Introduction
Darier disease (DD; OMIM 124200), also termed
keratosis follicularis and Darier-White disease, is an autosomal
dominant inherited skin disorder, characterized by abnormal
keratinization and loss of adhesion between epidermal cells, termed
acantholysis, and the development of warty papules and plaques in
seborrheic regions, including the central trunk, forehead, scalp
and flexures (1). It has a
prevalence of 1:30,000–100,000 worldwide. The lesions usually
present in the second decade, and penetrance is almost complete,
although the phenotypic expression is variable (2). Certain patients exhibit pits or
keratotic papules on the palms and distinctive nail abnormalities,
whereas patients with severe disease are handicapped by widespread
malodour keratotic plaques. Neurological and psychiatric
abnormalities have also been reported in patients with DD (1). The symptoms are often exacerbated by
heat, sweating, sunburn and stress (3,4).
The ATP2A2 gene encoding SERCA2, a calcium
pump of the sarco/endoplasmic reticulum, has been identified as the
defective gene in the disease (3).
ATP2A2 is positioned on chromosome 12q23–q24.1 and has 21
exons (5). SERCA pumps are
important in Ca2+ transport between the cytoplasm and
the endoplasmic reticulum (ER), using energy from adenosine
triphosphate (ATP) hydrolysis, and is, therefore, important in
signal transduction for gene expression and cell differentiation
(6,7).
The aim of the present study was to investigate the
genetic pedigrees of two families with DD, in order to identify
novel mutations.
Materials and methods
Patients
In the present study, two families, containing
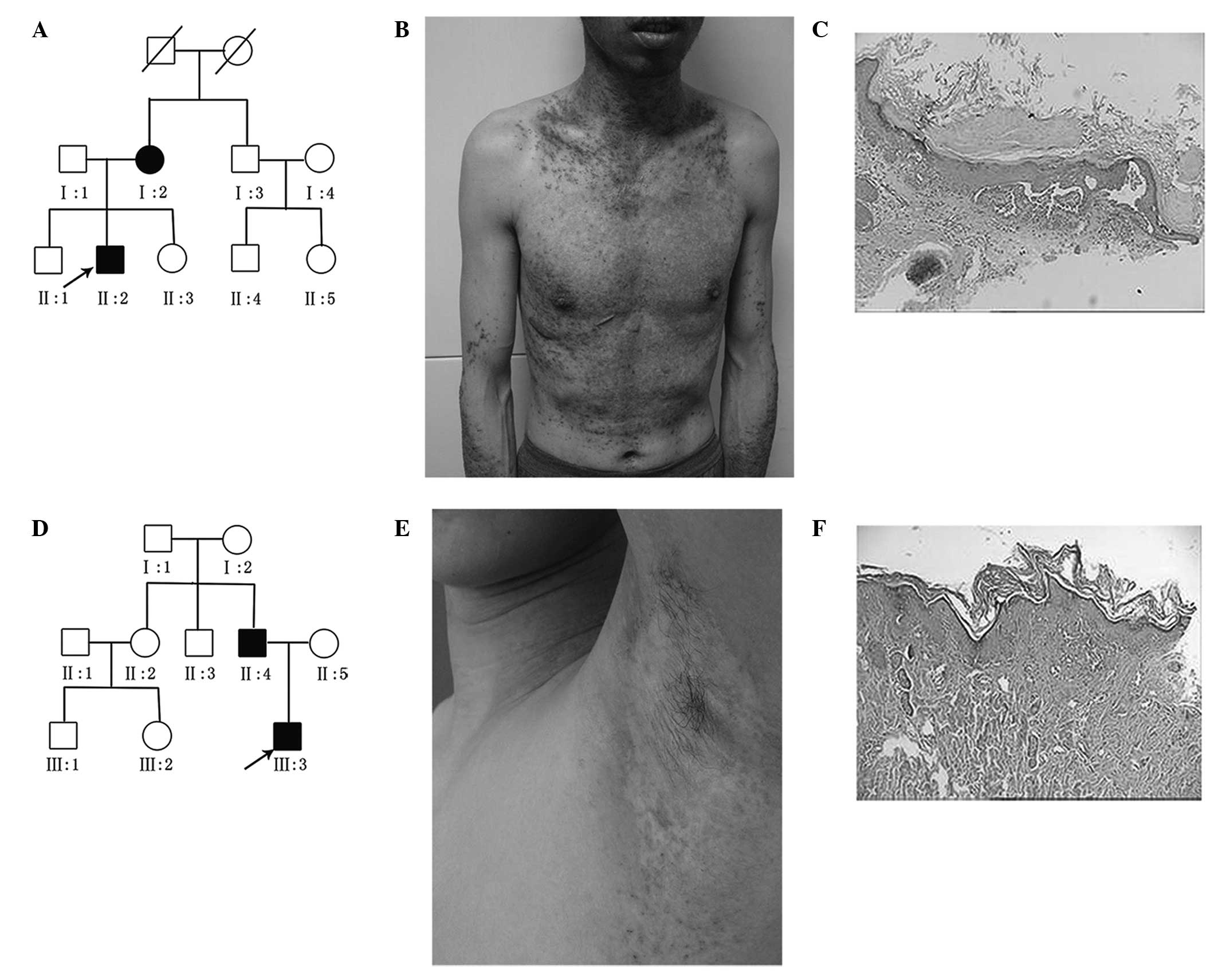
individuals with DD, were recruited. The proband in pedigree A
(Fig. 1A), a 36-year-old male,
exhibited widespread follicular papules at the age of 26 years
(Fig. 1B), and the proband’s
mother exhibited similar clinical manifestations. A 0.4 cm × 0.3 cm
biopsy was taken from the skin of all patients. The biopsies were
fixed in 10% buffered formalin (Shanghai Shiyi Chemicals Reagent
Co.,Ltd., Shanghai, China) and routinely processed and embedded in
