Introduction
Nitric oxide (NO) is an important signaling molecule
found in animals, including humans. Reduced NO production is
associated with important cardiovascular risk factors, such as
hyperlipidemia (1,2), diabetes, hypertension, smoking,
atherosclerosis and aging (3). The
bioavailability of NO in cells and tissues decreases with age. This
may be a result of a decrease in the expression of the constitutive
isoforms [endothelial nitric oxide synthase (eNOS) and neuronal NOS
(nNOS)], with age (4). Inducible
NOS (iNOS)-mediated NO formation has been shown to affect
longevity. In mice, iNOS overexpression may lead to increased
mortality, which is associated with cardiac hypertrophy and sudden
cardiac death as a result of bradyarrhythmias (5). iNOS expression is typically not
observed in the brains of young (1–3 months old) animals and its
expression increases with age (6).
Furthermore, it has been shown that in the brain superoxides
combine with NO to form peroxynitrite, thereby reducing the
bioavailability of NO (7).
Age-associated impairment of macrophage function is associated with
a substantial decrease in iNOS levels in the immune system
(8).
In the healthy nervous system, NO contributes, with
other molecules, to learning and memory; synaptic activity; neural
plasticity, including neurogenesis and cell survival; and cell
differentiation. NO is associated with neurodegeneration,
neuroinflammation and pathophysiological conditions, such as
Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s
disease, multiple sclerosis, Huntington’s disease (9,10)
and brain tumors.
NO activity has been extensively studied in tumor
biology (11–28). It appears to be involved in all
phases of carcinogenesis (12).
Tumoricidal and tumor-promoting effects of NO have been observed,
which depend on the type of cancer and the availability of NO
(13). eNOS-derived NO has been
shown to enhance angiogenesis in gliomas (14). Low grade gliomas demonstrate
constitutive expression of eNOS in vessel endothelial cells and
astrocytes, whereas malignant gliomas demonstrate overexpression of
eNOS in aberrant vessels (15).
Furthermore, studies have demonstrated that Enos-deficient
(eNOS−/−) mice may be resistant to chemical
carcinogenesis (16) and
platelet-derived growth factor-induced gliomagenesis (17). eNOS inhibition was shown to lead to
a decrease in tumor neovascularization, vascular permeability and
tumor growth, in a number of carcinoma models (18,19).
Furthermore, increased nNOS expression has been observed in glial
neoplasms. The highest nNOS values were observed in world health
organization (WHO) grade III and IV tumors, and in carcinoma and
melanoma metastasic tissues, whereas no or low nNOS expression was
observed in WHO grade I or II tumors, and in meningiomas (20,21).
iNOS expression is induced in different types of human brain
tumors, including gliomas (22),
where it is known to promote glioma stem cell proliferation and
tumor growth (23). Therefore, the
inhibition of iNOS expression may be a useful therapeutic approach
for the suppression of local cancer growth (11,24).
Studies have suggested that NO donors may exhibit
antiglioma (25) or antileukemic
(26) effects. Evidence from
previous studies indicates that NOS knockout may induce tumor
development and progression (27,28).
NO is referred to as a ‘double-edged sword’ for a number of
reasons: The three NOS isoforms show different spatial and temporal
expression patterns; the levels of NO production are variable,
depending on its (sub-)cellular source; NO may cause either cell
death or proliferation; and, depending on its concentration, NO may
exhibit anti- or pro-inflammatory activity.
As with NOS isoforms, studies have shown that
members of the matrix metalloproteinase (MMP) family and a
disintegrin and metalloproteinase domain-containing proteins
(ADAMs) are overexpressed in high-grade glioma. Glioma-associated
proteins include MMP-1, MMP-11, MMP-19 (29,30)
and ADAM12 (31). These
extracellular matrix degrading enzymes are associated with the
fatal invasive capacity of high grade gliomas (32,33).
Furthermore, ADAM12 determines the localization, and promotes the
activation, of MMP-14 (34).
It is hypothesized that ADAM12 and NO may act
synergistically. NO suppresses MMP-9 expression by destabilizing
its mRNA in rat mesangial cells (35), whereas inhibition of NOS has been
found to promote cytokine-induced MMP-9 expression in aortic smooth
muscle cells (36) and
exotoxin-mediated MMP expression in iNOS−/− mice
(37). Furthermore, NO may inhibit
hypoxia-induced expression of ADAM10 (38). Therefore, it is hypothesized that
NO may affect the expression of other ADAMs, including ADAM12. The
present study analyzed ADAM12 expression levels in the cortex and
hippocampus of wild-type mice, and in eNOS−/−,
nNOS−/− and iNOS−/− mice, at different stages
of development. For NO production, an age dependency is clear,
however concerning ADAM12 expression in aged animals, available
data are rare and limited to the musculature only (39).
ADAM12 belongs to a subgroup of the ADAM family,
consisting of ADAM8, ADAM9, ADAM10, ADAM17 and ADAM19. They are
cell-surface glycoproteins, containing metalloprotease, disintegrin
(Arg-Gly-Asp-binding motif), cysteine-rich and epidermal growth
factor (EGF)-like domains, and they are responsible for the release
of the extracellular parts of membrane-bound proteins (shedding).
Two splice variants of ADAM12 occur in humans: Membrane-anchored
(ADAM12L) and cytoplasmic secreted (ADAM12S) (40). ADAM12 is proteolytically active
in vitro and in vivo. ADAM12 may cause the shedding
of pro heparin-binding-EGF (proHB-EGF), insulin-like growth
factor-binding protein 3 (IGFBP-3), IGFBP-5 and oxytocinase
(41). Recently, ADAM12 has been
associated with ectodomain shedding of endothelial proteins in
tumor vasculature (42). A
consensus sequence that is required for, or facilitates, ADAM12
cleavage remains to be identified (43). The cytoplasmic tail of ADAM12 is
one of the longest observed among the ADAMs (179 amino acids). It
appears to be involved in the regulation of ADAM12 cellular
localization (44). The
cytoplasmic domain of ADAM12 contains a number of proline-rich
motifs that are putative Src homology 3 (SH3) binding sites, which
enable the recruitment of adapter molecules and subsequent
activation of cellular signaling pathways. In addition, the
cytoplasmic domain may contain one potential tyrosine
phosphorylation site (SH2-binding site) and several
serine/threonine phosphorylation sites (40). The following cytoplasmic binding
partners have been identified for ADAM12: Src, proto-oncogene
tyrosine-protein kinase Yes, growth factor receptor-bound protein
2, phosphoinositide 3-kinase, α-actinin-1, α-actinin-2, protein
kinase C Δ, fluorescence in situ hybridization and protein
kinase C, and casein kinase substrate in neurons protein 3
(40,41). Cytoplasmic binding sites may
influence the maturation, trafficking, membrane (raft) localization
and proteolytic activity of ADAMs.
In the early stages of mouse development, ADAM12
mRNA expression is prominent in mesenchymal cells, which develop
into skeletal muscle, bone and visceral organs (45). ADAM12 mRNA may be detected at
>10.5 DNA protein cross-link levels. It was originally reported
to exhibit a restricted expression pattern in adult tissues, and
the highest levels of ADAM12 expression have been observed in bone
tissue samples (46). More
recently, it has been shown that ADAM12 is ubiquitously expressed
in adult mice. Studies have demonstrated that ADAM12 expression in
mouse brain is predominantly, yet not exclusively, observed in
oligodendrocytes (47,48).
Materials and methods
Mice
Experiments were performed in accordance with the
recommendations of and was approved by the Commission for Animal
Care of the State of Saxony-Anhalt (Dessau, Germany) and German law
of the Protection of Animals (permit number: 42502-2783 UniMD). The
present study was conducted with laboratory-bred mice. Mice
(C57BL/6 and wild-type control strains) were obtained from the
homozygous institute breeding colony. nNOS−/− mice were
provided by the Hunag Lab (49,50),
eNOS−/− mice were provided by the Gödecke Lab (51), iNOS−/− mice were
obtained from Charles River Laboratories (Sulzfeld, Germany) as
developed in the C57BL/6 strain (Charles River Laboratories) and
their wild-type control strains. Animals were housed at 21°C,
exposed to a 12 h light and darkness cycle with access to food and
water, ad libitum. Tissue from fetal, neonatal (5 days
postnatal), adult (12 weeks old) and >1 year old mice was used,
whereby the >1 year group consisted of animals aged between 12
and 18 months old. (n=3 per age group).
