Introduction
Porphyromonas gingivalis (P.
gingivalis) is a Gram-negative, rod-shaped, anaerobic
pathogenic bacterium associated with several periodontal diseases
(1). The occurrence of
periodontitis in >47% of the US population, with a high
prevalence of mild (8.7%), moderate (30%) and severe (8.5%) cases,
is due to variables, such as oral hygiene, socioeconomic status and
other environmental, genetic and metabolic risk factors (2). P. gingivalis exhibits a strong
positive association with the diagnostic parameters of
periodontitis, including gingival recession, increased sulcular
pocket depth and bleeding upon probing (3). In addition, Hajishengallis et
al (4) demonstrated that
although P. gingivalis does not independently cause
periodontal disease in a germ-free murine model, low numbers of
P. gingivalis are able to disrupt host homeostasis through
actions involving commensal microorganisms and complement, leading
to inflammation and periodontal disease (4).
P. gingivalis produces multiple virulence
factors that allow successful colonization and support evasion of
host defenses, a number of which contribute to the inflammation and
destruction of host tissues (5).
Adhesins, such as fimbraie and hemagglutinins, promote attachment
(5,6) and proteolytic enzymes, such as
cysteine proteinases and hemagglutinins, are capable of degrading
multiple substrates in the gingival crevice, facilitating nutrient
acquisition and contributing to host tissue degradation (5,6).
The control of oral bacteria is mediated by a
diverse array of specific and non-specific innate immune molecules
present in saliva and on mucosal surfaces (7). There are number of functional
families consisting of >45 antimicrobial proteins and peptides,
including cationic peptides, metal ion chelators, histatins,
defensins, bacterial adhesions and agglutinators and enzymes
directed at the bacterial cell wall. However, the physiological
concentration of the majority of salivary antimicrobial proteins
and peptides is lower than the effective concentration in
vivo (7), which suggests that
there may be additional immune functions within the saliva.
The enzyme α-amylase catalyzes the hydrolysis of
internal α-1,4-glycosidic linkages within carbohydrate moieties,
including glucose, maltose and maltotriose units (8,9).
α-amylases are used in a number of industrial processes in the
food, fermentation, textiles, paper, detergent and pharmaceutical
industries. Fungal and bacterial amylases may have the potential
for use in the pharmaceutical and fine-chemical industries.
Advances in biotechnology have led to the expansion of amylase
application in numerous fields, such as biomedical and analytical
chemistry, and also textiles, food, brewing and distilling
industries (8,10).
The bisbenzamidine derivative, pentamidine, has
proved one of the most successful agents for targeting eukaryotic
parasites, and has been used clinically for >70 years (11,12).
In 1938, pentamidine isethionate was identified to have
anti-protozoal activity, and was approved in the United States for
the treatment of Pneumocystis carinii pneumonia and other
protozoal diseases (13).
Pentamidine has the ability to inhibit interaction at the
Ca2+/p53 site of the protein, and has been reported to
inhibit S100B activity (14). On
the basis of the previous studies, the present study aimed to take
advantage of the antimicrobial activity of α-amylase and
pentamidine to attenuate oral infection of P.
gingivalis.
Materials and methods
Reagents and chemicals
α-amylase from porcine pancreas was purchased from
Sigma-Aldrich (St. Louis, MO, USA). The bisbenzamidine derivative,
pentamidine was purchased from Sanofi S.A. (Paris, France). The
media (Terrific broth and Luria-Bertani broth) were obtained from
Thermo Fisher Scientific (Pittsburgh, PA, USA). The modified
BacTiter-Glo Microbial Cell Viability Assay kit (Promega
Corporation, Madison, WI, USA) was purchased for the determination
of minimum inhibition concentration (MIC). The polymerase chain
reaction (PCR) reagents were obtained from Bio-Rad Laboratories
(Hercules, CA, USA), and Invitrogen Life Technologies (Carlsbad,
CA, USA).