paraffin. Sections (4 µm) were cut and stained with
hematoxylin and eosin (Shanghai Shiyi Chemicals Reagent Co.,Ltd.).
Histological examination demonstrated hyperkeratosis with
parakeratosis, and suprabasal acantholysis with dyskeratotic cells,
including corps ronds and grains. The diagnosis of DD was based on
clinical and histopathological examinations (Fig.1C). The proband in pedigree B
(Fig. 1D), an 18-year-old man, had
DD with a 4-year history of papules and hyperkeratotic plaques over
the seborrheic area. Physical examination revealed scattered
erythema, and follicular papules on the neck and armpit (Fig. 1E). The proband’s father, a
43-year-old male, had been diagnosed with DD for >10 years. This
patient manifested greasy, hyperkeratotic papules on the face,
forehead, armpit, upper extremities and inguen. None of the
patients presented with any neuropsychiatric symptoms. The
diagnosis of DD was based on clinical and histopathological
examinations (Fig. 1F).
Genetic analysis
Ethical approval for the present study was obtained
from the Ethical Committee of Fuzhou Dermatology Hospital for human
studies (Fuzhou, China). Informed consent was obtained from the two
family members. A total of 5 ml median cubital vein, basilic vein
or cephalic vein blood was collected into a tube with 2% EDTA (0.5
ml), then genomic DNA was extracted from EDTA blood samples, using
DNA extraction regents (TIANamp Blood DNA Midi Kit DP332-01,
Tiangen, Shanghai, China). Polymerase chain reaction (PCR) was
performed in a 25 µl reaction volume, containing 12.5
µl PCR mix, 11 µl double distilled H2O,
0.5 µl forward primer, 0.5 µl reverse primer and 0.5
µl (200 ng) genomic DNA. The PCR amplification of
ATP2A2 was performed from the genomic DNA using 19 pairs of
primers, spanning all 21 exons and flanking splice sites of the
gene, as presented in Table I
(5). The amplification was
performed at 94°C for 5 min, followed by 32 cycles of 30 sec
denaturation at 94°C, 30 sec annealing at 50–62°C, 45 sec extension
at 72°C, and termination for 10 min at 72°C (Mastercycler ep
gradient S; Eppendorf, Hamburg, Germany).