Tissue preparation
In order to minimize circadian influences on the
expression of hypothalamic peptides, mice were sacrificed between
09:00-09:30 h. Mice were anesthetized using isoflurane (Baxter GmbH
Deutschland, Unterschleißheim, Germany). Subsequently, the mice
were subjected to perfusion through the left ventricle for 30 sec,
with 0.1 M phosphate-buffered saline (PBS; pH 7.4; Sigma-Aldrich,
Heidelberg, Germany) and then at 15 ml/min with 250 ml 4% buffered
paraformaldehyde (pH 7.4; PFA; Sigma-Aldrich). Brains were removed
and post-fixed in 4% buffered PFA overnight at room temperature.
The brains were subjected to cryoprotection for 2 days using 20%
sucrose in 0.4% buffered PFA (pH 7.4) and embedded in paraffin.
Brain coronal sections (10 µm) were prepared using a sliding
microtome (SM2010 R, Leica, Bensheim, Germany). Tissue samples were
obtained from mice (n=10 per age group) that had not been subjected
to perfusion and were directly frozen in liquid nitrogen (−80°C),
in order to conduct reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and immunoblot analyses.
Cell cultures
The following cell lines, representing the four main
cell types in the brain, were used in the experiments: N9 mouse
microglia cell line, [Ricciardi-Castagnioli lab (52)], C6 rat astroglia [CCL-107™;
American Type Culture Collection (ATCC), Manassas, VA, USA], OLN-93
rat oligodendroglial [Richter-Landsberg lab (53)] and rat neuronal PC12 (CRL-1721™;
ATCC). Cryopreserved cells were defrosted, resuspended in an RPMI
1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine
serum, 1% L-glutamine, 50 U/ml penicillin and 50 µg/ml
streptomycin (Life Technologies GmbH, Darmstadt, Germany), and
transferred to culture flasks (Sarstedt, Nümbrecht, Germany).
Following 3 days of incubation, cells were removed from the flasks
by gentle agitation and incubated on poly-D-lysine coated Ø35-mm
petri dishes (Sarstedt; ~50,000 cells/dish), for 6 days.
Subsequently, the respective experiments were performed, using the
following NOS inhibitors: L-N6-(1-Iminoethyl)lysine
dihydrochloride (L-NIL; 0.5 mM; specific inhibitor of iNOS),
Nω-Nitro-L-arginine methyl ester hydrochloride (L-NAME;
0.5 mM; inhibitor of nNOS and eNOS, to lesser extent also of iNOS)
and asymmetric dimethylarginine (ADMA; 10 µM; specific
inhibitor of eNOS) all for 24 h, all from Sigma-Aldrich. Cultures
were maintained at 37°C in a humidified 5% CO2
atmosphere and the medium was changed every other day.
Immunoblot analysis of ADAM12
Frozen tissue samples were pulverized in liquid
nitrogen and subsequently homogenized in lysis buffer (50 mM
Tris/HCl, pH 7.5, 5 mM EDTA, 100 mM NaCl, 0.5% Triton X-100, 10%
glycerol, 10 mM K2HPO4 and 0.5% NP-40; all
from Sigma-Aldrich), containing a protease inhibitor cocktail
(Boehringer, Mannheim, Germany), 1 mM sodium vanadate, 0.5%
deoxycholate, 0.1 mM phenylmethanesulfonylfluoride, 20 mM NaF, and
20 mM glycerol 2-phosphate (Sigma-Aldrich). Tissue homogenates were
centrifuged at 15,000 × g for 15 min and the resulting supernatant
was stored at −20°C.
Extracted proteins (30 µg per lane) were
separated using 10% SDS-PAGE and transferred to nitrocellulose
membranes (Schleicher and Schuell BioScience GmbH, Dassel,
Germany). Membranes were blocked using 1 × Roti-block solution
(Carl Roth, Karlsruhe, Germany) and then incubated with a primary
antibody against ADAM12 cytoplasmic domain (AB19032; 1:1,000
dilution, affinity isolated, polyclonal, anti-rabbit; Merck
Millipore, Darmstadt, Germany) diluted in PBS (0.1%) buffer with
Tween-20® in bovine serum albumin (BSA; 5%;
Sigma-Aldrich, Hofheim, Germany). Subsequently, the blots were
washed three times in PBS (0.3%) with Tween-20 and then polyclonal
horseradish peroxidase-conjugated anti-rabbit antibodies (#7074;
1:2,000 dilution with 1 × Roti-Block; Cell Signaling Technology,
Frankfurt, Germany) were applied. The SuperSignal West Dura
Extended Duration substrate (Pierce Biotechnology, Inc., Rockford,
IL, USA) was used in order to detect chemiluminescence. In order to
compare groups, densitometric quantification was performed on
equally processed blots that had been exposed to the same X-ray
film [CEA (Deutschland) GmbH, Hamburg, Germany].
RNA extraction and RT-qPCR analysis of
ADAM12 expression
Total RNA from cortical and hippocampal tissue
samples was extracted using a two-step protocol using a
TRIzol® extraction (Invitrogen Life Technologies,
Carlsbad, CA, USA) and the RNeasy kit™ (Qiagen, Hilden, Germany)
according to a previous study (54). Tissue samples were frozen in liquid
nitrogen and subsequently stored in 0.5 ml TRIzol at −80°C. Tissue
samples were then homogenized using DSTROY-S pistils (Biozym
Scientific GmbH, Oldendorf, Germany) by subjecting the sample to
3–5 freezing (using liquid nitrogen; −195°C) and thawing (using an
ice bath; 0°C) cycles. After complete homogenization, 0.2 ml
chloroform was added, and the mixture was extensively vortexed and
centrifuged using a microcentrifuge (12,000 × g, 4°C) for 15 min.
The supernatant was incubated with isopropanol (volume ratio 1:1)
at room temperature for 10 min, and the precipitated RNA was
obtained by centrifugation (12,000 × g, 4°C, 10 min). The RNA
pellet was resolved in 100 µl RNase-free water and
subsequently purified using the RNeasy kit, according to the
manufacturer’s instructions. Finally, the RNA was eluted in 50
µl RNase-free water, confirmed using gel electrophoresis,
and 5 µl of RNA was used to measure the concentration of the
RNA samples via ultraviolet spectroscopy (NanoDrop 2000c; Thermo
Fisher Scientific, Wilmington, DE, USA).
For the cell lines, RNA was isolated using the
innuPREP RNA isolation kit (Analytik Jena, Jena, Germany), as
described previously (55).
Total RNA (1 µg) was transcribed to a final
volume of 40 µl using 20 units of avian myeloblastosis virus
reverse transcriptase (Promega GmbH, Mannheim, Germany), containing
1 × reaction buffer, 0.5 mM dNTP (Roche Diagnostics GmbH, Mannheim,
Germany), 10 mM random hexanucleotides and 50 units of placenta
RNase inhibitor (Promega GmbH). The samples were incubated at 42°C
for 1 h. Subsequently enzymes were inactivated at 95°C for 10 min
and the reaction mixture was frozen at −80°C, prior to enzymatic
amplification.
ADAM12 transcript levels were determined using
RT-qPCR with an iCycler (Bio-Rad, Munich, Germany) and the
SensiMix™ dT kit (Bioline GmbH, Luckenwalde, Germany). The PCR
reaction mixture (25 µl) consisted of 12.5 µl of 2 ×
concentrated master mix (SensiMix™; Bioline GmbH), 2 µl
RT-reaction and 0.25 µM reverse and forward primers for
ADAM12, large ribosomal protein p0 (RPLP0) or 60S ribosomal protein
L13a (RPLP13a). The latter two primers were used for
standardization. The following PCR protocol was performed: Initial
denaturation and activation at 95°C for 10 min, followed by 40
cycles of denaturation at 94°C for 15 sec, annealing at 60°C for 20
sec and elongation at 72°C for 30 sec. Fluorescence intensity of
the double-stranded specific SYBR-Green I, reflecting the quantity
of PCR-product, was measured (CFX96 Thermocycler; Bio-Rad) at the
end of each elongation step. Correlation coefficients of all three
standard curves were >0.95. Final results are expressed as
artificial units. RPLP0 and RPLP13a expression levels were used to
normalize the cDNA contents. In order to verify the size of the PCR
product, samples were separated on 1.8% agarose gels and stained
with ethidium bromide. The following primers, which were designed
using the Invitrogen OligoPerfect Designer and obtained from
Invitrogen Life Technologies were used for the RT-qPCR analysis:
Forward: 5′-GCA CTC TCg CTT TCT GGA GGG TGT-3′ and reverse: 5′-TGA
CTT GGT TGC TTT GGC GGG ATT-3′ for mouse RPLP0 (344 bp), forward:
5′-CTG GTA CTT CCA CCC GAC CTC-3′ and reverse: 5′- GGA TCC CTC CAC
CCT ATG ACA-3′ for rat RPLP13a (131 bp), forward: 5′-CAG CAA CTC
CTG TGA CCT CC-3′ and reverse: 5′-GTA CCA ATG ACA GGT CGG CT-3′ for
mouse ADAM12 (333 bp) and forward: 5′-GCT TGC AGG AAC CAA GTG TG-3′
and reverse: 5′-CTG GTT ATC TGC TTG CCG GA-3′ for rat ADAM12 (226
bp).
