expression of Nlp, a potential oncogene in ovarian cancer and its
implication in carcinogenesis. Gynecol Oncol. 110:230–236. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu L, Song Y, Zhang Q, et al: Ninein-like
protein is overexpressed in head and neck squamous cell carcinoma
and contributes to cancer growth and resistance to apoptosis. Oncol
Rep. 22:789–798. 2009.PubMed/NCBI
|
|
13
|
Pihan GA, Wallace J, Zhou Y, et al:
Centrosome abnormalities and chromosome instability occur together
in pre-invasive carcinomas. Cancer Res. 63:1398–1404.
2003.PubMed/NCBI
|
|
14
|
Pihan GA, Purohit A, Wallace J, et al:
Centrosome defects can account for cellular and genetic changes
that characterize prostate cancer progression. Cancer Res.
61:2212–2219. 2001.PubMed/NCBI
|
|
15
|
Duensing S: A tentative classification of
centrosome abnormalities in cancer. Cell Biol Int. 29:352–359.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu L, Song Y, Zhang Q, et al: Ninein-like
protein is overexpressed in head and neck squamous cell carcinoma
and contributes to cancer growth and resistance to apoptosis. Oncol
Rep. 22:789–798. 2009.PubMed/NCBI
|
|
18
|
Qu D, Qu H, Fu M, et al: Increased
expression of Nlp, a potential oncogene in ovarian cancer and its
implication in carcinogenesis. Gynecol Oncol. 110:230–236. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
D’Assoro AB, Lingle WL and Salisbury JL:
Centrosome amplification and the development of cancer. Oncogene.
21:6146–6153. 2002. View Article : Google Scholar
|
|
20
|
Duensing S: A tentative classification of
centrosome abnormalities in cancer. Cell Biol Int. 29:352–359.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Yang H, Liu H, et al: Effect of
staurosporine on the mobility and invasiveness of lung
adenocarcinoma A549 cells: An in vitro study. BMC Cancer.
9:1742009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhi YH, Song MM, Wang PL, et al:
Suppression of matrix metallo-proteinase-2 via RNA interference
inhibits pancreatic carcinoma cell invasiveness and adhesion. World
J Gastroenterol. 15:1072–1078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng Y, Broder CC, Kennedy PE, et al:
HIV-1 entry cofactor: Functional cDNA cloning of a
seven-transmembrane, G protein-coupled receptor. Science.
272:872–877. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Juarez J, Bendall L and Bradstock K:
Chemokines and their receptors as therapeutic targets: The role of
the SDF-1/CXCR4 axis. Curr Pharm Des. 10:1245–1259. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiller D and Chu QD: CXCR4 and axillary
lymph nodes: review of a potential biomarker for breast cancer
metastasis. Int J Breast Cancer. 4209812011.
|
|
26
|
Chu QD, Panu L, Holm NT, et al: High
chemokine receptor CXCR4 level in triple negative breast cancer
specimens predicts poor clinical outcome. J Surg Res. 159:689–695.
2010. View Article : Google Scholar
|
|
27
|
Hernandez L, Magalhaes MA, Coniglio SJ, et
al: Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis.
Breast Cancer Res. 13:R1282011. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cojoc M, Peitzsch C, Trautmann F, et al:
Emerging targets in cancer management: role of the CXCL12/CXCR4
axis. OncoTargets Ther. 6:1347–1361. 2013.
|
|
29
|
Zhang Z, Ni C, Chen W, et al: Expression
of CXCR4 and breast cancer prognosis: a systematic review and
meta-analysis. BMC Cancer. 14:492014. View Article : Google Scholar : PubMed/NCBI
|