 | Table IPolymerase chain reaction primers for
amplification of ATP2A2 from genomic DNA. |
Table I
Polymerase chain reaction primers for
amplification of ATP2A2 from genomic DNA.
| Exon | Forward primer
sequence | Reverse primer
sequence | Annealing temperature
(°C) |
|---|
| 1 |
CGAGGCGGAGGCGAGGAG |
GGAGCCGAAGCCCACGCG | 62.0 |
| 2 + 3 |
ACCTCCCTCTTGACACATTG |
GACAACTCCTAACCACACTG | 55.1 |
| 4 |
CGTGCCATTTCTCTTCTAGG |
CTCAACACATCAGGAAAAACAG | 55.1 |
| 5 |
AGTGTCAGGCAGGTCTTTAC |
AGGAAGGGAGGTGCTAAAAC | 55.1 |
| 6 |
AGCCTCATTCTCTTCCTTCC |
ATGGAGCGAGACTAAAGCAC | 55.1 |
| 7 |
CTTGGTGTGGGTCGCAGAG |
CCTTTAGAATGATAGCCAGTG | 50.3 |
| 8 |
GTTGTATGGCTGGTTGCTTG |
GAACAAAGAACCACGACACG | 52.0 |
| 9 |
GGTTGTTTGCCTTTGTCCTAA |
ATAACAAACACAAATCCCTCTT | 50.3 |
| 10 |
GGCGACCATACCCTGCTC |
CCCACCCCACCCTTGAAC | 55.0 |
| 11 |
TCAGAGGAGGATAAAAATGGC |
CTGTAAGTTTGAGGAGATAAGG | 52.0 |
| 12 + 13 |
ATTGCCACCCAGTAGTATCC |
GAACTGTTTGACCTTTTGCTTG | 55.1 |
| 14 |
CTAGAACTTGCCACTTTTATTTA |
GAGGCTACTATGTGCTTGTG | 50.0 |
| 15 |
TTTCCTCCTGCTTCCCATTC |
GCAATCTGGAGAGCAAACTG | 55.0 |
| 16 |
TCATTTATTTTTCTGGAGGAGG |
CATCTCTGTCTTTTGCTACCC | 53.8 |
| 17 |
TGATCTTCGTCCTTGTGGGG |
TGATAGATACCGAAACCACAG | 53.8 |
| 18 |
GGGTTGGAGCCTGGACTTG |
TTTTGGGAAGGGAAGAACTGT | 55.0 |
| 19 + 20 |
TCCCCACCTCTCCTTGCTC |
CCTCCATCACCAGCCAGTAT | 57.5 |
| 21 |
GTTCCTTTTCATCTGTCGCTG |
TCTTTTTCCCCAACATCAGTC | 53.8 |
The purified PCR products were detected by
bidirectional direct sequencing using an ABI3130 automated
sequencing system (Applied Biosystems, Foster City, CA, USA). DNA,
extracted from blood samples obtained from 100 ethnicity-matched
healthy individuals, were used as controls. The ATP2A2
sequence, which was published in the NCBI database (http://www.ncbi.nlm.nih.gov), was used as a reference
sequence, which was compared with the experimental results to
identify mutations. The nucleotide numbering followed the
NM_001681.3 sequence and the amino acid numbering followed the
NP_001672.1 ATP2A2 protein sequence. To determine whether
the mutations were pathogenic, in silico analysis was
performed using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) (8). The categories of pathogenicity
included benign, possibly damaging and probably damaging. The
mutations were assigned a score between 0 and 1, with the highest
score reflecting the closest adherence to be damaging. The mutation
(R603I) was predicted to be ‘probably damaging’ with a score of 1
and the mutation (G749V) was predicted to be ‘probably damaging’
with a score of 0.998. Scale-invariant feature transform (SIFT)
(http://sift.jcvi.org/) (9), was also used which predicts the
effect of a specific amino acid change at the protein level. Amino
acids with probabilities <0.05 are predicted to be deleterious.