Immunohistochemistry
ADAM12 immunoreactive material was immunolocalized
using a rabbit polyclonal antiserum against the appropriate peptide
sequence (1:200 dilution; SA-378; Biomol GmbH, Hamburg, Germany)
and a nickel-amplified avidin-biotin technique, as previously
described (47). The sections were
incubated with methanol and H2O2 in order to
suppress endogenous peroxidases. Subsequently, they were repeatedly
washed with PBS and the primary antiserum was then applied. The
immunohistochemical protocol used an avidin-biotin method and a
vectastain-peroxidase kit (Camon, Wiesbaden, Germany), using
3,3′-diaminobenzidine as chromogen. The color reaction was enhanced
by adding 2 ml of a 0.5% nickel ammonium sulfate solution, which
yields a dark purplish-blue reaction product. Specific antibodies
were replaced with buffer solution or normal rabbit serum (both
from Sigma-Aldrich) for the control samples. Pre-absorption of the
antiserum was performed using a peptide in order to produce the
antiserum, according to the methods described in a previous study
(47).
Statistical analysis
Data from brain tissue experiments were analyzed
using Microcal Origin™ version 6.0 (OriginLab Corporation;
Northampton, MA, USA). A Mann-Whitney test was used for the
evaluation of ADAM12 expression.
For statistical analysis of the culture experiments,
GraphPad Prism® 6.0 program package (Graphpad Software,
Inc., La Jolla, CA, USA) was used. A Kolmogorov-Smirnov-Test was
conducted in order to check that the results were normally
distributed, and a one-way analysis of variance was performed in
order to compare groups. Significant interactions were investigated
using Tukey’s post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
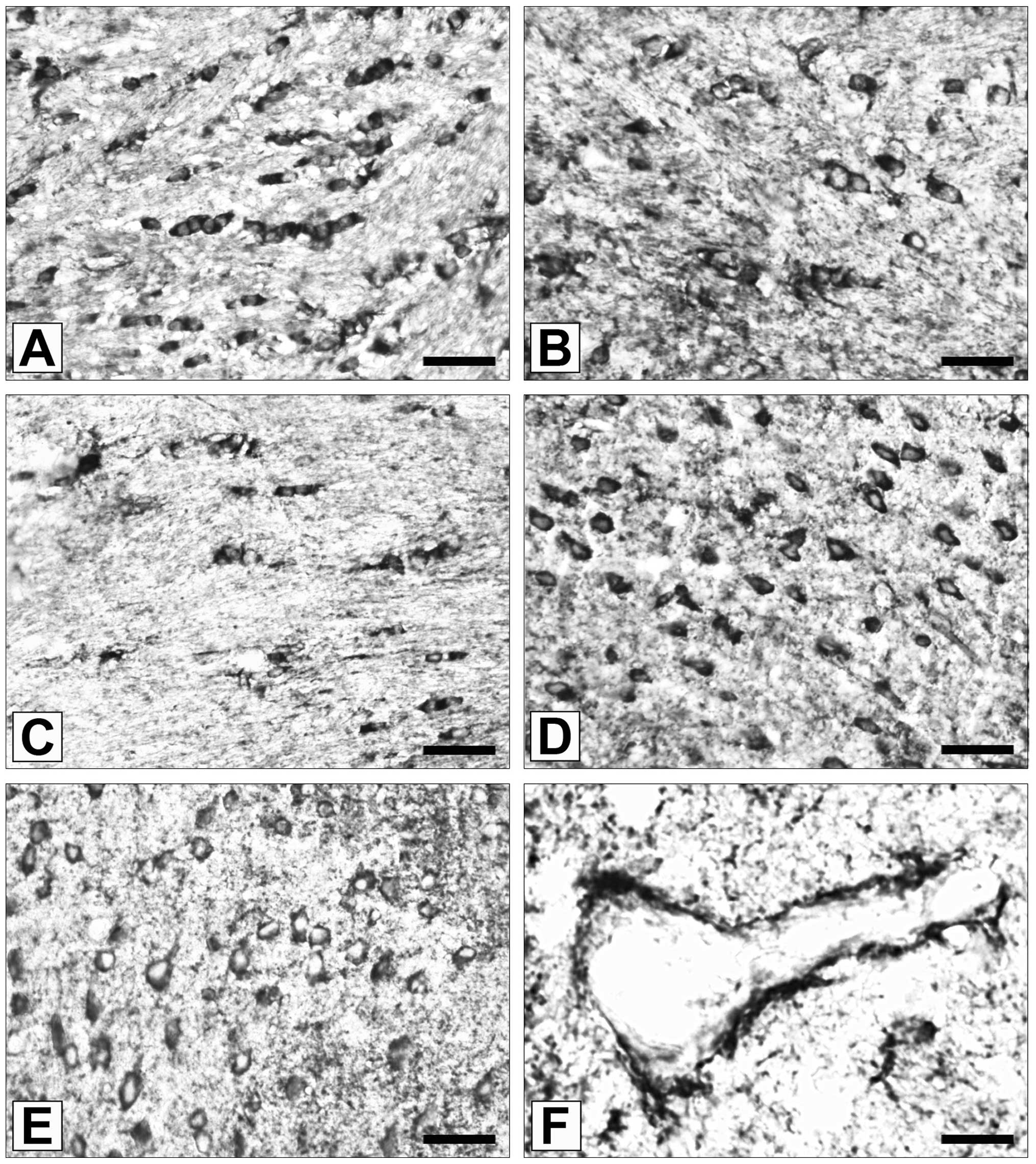
Localizations of ADAM12 expression
In the wild-type mouse brain samples, ADAM12
immunoreactivity was observed almost exclusively in
oligodendrocytes (Fig. 1A).
Oligodendrocytes were also the predominant cell type to express
ADAM12 in eNOS−/− and nNOS−/− mouse brain
samples (Fig. 1B and C; P>0.05
vs. wild-type). In addition to oligodendroglial cells, a number of
neurons were observed to be ADAM12 immunoreactive in
eNOS−/− (Fig. 1D) and
nNOS−/− (Fig. 1E) mouse
brain samples. By contrast, ADAM12 immunopositivity was observed
less frequently in neurons (P<0.001 vs. nNOS−/−;
P<0.002 vs. eNOS−/−), astrocytes and blood vessel
endothelial cells of wild type mice (Fig. 1F).