Bacterial culture
Porphyromonas gingivalis ATCC 33277 was
purchased from the German Collection of Microorganisms and Cell
Cultures (DSMZ; Braunschweig, Germany). The cells were cultured in
modified Gifu anaerobic medium (GAM) broth (Nissui, Tokyo, Japan),
in an aerobic jar and in the presence of a deoxygenating reagent
(AnaeroPack; Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan)
for 48 h at 37°C. Cell concentration was standardized by measuring
optical density at 650 nm using a Lumetron colorimeter (Photovolt
Corp., Indianapolis, IN, USA).
MIC and minimum bactericidal
concentration (MBC) assay
MICs were determined with modifications for each
organism according to methods described by Cole et al
(15). In brief, the appropriate
growth medium for P. gingivalis was used to prepare 5-ml
overnight cultures to an exponential phase. Bacteria were adjusted
to a concentration of 4.5×105 colony-forming units/ml,
added to various concentrations of antibiotic in 96-well plates,
and incubated at 37°C for a period of 18–24 h in a humidified
container. The MIC was defined as the lowest concentration that
prevented 50% growth of cells. MBCs were determined by plating the
wells with concentrations of 50–200% MIC. Following 24–48-h growth,
the MBC was determined as the lowest concentration that did not
permit visible growth on the surface of the agar. All MIC assays
were performed in triplicate.
Scanning electron microscopy (SEM)
P. gingivalis cultures were grown to the
mid-log phase, and 10 ml cell suspension [1×104 cells/ml
in modified GAM supplemented with α-amylase (12 ng/ml) or
pentamidine (100 ng/ml)] was incubated at 37°C for 2 h prior to
collection and fixed in 2.5% glutaraldehyde. The samples were
dehydrated with graded ethanol and t-butanol, dried using the
critical point method and coated with gold. Cells were observed
under a JSM-6510 LV scanning electron microscope (JEOL, Ltd.,
Tokyo, Japan).
Determination of differential gene
expression
The differential gene expression was determined by
quantitative (q) PCR. The bacterial culture (P. gingivalis)
grown to early exponential phase was adjusted to an optical density
600 of 0.1 and split into two groups. One half was left untreated,
while the other half was treated with α-amylase (12 ng/ml) and
pentamidine (100 ng/ml). Following anaerobic incubation for 2 h,
the cells were harvested, and total RNA was extracted using TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). cDNA was
synthesized with 1 µg total RNA using the SuperScript II
reverse transcriptase (Invitrogen Life Technologies). To identify
the expression value of genes associated with hemagglutination,
hemolysis, proteolysis, hemin uptake, chromosome replication,
energy production, iron storage and oxidative stress, qPCR was
performed using specific primers for the selected genes (Table I). The housekeeping gene
glyceraldehyde 3-phosphate dehydrogenase (gapA) was used as a
control gene. qPCR was conducted using the MiniOpticon Real-Time
PCR Detection system (Bio-Rad Laboratories) with a reaction mixture
containing 10 µl iQ SYBR Green Supermix (Bio-Rad
Laboratories), 1 µl cDNA and primers to a final
concentration of 250 nm in a final volume of 20 µl. To
confirm that a single PCR product was amplified, a melting curve
analysis was performed under the following conditions: 65°C to
95°C, with a heating rate of 0.2°C/sec. All quantifications were
normalized to the P. gingivalis 16S ribosomal RNA gene.
 | Table IPrimer sequences used in the current
study. |
Table I
Primer sequences used in the current
study.