Substitutions at position 603 from R to I and position 749 from G
to V are predicted to ‘affect protein function’ with a score of
0.00.
Results
The entire 21 exons and exon-intron boundaries of
the ATP2A2 gene were sequenced in the two families, in which
two novel heterozygous mutations were identified (Fig. 2). In family A, the mutation
analysis revealed a nucleotide alteration between G and T
(2246G>T) in exon 15 in the two affected individuals, which
caused an amino acid substitution from glycine to valine and was
designated as p.G749V. In family B, a heterozygous nucleotide
change in G1808T in exon 14 was identified in the two affected
individuals. This mutation caused an amino acid substitution from
arginine to isoleucine acid at position 603 (p.R603I). No
ATP2A2 variants were detected in the unaffected individuals,
in either the healthy relatives in the two families or in the 100
ethnicity-matched healthy controls. Furthermore, PolyPhen-2 and
SIFT were used to predict the pathogenicity of the mutations. The
two novel missense mutations were scored as ‘probably damaging’
using PolyPhen. The prediction scores of the SIFT for each mutation
were 0, therefore, p.R603I and p.G749V were considered deleterious
mutations, affecting the protein structure and/or function of
SCRCA2.
Discussion
In the present study, two novel mutations in the
ATP2A2 gene were identified following genetic analyses of
two families containing patients with DD. The G1808T mutation was
detected in codon 603, located within the ATP-binding domain. This
missense mutation resulted in the substitution of hydrophilic polar
amino acid, arganine, to the hydrophobic amino acid, isoleucine
acid, which may have affected the affinity of ATP, leading to
alterations in the steric structural and biochemical properties of
SERCA2.
The present study also identified G2246T as a second
mutation in DD, which was positioned in codon 749. An amino acid
substitution from glycine to arginine (G749R) has been previously
reported (3), however, the present
study demonstrated that a different amino acid substitution at the
same amino acid position in the ATP2A2 gene may have also
lead to a steric clash and disrupt the function of SERCA2, leading
to DD. These amino acid changes were not observed in the remaining
unaffected family members or in the 100 healthy control
individuals, therefore, these changes were unlikely to represent
silent polymorphisms.
SERCA2, encoded by the ATP2A2 gene,
transports Ca2+ between the cytoplasm and the ER and is
important in maintaining calcium homeostasis (3,5,10,11).
It has been recognized that extracellular calcium
([Ca2+]e) is important in epidermal
differentiation and intra-epidermal cohesion (12). The differentiation of keratinocytes
is triggered by increasing the [Ca2+]e
>0.1 mM (13). Another previous
study demonstrated that the calcium concentration in the basal
layer in the skin of patients with DD skin is lower than in normal
skin in vivo (11, 14).
In addition, the keratinocytes of patients with DD
exhibit depleted ER Ca2+ stores due to the loss of
SERCA2 Ca2+ transport (15,16).
This depletion of Ca2+ ER stores potentially triggers
the ER stress response, and persistent ER stress, which cannot be
overcome due to the defective upregulation of SERCA2, may result in
apoptotic keratinocytes, as observed in DD (12). A previous study demonstrated that
SERCA2 mutant proteins, which are not degraded by the proteasome,
form insoluble aggregates and cause ER stress and apoptosis
(17).
Previous studies have also provided evidence to
support the role of SERCA2 in the assembly of the desmosomal
complex. In normal human keratinocytes, the inhibition of SERCA by
thapsigargin, an inhibitor of SERCA2, impairs the trafficking of
desmoplakin to the cell surface, and the trafficking of desmoplakin
is inhibited in Darier keratinocytes (12). Defective addressing to the
desmosomal plaque of desmoplakin, which links the cytoskeleton to
the desmosomal complexes, may affect the stability and cell surface
expression of desmosomal proteins, impairing cell-to-cell adhesion
in the DD epidermis and leading to acantholysis (12,17).
The p.R603I and p.G749V mutations identified in the
present study were located within the ATP-binding and stalk domain
regions of SERCA2, respectively. These functional domains have been
demonstrated to be highly conserved in different species and likely
to be critical for the normal function of SERCA2 (Fig. 3) (5). Therefore, it is possible that these
missense mutations, which altered polarity and hydrophobicity, may
have interfered with the function of SERCA2.