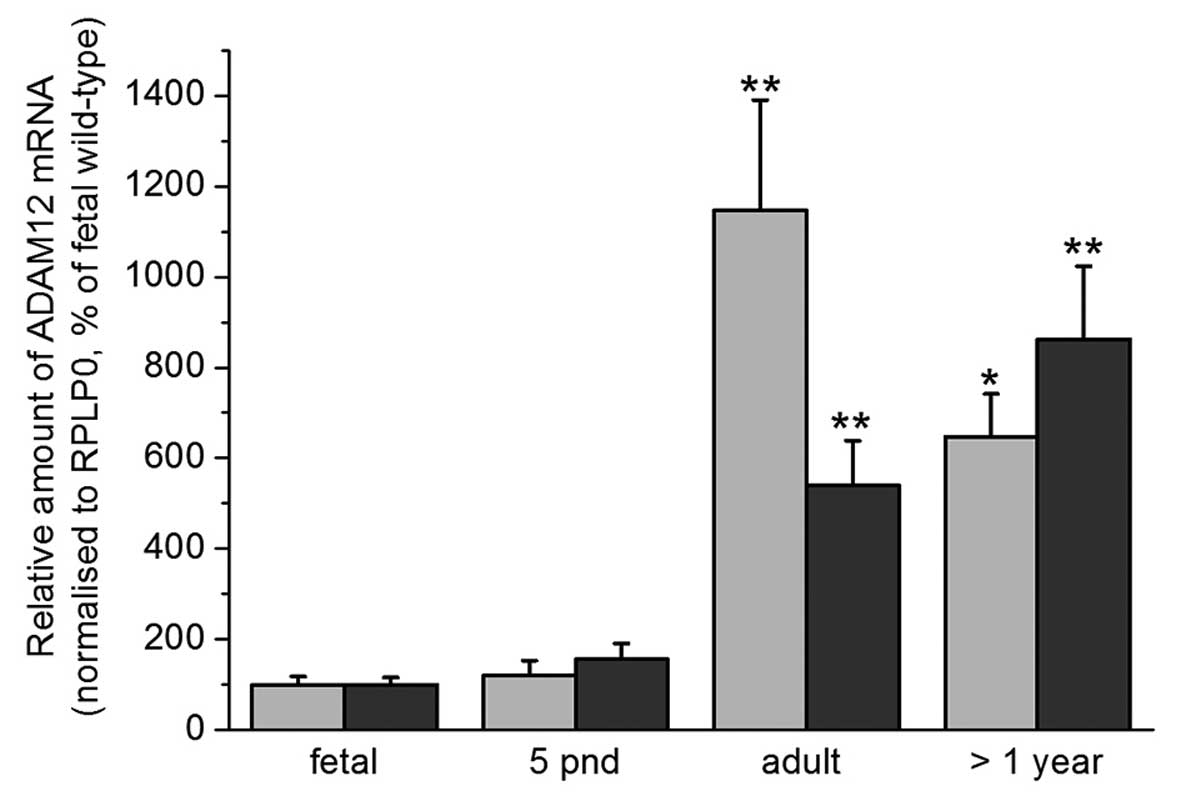
ADAM12 mRNA expression in the cortex and
hippocampus of wild type mice
RT-qPCR analysis was conducted in order to
investigate the expression levels of ADAM12 in the cortex and
hippocampus of wild-type mice. An increase in ADAM12 mRNA
expression was observed with increasing age. The highest levels of
ADAM12 mRNA expression were observed in the hippocampus of adult
mice (Fig. 2).
Expression of ADAM12 in the cortex of
wild-type and NOS−/− mice
In order to verify the age-associated changes in
ADAM12 mRNA expression levels, immunoblot analyses were performed.
Mature (90 kDa) ADAM12 was detected in the cortex of wild-type
C57/BL6 mice, at all ages. In accordance with the mRNA expression
data, protein expression was low in the cortex of fetal and
neonatal mice. By contrast, a markedly increased level of ADAM12
expression was observed in adult and >1 year old mice. A similar
increase in ADAM12 expression with age was observed in the cortex
of nNOS−/−, iNOS−/− and eNOS−/−
mice. However, in adult mice of the latter genotype, a tendency for
ADAM12 expression levels to remain below those of wild-type mice
was observed (P>0.05). Likewise, iNOS−/− mice >1
year old exhibited a tendency towards reduced levels of ADAM12
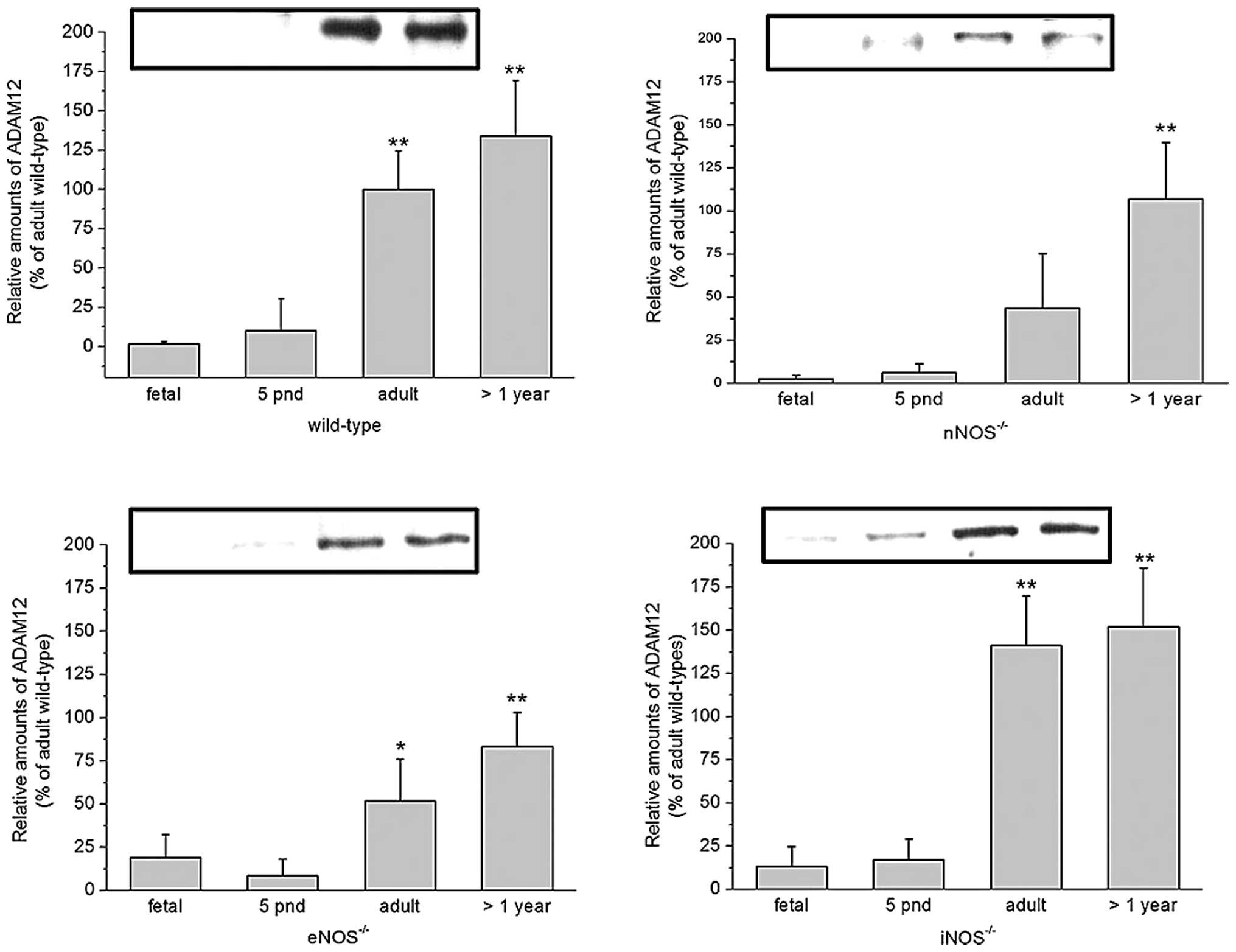
expression compared with wild-type mice (P<0.1) (Fig. 3).