| Target gene | Gene
identification | Primer sequence
(5′-3′) |
|---|
| 16S ribosomal
RNA | |
F:TGTTACAATGGGAGGGACAAAGGG
R:TTACTAGCGAATCCAGCTTCACGG |
| gapA | GAPDH, type I |
F:GGCAAACTGACGGGTATGTC
R:ATGAAGTCGGAGGAAACCAC |
| atpA | ATP synthase subunit
A |
F:ATCAGGACGGGAAAGACCAC
R:ACGATGGGGTTGAAAGTGTC |
| cydA | Cytochrome d
ubiquinol oxidase, subunit I |
F:TGGATTCTTATCGCCAATGC
R:ATACGCCCAAAGCAAATACG |
| dnaG | DNA primase |
F:GACACAGGGCTTTCCATCC
R:GCGAGCAATCTCTTTCTTGG |
| dps | Dps family
protein |
F:CAGAAGTGAAGGAAGAGCACGAA
R:GTAGGCAGACAGCATCCAAACG |
| Rbr | Rubrerythrin |
F:TCCACGGCTGAGAACTTGCG
R:TGCTCGGCTTCCACCTTTGC |
| ftn | Ferritin |
F:CGTGGCGGCGAGGTGAAG
R:CGGAAGCAGCCCTTACGACAG |
| sodB | Superoxide dismutase,
Fe-Mn |
F:GCCAAACCCTCAACCACAATCTC
R:GCCATACCCAGCCCGAACC |
| hagA | Hemagglutinin protein
HagA |
F:ACAGCATCAGCCGATATTCC
R:CGAATTCATTGCCACCTTCT |
| hagB | Hemagglutinin protein
HagB |
F:TGTCACTTGACACTGCTACCAA
R:ATTCAGAGCCAAATCCTCCA |
| rgpA | Arginine-specific
cysteine proteinase |
F:GCCGAGATTGTTCTTGAAGC
R:AGGAGCAGCAATTGCAAAGT |
| rgpB | Arginine-specific
cysteine proteinase |
F:CGCTGATGAAACGAACTTGA
R:CTTCGAATACCATGCGGTTT |
| kgp | Lysine-specific
cysteine proteinase |
F:GCTTGATGCTCCGACTACTC
R:GCACAGCAATCAACTTCCTAAC |
Results
Minimum inhibition and bactericidal
concentration assay
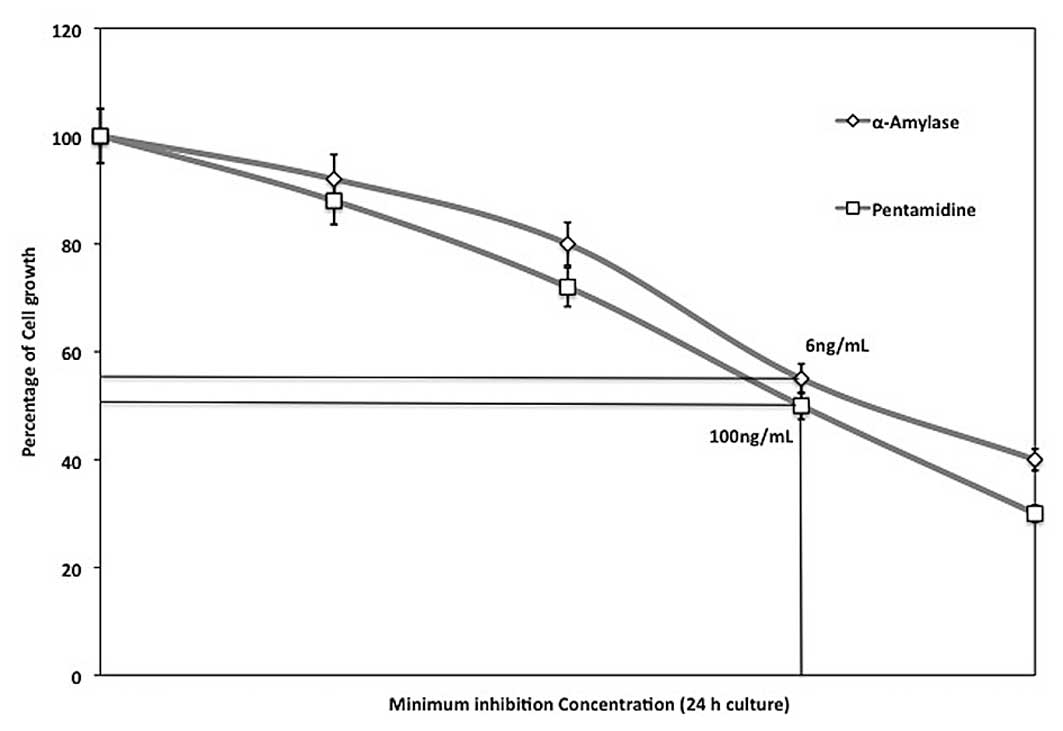
The minimum inhibitory activity against P.