In conclusion, the present study identified two
novel missense mutations, p.R603I and p.G749V, in the ATP2A2
gene in two families containing individuals diagnosed with DD.
These results contribute to the expanding database of ATP2A2
gene mutations. Supplemental functional trials are imperative to
confirm the relevance between the mutations and the disease.
Acknowledgments
The authors would like to thank the patients with DD
and the healthy individuals for their participation in the study.
The current study was supported by the Jiangsu Technology Project
of Chinese Medicine Pharmacy (grant no. LZ11082); the Fifth
High-level Talent Project of “Six Talent Peaks” of Jiangsu Province
(grant no. 2008.101); and the Fujian Science and Technology Project
(grant no. 2011-5-81-1).
References
|
1
|
Burge SM and Wilkinson JD: Darier-White
disease: a review of the clinical features in 163 patients. J Am
Acad Dermatol. 27:40–50. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Munro CS: The phenotype of Darier’s
disease: penetrance and expressivity in adults and children. Br J
Dermatol. 127:126–130. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakuntabhai A, Ruiz-Perez V, Carter S, et
al: Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier
disease. Nat Genet. 21:271–277. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Green EK, Gordon-Smith K, Burge SM, et al:
Novel ATP2A2 mutations in a large sample of individuals with Darier
disease. J Dermatol. 40:259–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sakuntabhai A, Burge S, Monk S and
Hovnanian A: Spectrum of novel ATP2A2 mutations in patients with
Darier’s disease. Hum Mol Genet. 8:1611–1619. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clapham DE: Calcium signaling. Cell.
80:259–268. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dolmetsch RE, Xu K and Lewis RS: Calcium
oscillations increase the efficiency and specificity of gene
expression. Nature. 392:933–936. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar P, Henikoff S and Ng PC: Predicting
the effects of coding non-synonymous variants on protein function
using the SIFT algorithm. Nat Protoc. 4:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dhitavat J, Fairclough RJ, Hovnanian A and
Burge SM: Calcium pumps and keratinocytes: lessons from Darier’s
disease and Hailey-Hailey disease. Br J Dermatol. 150:821–828.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leinonen PT, Hägg PM, Peltonen S, et al:
Reevaluation of the normal epidermal calcium gradient and analysis
of calcium levels and ATP receptors in Hailey-Hailey and Darier
epidermis. J Invest Dermatol. 129:1379–1387. 2009. View Article : Google Scholar
|
|
12
|
Savignac M, Edir A, Simon M and Hovnanian
A: Darier disease: a disease model of impaired calcium homeostasis
in the skin. Biochim Biophys Acta. 1813:1111–1117. 2011. View Article : Google Scholar
|
|
13
|
Elias PM, Ahn SK, Denda M, et al:
Modulations in epidermal calcium regulate the expression of
differentiation-specific markers. J Invest Dermatol. 119:1128–1136.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dhitavat J, Cobbold C, Leslie N, Burge S
and Hovnanian A: Impaired trafficking of the desmoplakins in
cultured Darier’s disease keratinocytes. J Invest Dermatol.
121:1349–1355. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Foggia L, Aronchik I, Aberg K, Brown B,
Hovnanian A and Mauro TM: Activity of the hSPCA1 Golgi Ca2+ pump is
essential for Ca2+-mediated Ca2+ response and cell viability in
Darier disease. J Cell Sci. 119:671–679. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leinonen PT, Myllylä RM, Hägg PM, et al:
Keratinocytes cultured from patients with Hailey-Hailey disease and
Darier disease display distinct patterns of calcium regulation. Br
J Dermatol. 153:113–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Bruce AT, Tu C, et al: Protein
aggregation of SERCA2 mutants associated with Darier disease
elicits ER stress and apoptosis in keratinocytes. J Cell Sci.
124:3568–3580. 2011. View Article : Google Scholar : PubMed/NCBI
|