1. Introduction
Hypoxia-inducible factors (HIFs) are commonly
expressed in humans and other mammals as transcriptional factors
(1,2). They positively regulate the
expression levels of >100 target genes, which encode protein
products involved in the response to hypoxia (3–5).
Tumor hypoxia was first described in the 1950s and currently there
is increasing evidence to demonstrate that hypoxia is regulated by
HIFs and is a common feature in several types of cancer (6–9). In
1992, Semenza et al (10)
identified a nuclear factor, which binds to the 3′-flanking
sequence of the human erythropoietin gene (EPO) and promotes the
expression of EPO under anoxic conditions. This factor, termed HIF,
is able to increase the number of erythrocytes and increase the
efficiency of oxygen transportation (10). At present, three types of HIF have
been identified, namely HIF-1 (10), HIF-2 (11) and HIF-3 (12). HIF-3α is the most recently
identified member of the HIF family. Mouse HIF-3α (mHIF-3α) was
initially identified by Gu et al in 1998 (12) and human HIF-3α (hHIF-3α) was
identified in 2001 (13). HIF-3α
has been investigated to a lesser degree compared with HIF-1 and
HIF-2. HIF-1α and HIF-2α are often overexpressed in cancer tissue,
leading to progression of aggressive tumors, tumor resistance to
chemotherapy and radiation, and poor prognosis of the disease
(14–17). The role of HIF-3α in tumors types
remains to be elucidated, however, previous studies have indicated
that HIF-3α may suppress the expression of genes, which are
typically inducible by HIF-1α and HIF-2α in tumor cells (13,18).
Therefore, HIF-3α has transcriptional regulatory functions and is a
negative regulator of gene expression during hypoxia (13,18).
Furthermore, HIF-3α is a true transcription factor since it
actively stimulates the expression of a number of target genes
(19). This review comprehensively
discusses the current knowledge of the gene structure, regulation
of expression and biological function of HIF-3.
2. Structure of human HIF-3
hHIF-3 is a heterodimer, which consists of
hypoxia-regulated-α (HIF-α) and oxygen-insensitive β subunits, and
is a member of the aryl-hydrocarbon receptor nuclear translator
(ARNT) family (20). hHIF-3α is
located at chromosome 19q13.13–13.2 (12), which differs from the locus of
HIF-1α (14q21–24) and HIF-2α (2p16–21) (10,11).
The first full-length hHIF-3α cDNA, now termed HIF-3α1, encodes a
668 amino acid protein with a relative molecular mass of 73 kDa
(13). The N-terminus of HIF-3α is
a basic-helix-loop-helix (bHLH) region, which is responsible for
DNA binding (Fig. 1). Following
the bHLH region is a Per/Arnt/Sim (PAS) region, which consists of
~300 aminophenoal residues. The PAS region contains PAS-A, PAS-B
and two replication regions, which form dimmers with the bHLH
region of HIF-1β (13). Following
the PAS region is an oxygen-dependent degradation (ODD) domain.
This oxygen regulatory region is involved in the degradation of
HIF-3α (13). The C-terminus of
HIF-3α is a transactivation domain (TAD). HIF-1α and HIF-2α have
two TADs, which are located at the N and C-terminus, however,
HIF-3α has only one TAD at the N-terminus (13). The TAD in HIF-3α shares 58% and 52%
identity with the TADs in the N-terminus of HIF-1α and HIF-2α,
respectively (12). Multiple
splice variants of hHIF-3α, namely hHIF-3α1-10, have been reported
(21,22). hHIF-3α has 19 exons spanning 43 kb
in chromosome 19q13.2. Three unique exons, namely exons 1a, 1b and
1c, are likely to contain transcription initiation sites for the
variants. Exon 2 encodes the bHLH domain and exons 3–9 contain the
coding sequence for the PAS domain. HIF-3α2 consists of 632 amino
acids and begins from exon 1a and ends at exon 13a, skipping exons
1b and 1c (22). hHIF-3α3 begins
at exon 1b and ends at exon 17, skipping exons 1a, 1c, 15 and 16.
hHIF-3α3 has 648 amino acids and contains ODD and LXXLL motifs,
however lacks any recognizable DNA-binding sequences, including the
bHLH or LZIP domains (22).