gingivalis ATCC 33277 cell growth was measured using
differential concentrations of α-amylase (2, 4, 6 and 8 ng/ml) and
pent-amidine (50, 75, 100 and 125 ng/ml). The concentrations that
prevented 50% growth of cells observed for 24 h were considered to
be the MIC and were determined as 6 and 100 ng/ml α-amylase and
pentamidine, respectively (Fig.
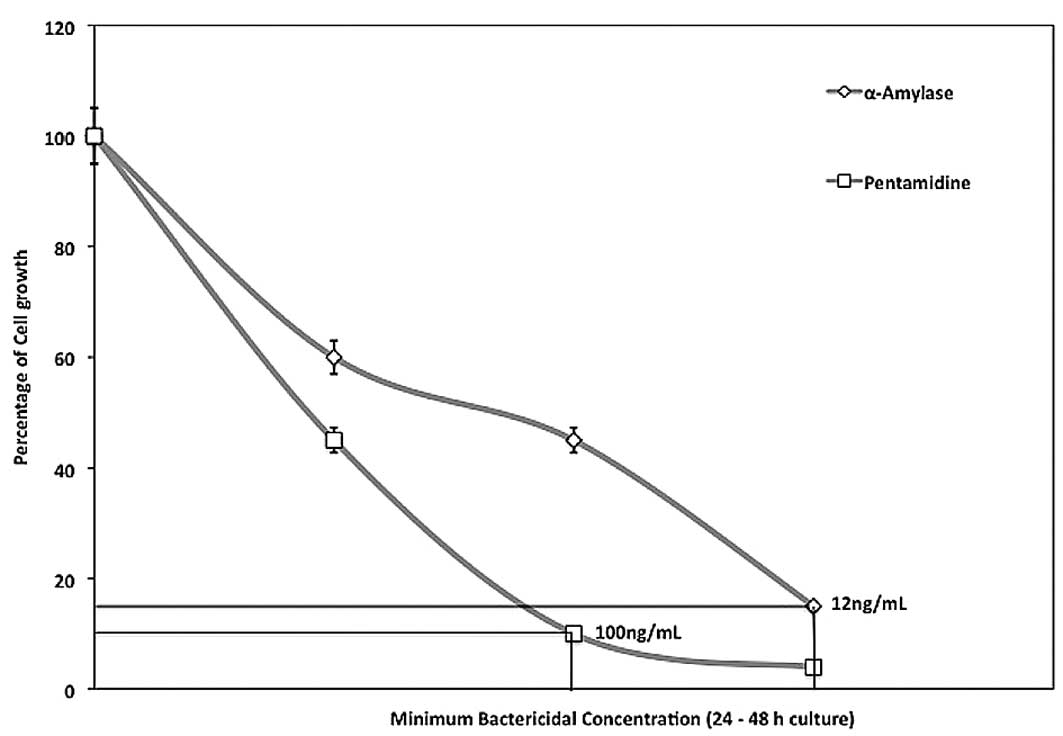
1). The MBC was tested using concentrations of 50–200% MIC and
cells were cultured for 24–48 h. The MBCs were determined as 12 and
100 ng/ml for α-amylase and pentamidine, respectively (Fig. 2). This is the lowest concentration
that did not permit visible growth on the surface of the agar,
suggesting that P. gingivalis cells underwent significant
cellular damage.