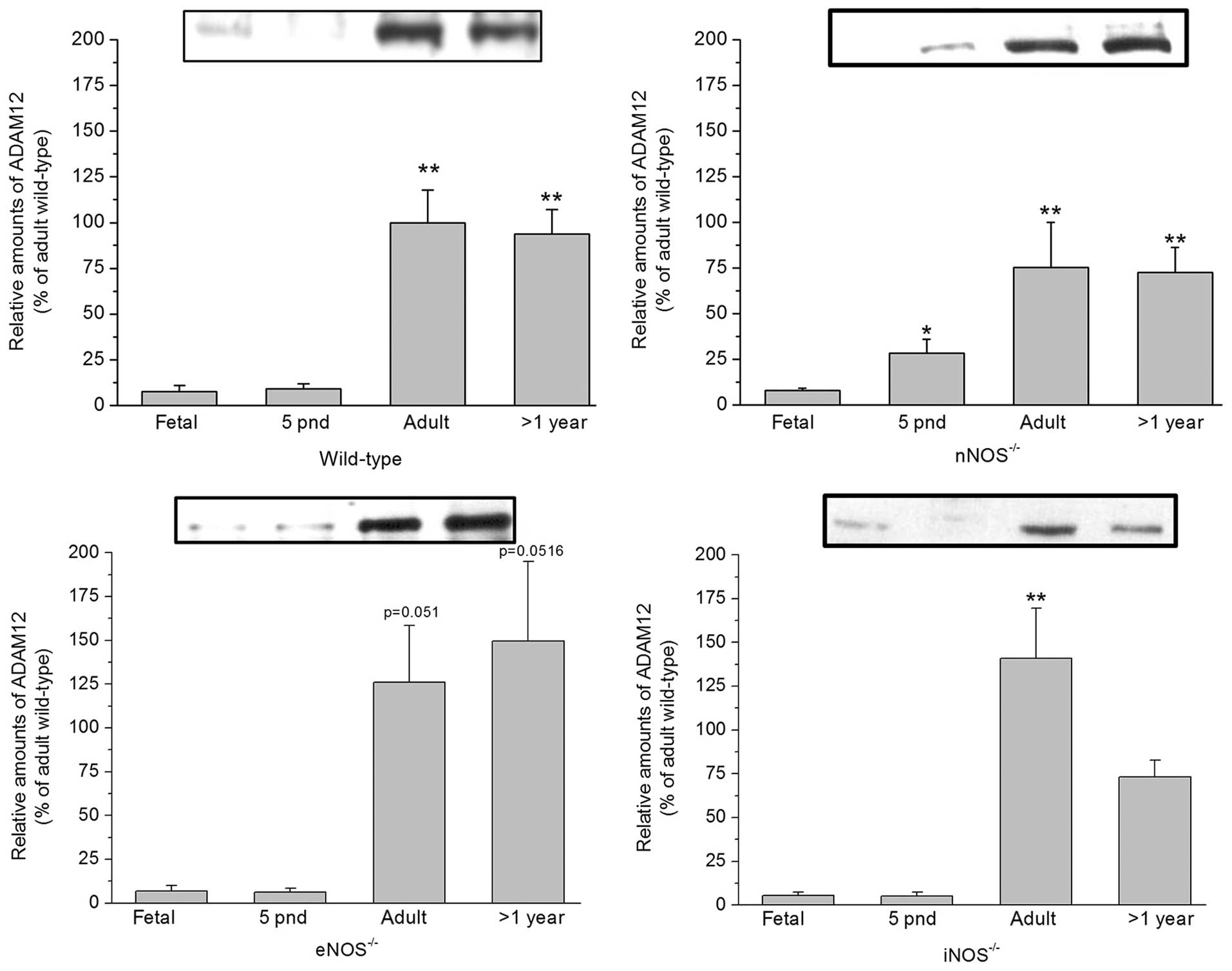 | Figure 3ADAM12 expression in the cortex of
wild-type (top left), nNOS−/− (top right),
eNOS−/− (bottom left) and iNOS−/− (bottom
right) mice at different ages, assessed using immunoblot analysis.
Data are presented relative to that of adult wild-type mice,
standardized to 100% (mean ± standard error, n≥6).
*P<0.05 and **P<0.01 vs. fetal).
ADAM12, a disintegrin and metalloproteinase domain-containing
protein 12; nNOS−/−, neuronal nitric oxide synthase
deficient; iNOS−/−, inducible nitric oxide synthase
deficient; eNOS−/−, endothelial nitric oxide synthase
deficient; pnd, postnatal days. |
Expression of ADAM12 in the hippocampus
of wild-type and NOS−/− mice
As with ADAM12 expression levels in the cortex,
mature (90 kDa) ADAM12 expression was observed in the hippocampus
of mice of all ages and genotypes. A marked increase in ADAM12
expression was observed in adult and mice >1 year old, compared
with that in fetal and neonatal mice. There was a tendency towards
increased levels of mature ADAM12 in iNOS−/− mice and
reduced expression in eNOS−/− and nNOS−/−
mice when compared with wild-type mice (P<0.1; Fig. 4).
 | Figure 4ADAM12 expression in the hippocampus
of wild-type (top left), nNOS−/− (top right),
eNOS−/− (bottom left) and iNOS−/− (bottom
right) mice at different age, as assessed using immunoblot
analysis. Data are presented relative to that of adult wild-type
mice, standardized to 100% (mean ± standard error, n≥6,
*P<0.05 and **<0.01 vs. fetal). ADAM12,
a disintegrin and metalloproteinase domain-containing protein 12;
nNOS−/−, neuronal nitric oxide synthase deficient;
iNOS−/−, inducible nitric oxide synthase deficient;
endothelial nitric oxide synthase deficient, eNOS−/−;
pnd, postnatal days. |
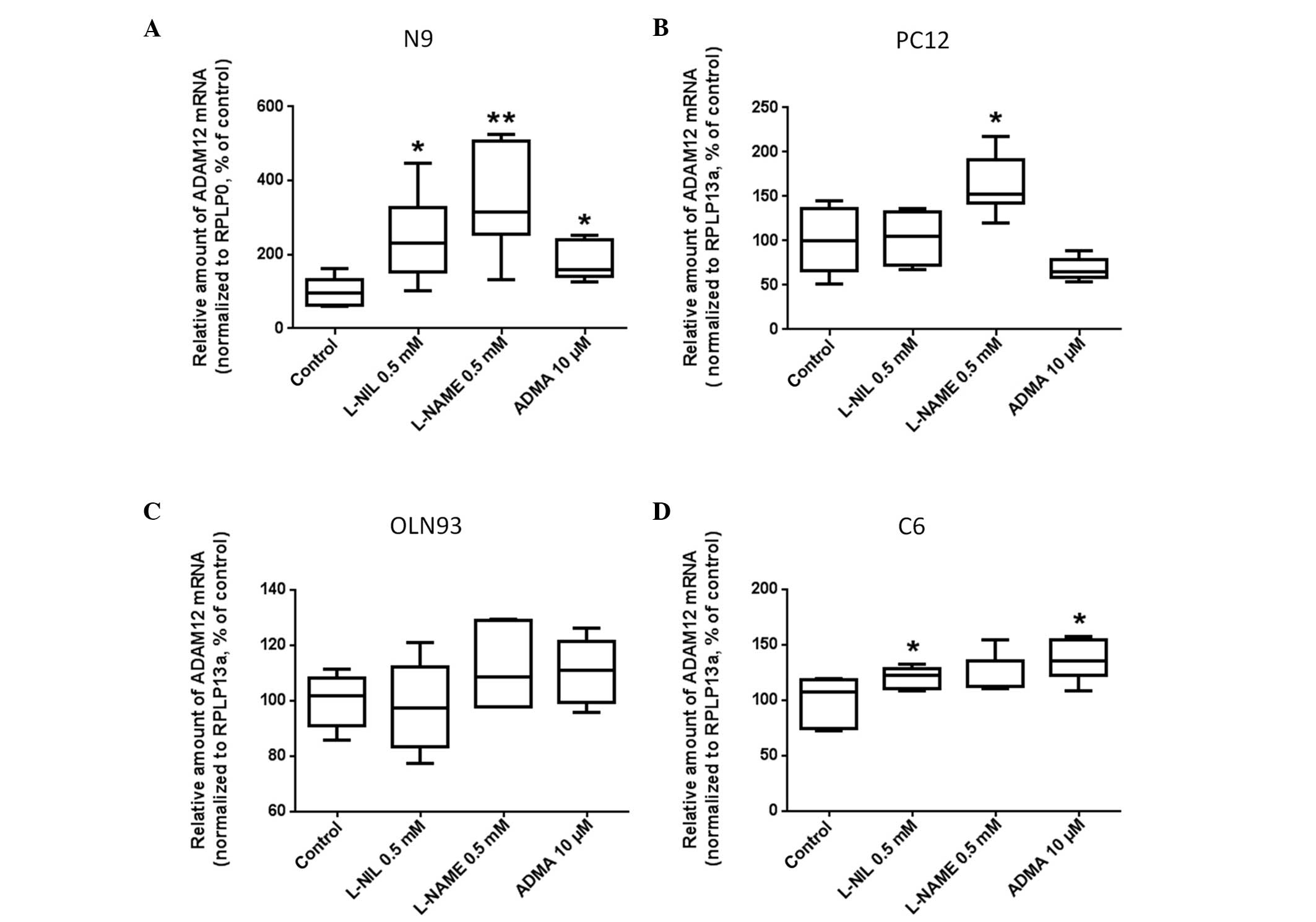
Effects of pharmacological inhibition of
NOS on ADAM12 expression in glial cell lines
The N9 microglial cell line was exposed to the
following NOS inhibitors: L-NIL, L-NAME and ADMA for 24 h.
Subsequently, ADAM12 mRNA expression in N9 cells was determined
using RT-qPCR. NOS inhibition using L-NIL and L-NAME (0.5 mM) led
to a significant increase in ADAM12 mRNA expression levels compared
with the controls (300±50%, P<0.05 and 400±50%, P<0.03,
respectively; Fig. 5A). NOS
inhibition using ADMA at 10 µM led to a significant, 2-fold
increase in ADAM12 mRNA expression in N9 cells, compared with that
in the control cells (P<0.02).