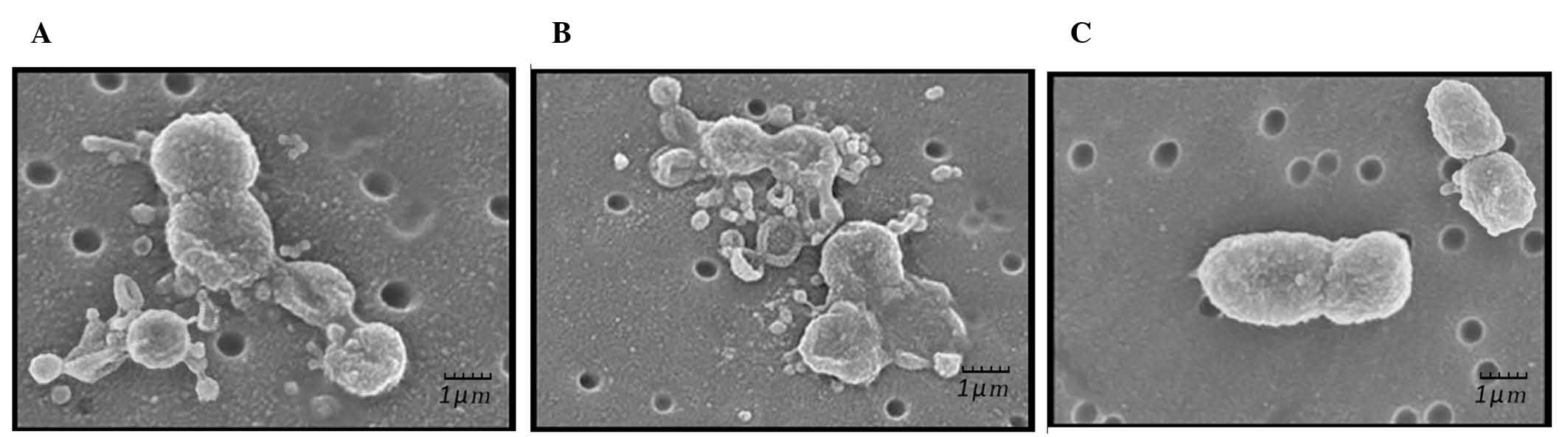
SEM analysis
SEM results demonstrated that P. gingivalis
cells treated with α-amylase (Fig.
3A) or pentamidine (Fig. 3B)
exhibited various stages of lysis. Cellular debris and detached
pieces of membrane lay adjacent to the cells. A number of cells
were distorted with irregular morphology and loss of cellular
content. In addition, the cells were more closely aggregated and
increased numbers of external blebs were present on and around the
bacteria compared with those in controls (Fig. 3A). Similar to α-amylase-treated
bacteria, pentamidine-treated P. gingivalis (Fig. 3B) was also distorted with irregular
morphology and presented various stages of lysis with loss of
intracellular content. Untreated P. gingivalis (Fig. 3C) cells exhibited an external
structure typical of a healthy Gram-negative coccobacillus 7
bacterium with multiple blebs present on the cell surface.
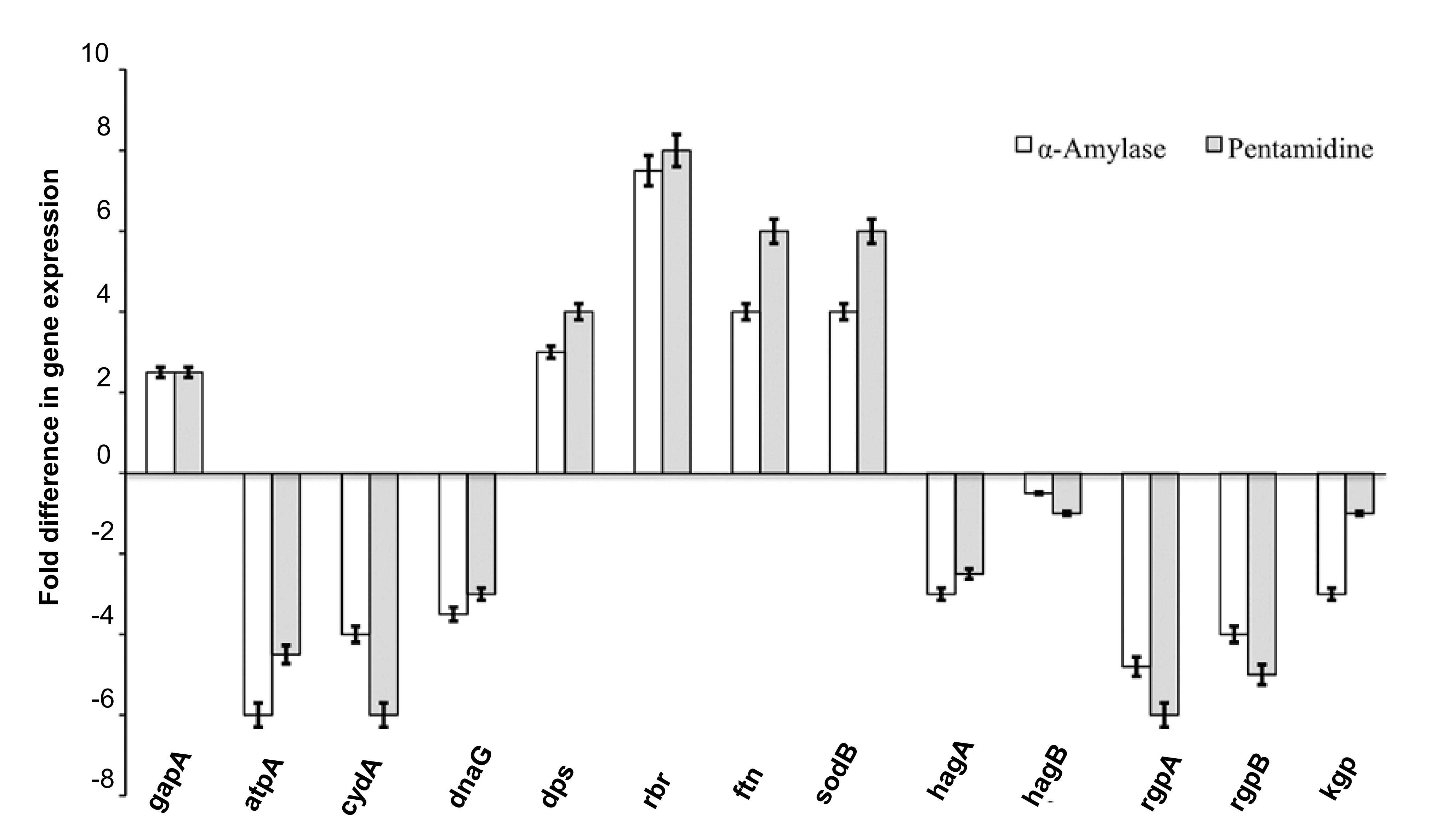
Determination of differential gene
expression
Following determination of the MBC, differential
gene expression was investigated by exposing the culture to the
MBC. The genes associated with iron storage (dps, rbr and ftn) and
oxidative stress (sodB) were indicated to have upregulated
expression levels, while levels of genes encoding gingipains (rgpA,
rgpB and kgp) and hemagglutinins (hagA and hagB) were downregulated
by α-amylase and pentamidine (Fig.
4). The inhibitors also downregulated the expression of genes
associated with energy production (atpA and cydA) and chromosome
replication (dnaG). The expression levels of the control gene gapA
were not significantly affected.
Discussion
In the current study, two potent inhibitors of the
P. gingivalis species (α-amylase and pentamidine) were
investigated. These amylases, which are endogenous to saliva and
oral mucosa are antimicrobial for P. gingivalis, and induce
structural damage. α-amylase and pentamidine demonstrated
significant inhibitory activity against P. gingivalis cell
growth, and had MICs of 6 and 100 ng/ml, respectively. Similarly,
the MBCs of α-amylase and pentamidine were determined as 12 and 100
ng/ml, respectively. SEM analysis suggested that the cell membrane
structure of bacterial cells was compromised, likely resulting from
damage caused by the inhibitors. However, the nature and mechanisms
of action of the inhibitors remain unclear.