The effects of pharmacological NOS inhibition on
ADAM12 expression in the PC12 neuronal cell line was then
investigated. In the PC12 cell line, inhibition of NOS using
L-NAME, significantly increased ADAM12 mRNA expression, compared
with that of the controls (173±25%, P<0.05). By contrast, no
differences were observed in ADAM12 mRNA expression levels between
PC12 cells treated with L-NIL or ADMA, and those in the control
cells (Fig. 5B).
ADAM12 mRNA expression in rat OLN93 cells was not
affected by L-NAME, L-NIL nor ADMA treatment, compared with
expression the control cells (Fig.
5C). Rat C6 glia cells exhibited increased ADAM12 mRNA
expression in response to L-NIL and ADMA treatment, although not in
response to L-NAME treatment (Fig.
5D). The efficacy of the NOS inhibitors was confirmed in pilot
experiments by measuring the alterations in NO concentration using
an NO-sensitive electrode (ISO-NO-METER; World Precision
Instruments, Berlin, Germany) as described in (56).
Discussion
ADAMs exhibit a multi-domain structure and
ectopeptidase activity, and are involved in the regulation of
cell-matrix interactions and cell-cell communication. ADAM
expression and activity has been linked to important functions,
such as cell differentiation, proliferation, development and
inflammation, and to tumor cell growth, invasion and metastasis.
ADAM12 is a proteolytically active ADAM, and two isoforms are
produced as a results of alternative splicing: The membrane-bound,
ADAM12-L, and the shorter soluble form, ADAM12-S (57). Increased expression levels of
ADAM12-L and ADAM12-S, have been observed in several types of
cancer, including carcinoma of the breast, bladder and lung, and in
glioblastoma (58). Urinary
concentrations of ADAM12 have been shown to correlate with cancer
stage, making it a potential biomarker with which to monitor tumor
progression (59–61). ADAM12 facilitates tumor progression
by stimulating cell proliferation and survival pathways (62–64).
This is achieved by the proteolytic activation and release
(ectodomain shedding) of membrane-bound growth factors, including
HB-EGF, erythroblastic leukemia viral oncogene homolg 4, tumor
necrosis factor α and tumor growth factor α (40,65).
In the present study, an increase in the expression
of ADAM12 in the cortex and hippocampus samples from mice was
observed was increasing age. In addition, ADAM12 expression levels
were significantly higher in eNOS−/−, nNOS−/−
and iNOS−/− adult, and mice >1 year old, compared
with those in wild-type mice. The results of the present study
suggest there may be an association between NOS activity and ADAM12
expression levels. In support of this view, an increased expression
of ADAM12 was observed in response to NOS inhibition in murine N9
microglia, rat C6 astroglia and, to a lesser extent, in rat PC12
neuronal cells. By contrast, no induction of ADAM12 expression was
observed in OLN93 rat oligodendrocytes. Consistent with the
predominant and apparently high constitutive expression of ADAM12
in oligodendrocytes (47,48), it may be hypothesized that ADAM12
expression may not be subject to significant regulation, at least
by NO, in oligodendrocytes. The association between ADAM12 and NOS
isotype expression observed in the current study together with the
known effects of NO on neural cell development (65), suggest that ADAM12 and NO are
interdependently associated with neuronal development and
function.
At the dosages applied in the present study, the NOS
inhibitor L-NIL would be expected to inhibit iNOS activity, whilst
eNOS activity remains largely unaffected. L-NAME would be expected
to inhibit nNOS, eNOS and, to a lesser extent, iNOS activity. By
contrast, ADMA (10 µM) would be expected to inhibit eNOS
activity, and not iNOS activity (66).
NO affects glutamatergic neurotransmission and is
associated with the storage, uptake and/or release of a number of
neurotransmitters in the central nervous system, such as
acetylcholine, dopamine, noradrenaline, γ-aminobutyric acid,
taurine and glycine (64,65,67),
as well as certain neuropeptides (68). Since NO is a highly diffusible
molecule, it is capable of mediating synaptic and non-synaptic
communication processes. Furthermore, NO is a free radical due to
its unpaired electron (•NO). Therefore, it exhibits toxic effects
at higher concentrations (69–72).
NO toxicity is accentuated in the presence of oxidative radicals
such as O2−, which may be generated by NOS when
L-arginine substrate concentrations are low (73,74).
The aging process is also associated with increased nitrosative and
oxidative damage (75). NO is
involved in brain development by influencing synaptic plasticity
and mediating the change from cell proliferation to differentiation
during neurogenesis (76).
ADAM12 has been shown to cause shedding of the
Δ-like protein 1 ectodomain, thereby facilitating the activation of
Notch signaling (77). In human
umbilical vein endothelial (HUVEC) cells, the activation of Notch
induces the expression of the NO receptor, soluble guanylyl cyclase
heterodimer, guanylate cyclase 1 soluble α 3 (GUCY1α3) and GUCY1β3
(78). Phosphatidylinositol-3
kinase/protein kinase B-dependent phosphorylation/activation of
eNOS was observed in HUVEC cells. Therefore, Notch signaling may
induce NO production and NO-receptor expression in HUVEC cells. By
contrast, downregulation of Notch1 has been shown to induce iNOS
expression in a model of myocardial ischemia/reperfusion (79). iNOS-mediated NO production,
together with the induction of the subunit of NADPH oxidase,
gp91phox, and the resulting increase in superoxide anion
production, is likely to contribute to increased infarct size and
fibrosis via the enhanced formation of peroxynitrite (79).
It has recently been established that Notch
signaling regulates the expression of ADAM12 (80). Therefore, it is hypothesized that
there is an association between ADAM12 expression, Notch signaling
and NOS/NO. The induction of ADAM12 in response to NOS-inhibition
that was observed in the present study is in accordance with this
hypothesis. However, the role of Notch signaling for ADAM12
expression requires further investigation.
In conclusion, the results of the present study
demonstrate that cortical and hippocampal ADAM12 expression
increases with mouse development and age. NOS-deficiency leads to
increased ADAM12 expression in adult and 1-year old mice, compared
with that in fetal and neonatal mice. However, the physiological
and pathophysiological mechanisms underlying these patterns require
further investigations.
Acknowledgments
The authors would like to thank Doris Treczak, Manja
Möller, Ines Schultz and Leona Bück for their technical assistance.
This study was supported by a grant from the Bundesministerium für
Forschung und Technik (grant no. 01ZZ0407/PFG1).
References
|
1
|
Schild L, Dombrowski F, Lendeckel U,
Schulz C, Gardemann A and Keilhoff G: Impairment of endothelial
nitric oxide synthase causes abnormal fat and glycogen deposition
in liver. Biochim Biophys Acta. 1782:180–187. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schild L, Jaroscakova I, Lendeckel U, Wolf
G and Keilhoff G: Neuronal nitric oxide synthase controls enzyme
activity pattern of mitochondria and lipid metabolism. FASEB J.
20:145–147. 2006.
|
|
3
|
Torregrossa AC, Aranke M and Bryan NS:
Nitric oxide and geriatrics: Implications in diagnostics and
treatment of the elderly. J Geriatr Cardiol. 8:230–242. 2011.
|
|
4
|
Cau SB, Carneiro FS and Tostes RC:
Differential modulation of nitric oxide synthases in aging:
Therapeutic opportunities. Front Physiol. 3:2182012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mungrue IN, Gros R, You X, Pirani A, Azad
A, Csont T, Schulz R, Butany J, Stewart DJ and Husain M:
Cardiomyocyte overex-pression of iNOS in mice results in
peroxynitrite generation, heart block, and sudden death. J Clin
Invest. 109:735–743. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Licinio J, Prolo P, McCann SM and Wong ML:
Brain iNOS: Current understanding and clinical implications. Mol
Med Today. 5:225–232. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Loo B, Labugger R, Skepper JN,
Bachschmid M, Kilo J, Powell JM, Palacios-Callender M, Erusalimsky
JD, Quaschning T, Malinski T, et al: Enhanced peroxynitrite
formation is associated with vascular aging. J Exp Med.
192:1731–1744. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mahbub S, Deburghgraeve CR and Kovacs EJ:
Advanced age impairs macrophage polarization. J Interferon Cytokine
Res. 32:18–26. 2012. View Article : Google Scholar :
|
|
9
|
Bernstein HG, Keilhoff G, Steiner J,
Dobrowolny H and Bogerts B: Nitric oxide and schizophrenia: Present
knowledge and emerging concepts of therapy. CNS Neurol Disord Drug
Targets. 10:792–807. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Keilhoff G: nNOS deficiency-induced cell
proliferation depletes the neurogenic reserve. Neurosci Lett.