The results of the present study are in agreement
with growing evidence that pentamidine kills bacteria in a
dose-dependent manner and induces cellular damage. For example,
E. coli and S. aureus treated with sphingosine,
phytosphingosine or dihydrosphingosine exhibit extensive and
differential intracellular and extracellular damage (16). Bibel et al (17) also demonstrated that sphinganine
(dihydrosphingosine) treatment of S. aureus results in
ultrastructural damage similar to antibiotic treatment, including
lesions of the cell wall, membrane evaginations and leakage. In
addition, treatment of Helicobacter pylori with oleic or
linoleic acid results in altered morphology, with a disruption of
cellular membranes and cell lysis (18). The present study indicates that
there may be different mechanisms underlying the actions of
different inhibitors. Antimicrobial activity and ultrastructural
damage are dependent upon the specific lipid treatment. These data,
combined with a previous observation that fatty acids and sphingoid
bases exhibit differential activity across bacterial species
(19), suggest that the
antimicrobial activity of fatty acids and sphingoid bases is a
specific interaction that depends upon characteristics of the
bacterium and a particular lipid. The current study indicates that
the mechanisms for the antimicrobial activity of α-amylase and
pentamidine against bacteria involve membrane disruption by
detergent activity and incorporation of lipids into the bacterial
plasma membrane.
Surface-accumulated hemin is transported into
bacterial cells so that it can be utilized. To evaluate the effect
of α-amylase and pentamidine on P. gingivalis, the
expression levels of selected genes were analyzed to determine
whether they were up- or downregulated. The genes associated with
iron storage (dps, rbr and ftn) and oxidative stress (sodB)
presented upregulated expression levels. Notably, pentamidine
increased the level of cell-associated hemin, which suggests that
surplus hemin is accumulated on the bacterial cell surface
regardless of energy-driven transport in the presence of
pentamidine. A small decrease in the level of kgp was observed, the
suppressed formation of µ-oxo bisheme may be explained by
the fact that RgpA or RgpB, or the two together, with kgp activity
are required by P. gingivalis to produce µ-oxo
bisheme (20). The formation of
µ-oxo bisheme represents an oxidative buffer mechanism for
inducing an anaerobic miroenvironment and protects from
hemin-mediated cell damage (20,21).
Therefore, excessive accumulation of hemin in the vicinity of the
bacterial cell surface without formation of µ-oxo bisheme by
the bacterium may cause oxidative stress on P. gingivalis.
This expectation was confirmed by qPCR, which indicated
upregulation of the genes involved in oxidative stress, such as
dps, rbr, ftn and sodB. An oxidative-stress-like phenomenon is one
of the shared downstream events leading to bacterial cell death
initiated by bactericidal antibiotics (22). Additionally, during bacterial cell
death, genes for energy production, chromosome replication and
nucleotide metabolism have been demonstrated to be inactivated
(23). Therefore, the observation
of the oxidative-stress-like response of P. gingi-valis and
decreased expression of the genes required for ATP synthesis and
chromosome replication of the bacterium grown with α-amylase and
pentamidine in the current study may also support the idea that
these inhibitors have a bactericidal effect.
In conclusion, the present study demonstrated that
the use of the growth inhibitors α-amylase and pentamidine for
controlling bacterial infection and aiding the innate immune
system, in addition to promoting gene expression, may be an
effective strategy for the prevention and treatment of oral cavity
infection. Following comparison of these two inhibitors,
pentamidine was demonstrated to be more effective at inhibiting the
growth of P. gingivalis cells. However, further
investigation is required to investigate its suitability for use in
humans.
References
|
1
|
Darveau RP, Tanner A and Page RC: The
microbial challenge in periodontitis. Periodontol 2000. 14:12–32.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eke PI, Dye BA, Wei L, et al CDC
Periodontal Disease Surveillance Workgroup: Prevalence of
periodontitis in adults in the United States: 2009 and 2010. J Dent
Res. 91:914–920. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hutter G, Schlagenhauf U, Valenza G, et
al: Molecular analysis of bacteria in periodontitis: evaluation of
clone libraries, novel phylotypes and putative pathogens.
Microbiology. 149:67–75. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hajishengallis G, Liang S, Payne MA, et
al: Low-abundance biofilm species orchestrates inflammatory
periodontal disease through the commensal microbiota and
complement. Cell Host Microbe. 10:497–506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holt SC, Kesavalu L, Walker S and Genco
CA: Virulence factors of Porphyromonas gingivalis. Periodontol
2000. 20:168–238. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lamont RJ and Jenkinson HF: Subgingival
colonization by Porphyromonas gingivalis. Oral Microbiol Immunol.
15:341–349. 2000. View Article : Google Scholar
|
|
7
|
Gorr SU: Antimicrobial peptides in
periodontal innate defense. Front Oral Biol. 15:84–98. 2012.
|
|
8
|
Gupta R, Gigras P, Mohapatra H, et al:
Microbial α-amylases: a biotechnological perspective. Process
Biochem. 38:1599–1616. 2003. View Article : Google Scholar
|
|
9
|
Rajagopalan G and Krishnan C:
Alpha-amylase production from catabolite derepressed Bacillus
subtilis KCC103 utilizing sugarcane bagasse hydrolysate. Bioresour
Technol. 99:3044–3050. 2008. View Article : Google Scholar
|
|
10
|
Pandey A, Nigam P, Soccol CR, et al:
Advances in microbial amylases. Biotechnol Appl Biochem. 31(Pt 2):
135–152. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burchmore RJ, Ogbunude PO, Enanga B and
Barrett MP: Chemotherapy of human African trypanosomiasis. Curr
Pharm Des. 8:256–267. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilson WD, Tanious F, Mathis A, et al:
Antiparasitic compounds that target DNA. Biochimie. 90:999–1014.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pearson RD and Hewlett EL: Pentamidine for
the treatment of Pneumocystis carinii pneumonia and other protozoal
diseases. Ann Intern Med. 103:782–786. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Charpentier TH, Wilder PT, Liriano MA, et
al: Divalent metal ion complexes of S100B in the absence and
presence of pent-amidine. J Mol Biol. 382:56–73. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cole AM, Weis P and Diamond G: Isolation
and characterization of pleurocidin, an antimicrobial peptide in
the skin secretions of winter flounder. J Biol Chem.
272:12008–12013. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fischer CL, Walters KS, Drake DR, et al:
Sphingoid bases are taken up by Escherichia coli and Staphylococcus
aureus and induce ultrastructural damage. Skin Pharmacol Physiol.
26:36–44. 2013. View Article : Google Scholar :
|
|
17
|
Bibel DJ, Aly R, Shah S and Shinefield HR:
Sphingosines: antimicrobial barriers of the skin. Acta Derm
Venereol. 73:407–411. 1993.PubMed/NCBI
|
|
18
|
Khulusi S, Ahmed HA, Patel P, Mendall MA
and Northfield TC: The effects of unsaturated fatty acids on
Helicobacter pylori in vitro. J Med Microbiol. 42:276–282. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fischer CL, Drake DR, Dawson DV, et al:
Antibacterial activity of sphingoid bases and fatty acids against
Gram-positive and Gram-negative bacteria. Antimicrob Agents
Chemother. 56:1157–1161. 2012. View Article : Google Scholar :
|
|
20
|
Smalley JW, Birss AJ, Szmigielski B and
Potempa J: The HA2 haemagglutinin domain of the lysine-specific
gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo
bishaem formation from monomeric iron(III) protoporphyrin IX.
Microbiology. 152:1839–1845. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lewis JP, Dawson JA, Hannis JC, Muddiman D
and Macrina FL: Hemoglobinase activity of the lysine gingipain
protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol.
181:4905–4913. 1999.PubMed/NCBI
|
|
22
|
Wright GD: On the road to bacterial cell
death. Cell. 130:781–783. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Asakura Y and Kobayashi I: From damaged
genome to cell surface: transcriptome changes during bacterial cell
death triggered by loss of a restriction-modification gene complex.
Nucleic Acids Res. 37:3021–3031. 2009. View Article : Google Scholar : PubMed/NCBI
|