505:248–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian K, Ghassemi F, Sotolongo A, Siu A,
Shauger L, Kots A and Murad F: NOS-2 signaling and cancer therapy.
IUBMB Life. 64:676–683. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choudhari SK, Chaudhary M, Bagde S,
Gadbail AR and Joshi V: Nitric oxide and cancer: A review. World J
Surg Oncol. 11:1182013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fukumura D, Kashiwagi S and Jain RK: The
role of nitric oxide in tumour progression. Nat Rev Cancer.
6:521–534. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Yan W, Lu X, Qian C, Zhang J, Li
P, Shi L, Zhao P, Fu Z, Pu P, et al: Overexpression of osteopontin
induces angiogenesis of endothelial progenitor cells via the
avβ3/PI3K/AKT/eNOS/NO signaling pathway in glioma cells. Eur J Cell
Biol. 90:642–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bulnes S, Argandoña EG, Bengoetxea H, Leis
O, Ortuzar N and Lafuente JV: The role of eNOS in vascular
permeability in ENU-induced gliomas. Acta Neurochir Suppl.
106:277–282. 2010.
|
|
16
|
Lim KH, Ancrile BB, Kashatus DF and
Counter CM: Tumour maintenance is mediated by eNOS. Nature.
452:646–649. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Charles N, Ozawa T, Squatrito M, Bleau AM,
Brennan CW, Hambardzumyan D and Holland EC: Perivascular nitric
oxide activates notch signaling and promotes stem-like character in
PDGF-induced glioma cells. Cell Stem Cell. 6:141–152. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gratton JP, Lin MI, Yu J, Weiss ED, Jiang
ZL, Fairchild TA, Iwakiri Y, Groszmann R, Claffey KP, Cheng YC, et
al: Selective inhibition of tumor microvascular permeability by
cavtratin blocks tumor progression in mice. Cancer Cell. 4:31–39.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giliano NY, Konevega LV and Noskin LA:
Dynamics of intracellular superoxide and NO content in human
endotheliocytes and carcinoma cells after treatment with NO
synthase inhibitors. Bull Exp Biol Med. 149:78–81. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Broholm H, Rubin I, Kruse A, Braendstrup
O, Schmidt K, Skriver EB and Lauritzen M: Nitric oxide synthase
expression and enzymatic activity in human brain tumors. Clin
Neuropathol. 22:273–281. 2003.PubMed/NCBI
|
|
21
|
Kostourou V, Cartwright JE, Johnstone AP,
Boult JK, Cullis ER, Whitley G and Robinson SP: The role of
tumour-derived iNOS in tumour progression and angiogenesis. Br J
Cancer. 104:83–90. 2011. View Article : Google Scholar :
|
|
22
|
Lam-Himlin D, Espey MG, Perry G, Smith MA
and Castellani RJ: Malignant glioma progression and nitric oxide.
Neurochem Int. 49:764–768. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eyler CE, Wu Q, Yan K, MacSwords JM,
Chandler-Militello D, Misuraca KL, Lathia JD, Forrester MT, Lee J,
Stamler JS, et al: Glioma stem cell proliferation and tumor growth
are promoted by nitric oxide synthase-2. Cell. 146:53–66. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tatemichi M, Ogura T and Esumi H: Impact
of inducible nitric oxide synthase gene on tumor progression. Eur J
Cancer Prev. 18:1–8. 2009. View Article : Google Scholar
|
|
25
|
Kogias E, Osterberg N, Baumer B, Psarras
N, Koentges C, Papazoglou A, Saavedra JE, Keefer LK and Weyerbrock
A: Growth-inhibitory and chemosensitizing effects of the glutathi
one-S-transferase-π-activated nitric oxide donor PABA/NO in
malignant gliomas. Int J Cancer. 130:1184–1194. 2012. View Article : Google Scholar
|
|
26
|
Bian H, Feng J, Li M and Xu W: Novel
antileukemic agents derived from tamibarotene and nitric oxide
donors. Bioorg Med Chem Lett. 21:7025–7029. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mocellin S, Bronte V and Nitti D: Nitric
oxide, a double edged sword in cancer biology: Searching for
therapeutic opportunities. Med Res Rev. 27:317–352. 2007.
View Article : Google Scholar
|
|
28
|
Cheng H, Wang L, Mollica M, Re AT, Wu S
and Zuo L: Nitric oxide in cancer metastasis. Cancer Lett. 353:1–7.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stojic J, Hagemann C, Haas S, Herbold C,
Kühnel S, Gerngras S, Roggendorf W, Roosen K and Vince GH:
Expression of matrix metalloproteinases MMP-1, MMP-11 and MMP-19 is
correlated with the WHO-grading of human malignant gliomas.
Neurosci Res. 60:40–49. 2008. View Article : Google Scholar
|
|
30
|
Pullen NA and Fillmore HL: Induction of
matrix metal-loproteinase-1 and glioma cell motility by nitric
oxide. J Neurooncol. 96:201–209. 2010. View Article : Google Scholar
|
|
31
|
Kodama T, Ikeda E, Okada A, Ohtsuka T,
Shimoda M, Shiomi T, Yoshida K, Nakada M, Ohuchi E and Okada Y:
ADAM12 is selectively overexpressed in human glioblastomas and is
associated with glioblastoma cell proliferation and shedding of
heparin-binding epidermal growth factor. Am J Pathol.
165:1743–1753. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fillmore HL, VanMeter TE and Broaddus WC:
Membrane-type matrix metalloproteinases (MT-MMPs): Expression and
function during glioma invasion. J Neurooncol. 53:187–202. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Van Meter TE, Broaddus WC, Rooprai HK,
Pilkington GJ and Fillmore HL: Induction of membrane-type-1 matrix
metalloproteinase by epidermal growth factor-mediated signaling in
gliomas. Neuro Oncol. 6:188–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Albrechtsen R, Kveiborg M, Stautz D,
Vikeså J, Noer JB, Kotzsh A, Nielsen FC, Wewer UM and Fröhlich C:
ADAM12 redistributes and activates MMP-14, resulting in gelatin
degradation, reduced apoptosis and increased tumor growth. J Cell
Sci. 126:4707–4720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Akool el-S, Kleinert H, Hamada FM, et al:
Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA
by inhibiting the expression of mRNA-stabilizing factor HuR. Mol
Cell Biol. 23:4901–4916. 2003. View Article : Google Scholar :
|
|
36
|
Knipp BS, Ailawadi G, Ford JW, Peterson
DA, Eagleton MJ, Roelofs KJ, Hannawa KK, Deogracias MP, Ji B,
Logsdon C, et al: Increased MMP-9 expression and activity by aortic
smooth muscle cells after nitric oxide synthase inhibition is
associated with increased nuclear factor-kappaB and activator
protein-1 activity. J Surg Res. 116:70–80. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Okamoto T, Gohil K, Finkelstein EI, Bove
P, Akaike T and van der Vliet A: Multiple contributing roles for
NOS2 in LPS-induced acute airway inflammation in mice. Am J Physiol
Lung Cell Mol Physiol. 286:L198–L209. 2004. View Article : Google Scholar
|
|
38
|
Barsoum IB, Hamilton TK, Li X, Cotechini
T, Miles EA, Siemens DR and Graham CH: Hypoxia induces escape from
innate immunity in cancer cells via increased expression of ADAM10:
Role of nitric oxide. Cancer Res. 71:7433–7441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jørgensen LH, Jensen CH, Wewer UM and
Schrøder HD: Transgenic overexpression of ADAM12 suppresses muscle
regeneration and aggravates dystrophy in aged mdx mice. Am J
Pathol. 171:1599–1607. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wever UM, Albrechtsen R and Engvall E:
ADAM12 The long and the short of it. The ADAM Family of Proteases.
Hooper NM and Lendeckel U: 4. Springer, Dordrecht; The Netherlands:
pp. 123–146. 2005
|
|
41
|
White JM: ADAMs: Modulators of cell-cell
and cell-matrix interactions. Curr Opin Cell Biol. 15:598–606.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fröhlich C, Klitgaard M, Noer JB, Kotzsch
A, Nehammer C, Kronqvist P, Berthelsen J, Blobel C, Kveiborg M,
Albrechtsen R, et al: ADAM12 is expressed in the tumour vasculature
and mediates ectodomain shedding of several membrane-anchored
endothelial proteins. Biochem J. 452:97–109. 2013.PubMed/NCBI
|
|
43
|
Jacobsen J, Visse R, Sørensen HP, Enghild
JJ, Brew K, Wewer UM and Nagase H: Catalytic properties of ADAM12
and its domain deletion mutants. Biochemistry. 47:537–547. 2008.
View Article : Google Scholar
|
|
44
|
Hougaard S, Loechel F, Xu X, Tajima R,
Albrechtsen R and Wewer UM: Trafficking of human ADAM 12-L:
Retention in the trans-Golgi network. Biochem Biophys Res Commun.
275:261–267. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kurisaki T, Masuda A, Osumi N, Nabeshima Y
and Fujisawa-Sehara A: Spatially- and temporally-restricted
expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse
embryo. Mech Dev. 73:211–215. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yagami-Hiromasa T, Sato T, Kurisaki T,
Kamijo K, Nabeshima Y and Fujisawa-Sehara A: A
metalloprotease-disintegrin participating in myoblast fusion.
Nature. 377:652–656. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bernstein HG, Keilhoff G, Bukowska A,
Ziegeler A, Funke S, Dobrowolny H, Kanakis D, Bogerts B and
Lendeckel U: ADAM (a disintegrin and metalloprotease) 12 is
expressed in rat and human brain and localized to oligodendrocytes.
J Neurosci Res. 75:353–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kanakis D, Lendeckel U, Theodosiou P,
Dobrowolny H, Mawrin C, Keilhoff G, Bukowska A, Dietzmann K,
Bogerts B and Bernstein HG: ADAM 12: A putative marker of
oligodendro-gliomas? Dis Markers. 34:81–91. 2013. View Article : Google Scholar :
|
|
49
|
Huang PL, Dawson TM, Bredt DS, Snyder SH
and Fishman MC: Targeted disruption of the neuronal nitric oxide
synthase gene. Cell. 75:1273–1286. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brenman JE, Xia H, Chao DS, Black SM and
Bredt DS: Regulation of neuronal nitric oxide synthase through
alternative transcripts. Dev Neurosci. 19:224–231. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gödecke A, Decking UK, Ding Z, et al:
Coronary hemodynamics in endothelial NO synthase knockout mice.
Circ Res. 82:186–194. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Corradin SB, Mauël J, Donini SD,
Quattrocchi E and Ricciardi-Castagnoli P: Inducible nitric oxide
synthase activity of cloned murine microglial cells. Glia.
7:255–262. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Richter-Landsberg C and Heinrich M:
OLN-93: A new permanent oligodendroglia cell line derived from
primary rat brain glial cultures. J Neurosci Res. 45:161–173. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wex T, Treiber G, Lendeckel U and
Malfertheiner P: A two-step method for the extraction of
high-quality RNA from endoscopic biopsies. Clin Chem Lab Med.
41:1033–1037. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Härdtner C, Mörke C, Walther R, Wolke C
and Lendeckel U: High glucose activates the alternative
ACE2/Ang-(1–7)/Mas and APN/Ang IV/IRAP RAS axes in pancreatic
β-cells. Int J Mol Med. 32:795–804. 2013.
|
|
56
|
Schild L, Reinheckel T, Reiser M, Horn TF,
Wolf G and Augustin W: Nitric oxide produced in rat liver
mitochondria causes oxidative stress and impairment of respiration
after transient hypoxia. FASEB J. 17:2194–2201. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gilpin BJ, Loechel F, Mattei MG, Engvall
E, Albrechtsen R and Wewer UM: A novel, secreted form of human ADAM
12 (meltrin alpha) provokes myogenesis in vivo. J Biol Chem.
273:157–166. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kveiborg M, Albrechtsen R, Couchman JR and
Wewer UM: Cellular roles of ADAM12 in health and disease. Int J
Biochem Cell Biol. 40:1685–1702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Roy R, Wewer UM, Zurakowski D, Pories SE
and Moses MA: ADAM 12 cleaves extracellular matrix proteins and
correlates with cancer status and stage. J Biol Chem.
279:51323–51330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roy R, Zurakowski D, Pories S, Moss ML and
Moses MA: Potential of fluorescent metalloproteinase substrates for
cancer detection. Clin Biochem. 44:1434–1439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fröhlich C, Albrechtsen R, Dyrskjøt L,
Rudkjaer L, Ørntoft TF and Wewer UM: Molecular profiling of ADAM12
in human bladder cancer. Clin Cancer Res. 12:7359–7368. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Roy R, Rodig S, Bielenberg D, Zurakowski D
and Moses MA: ADAM12 transmembrane and secreted isoforms promote
breast tumor growth: A distinct role for ADAM12-S protein in tumor
metastasis. J Biol Chem. 286:20758–20768. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ohlig S, Farshi P, Pickhinke U, van den
Boom J, Höing S, Jakuschev S, Hoffmann D, Dreier R, Schöler HR,
Dierker T, et al: Sonic hedgehog shedding results in functional
activation of the solubilized protein. Dev Cell. 20:764–774. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Pögün S and Kuhar MJ: Regulation of
neurotransmitter reuptake by nitric oxide. Ann NY Acad Sci.
738:305–315. 1994.PubMed/NCBI
|
|
65
|
Boehning D and Snyder SH: Novel neural
modulators. Annu Rev Neurosci. 26:105–131. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mukherjee P, Cinelli MA, Kang S and
Silverman RB: Development of nitric oxide synthase inhibitors for
neurodegeneration and neuropathic pain. Chem Soc Rev. 43:6814–6838.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Pepicelli O, Brescia A, Gherzi E, Raiteri
M and Fedele E: GABA(A), but not NMDA, receptors modulate in vivo
NO-mediated cGMP synthesis in the rat cerebral cortex.
Neuropharmacology. 46:480–489. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bernstein HG, Keilhoff G, Seidel B,
Stanarius A, Huang PL, Fishman MC, Reiser M, Bogerts B and Wolf G:
Expression of hypothalamic peptides in mice lacking neuronal nitric
oxide synthase: Reduced beta-END immunoreactivity in the arcuate
nucleus. Neuroendocrinology. 68:403–411. 1998. View Article : Google Scholar
|
|
69
|
Bredt DS and Snyder SH: Nitric oxide, a
novel neuronal messenger. Neuron. 8:3–11. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Nathan C and Xie QW: Regulation of
biosynthesis of nitric oxide. J Biol Chem. 269:13725–13728.
1994.PubMed/NCBI
|
|
71
|
Gross SS and Wolin MS: Nitric oxide:
Pathophysiological mechanisms. Annu Rev Physiol. 57:737–769. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Blaise GA, Gauvin D, Gangal M and Authier
S: Nitric oxide, cell signaling and cell death. Toxicology.
208:177–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pou S, Keaton L, Surichamorn W and Rosen
GM: Mechanism of superoxide generation by neuronal nitric-oxide
synthase. J Biol Chem. 274:9573–9580. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yanik M, Vural H, Tutkun H, Zoroğlu SS,
Savaş HA, Herken H, Koçyiğit A, Keleş H and Akyol O: The role of
the arginine-nitric oxide pathway in the pathogenesis of bipolar
affective disorder. Eur Arch Psychiatry Clin Neurosci. 254:43–47.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jung J, Na C and Huh Y: Alterations in
nitric oxide synthase in the aged CNS. Oxid Med Cell Longev.
2012:7189762012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gibbs SM: Regulation of neuronal
proliferation and differentiation by nitric oxide. Mol Neurobiol.
27:107–120. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Dyczynska E, Sun D, Yi H, Sehara-Fujisawa
A, Blobel CP and Zolkiewska A: Proteolytic processing of delta-like
1 by ADAM proteases. J Biol Chem. 282:436–444. 2007. View Article : Google Scholar
|
|
78
|
Chang AC, Fu Y, Garside VC, Niessen K,
Chang L, Fuller M, Setiadi A, Smrz J, Kyle A, Minchinton A, et al:
Notch initiates the endothelial-to-mesenchymal transition in the
atrioventricular canal through autocrine activation of soluble
guanylyl cyclase. Dev Cell. 21:288–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Pei H, Yu Q, Xue Q, Guo Y, Sun L, Hong Z,
Han H, Gao E, Qu Y and Tao L: Notch1 cardioprotection in myocardial
ischemia/reperfusion involves reduction of oxidative/nitrative
stress. Basic Res Cardiol. 108:3732013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li H, Solomon E, Duhachek Muggy S, Sun D
and Zolkiewska A: Metalloprotease-disintegrin ADAM12 expression is
regulated by Notch signaling via microRNA-29. J Biol Chem.
286:21500–21510. 2011. View Article : Google Scholar : PubMed/NCBI
|