hHIF-3α4 encodes a protein of 363 amino acids, which lacks NAD, CAD
and ODD domains. Compared with other hHIF-3α variants, hHIF-3α4
contains no LXXLL or LZIP motifs (22). hHIF-3α5 and hHIF-3α6 start at exon
1b and lack exon 3. hHIF-3α5 contains a short exon 14c and ends at
exon 15, and it encodes a protein containing partial PASa, PASb and
PAC domains. hHIF-3α6, similar to hHIF-3α4, contains intron 7 and
ends at intron 8, and it contains only a partial PASb domain at the
C-terminus (22). Similar to the
HIF-α subunit, HIF-1β contains bHLH, PAS and TAD domains (Fig. 1). However, HIF-1β lacks the ODD
domain, therefore, it is constitutively expressed in all tissues
under aerobic conditions (23–25).
3. Structure of mouse HIF-3α
The open reading frame of mHIF-3α spans 1.98 kb,
containing 15 exons, and encodes a protein of 662 amino acids
(12). mHIF-3α has also been
reported to produce alternatively spliced variants, the mouse
inhibitory PAS domain protein (IPAS) (26,27)
and neonatal and embryonic PAS protein (NEPAS) (28). Mouse IPAS is a hypoxia-inducible
short splice variant of mHIF-3α and shares three exons (2, 4 and 5)
with HIF-3. Mouse IPAS lacks NTAD, CTAD and ODD domains (26) and is known to bind to HIF-1α,
however not HIF-β (27). Similar
to IPAS, NEPAS mRNA is derived from HIF-3α and contains the first
exon (1a) of IPAS followed by the 2nd to 15th exon of HIF-3α
(28). NEPAS encodes a polypeptide
of 664 amino acids, containing the NTAD and ODD domains (28). Unlike IPAS, NEPAS is able to
dimerize with HIF-β (28).
4. Expression and regulation of HIF-3α
The expression profiles of HIF-1α and HIF-2α have
been well documented (29).
However, the expression profiles of HIF-3α variants are only
recently investigated. HIF-3α is expressed in human kidney
(13) and lung epithelial cells
(30). Northern blot analysis
demonstrated that the mRNA expression of hHIF-3α is high in the
heart, placenta and skeletal muscle, however, is low in the lung,
liver and kidney (22).
Immunofluorescence analysis demonstrated that HIF-3α is present in
the cytoplasm and nucleus under normoxic conditions, and that
exposure to hypoxia increases the nuclear fraction of HIF-3α
(31). mHIF-3α is expressed in the
adult thymus, lung, heart and kidney (12). In mice, IPAS is predominantly
expressed in corneal epithelium and Purkinje cells of the
cerebellum (26). NEPAS is
expressed almost exclusively in the late embryonic and early
postnatal stages, with the expression predominantly located in the
lungs and heart (28). By
contrast, IPAS is not detected during embryonic development
(28).
The expression of HIF-3 is predominantly regulated
at the transcriptional and post-transcriptional levels, which are
described in the following sections.
Regulation of HIF-3α expression at the
transcriptional level Hypoxia
Hypoxia increases the mRNA expression levels of
HIF-3α. Heidbreder et al (32) revealed that the mRNA expression
levels of HIF-3α were significantly increased in the lung and other
organs following a 2-h hypoxic exposure in rats. This was confirmed
by two other studies (33,34). Zhang et al (35) demonstrated that hypoxia increases
the mRNA and protein expression levels of HIF-3α in zebrafish.
HIF-1 and HIF-2
HIF-3α is a target gene of HIF-1 and modulates the
expression of hypoxic genes (31).
Tanaka et al (31) revealed
that siRNA-mediated knockdown of HIF-1α in human renal cell
carcinoma notably dampened the 2, 2′-dipyridy-stimulated induction
of HIF-3α protein. In addition, immunohistochemical analysis
revealed co-localization of HIF-1α and HIF-3α in cells (31). Pasanen et al (21) demonstrated that HIF-3α2 and HIF-3α4
are inducible during hypoxia and the inductions require HIF-1α. A
similar previous study revealed that HIF-1α binds to the hypoxia
response element (HRE) in the IPAS promoter and induces the
expression of IPAS (36). However,
the stabilized form of HIF-1α does not affect the mRNA expression
levels of HIF-3α in zebrafish embryos (35) and 3T3-L1 cells (37). This contradictory result may be due
to the different cell lines or animal models used in the
experiments. In addition to HIF-1α, Hatanaka et al (37) observed that the promoter activity
of HIF-3α is specifically activated by HIF-2α. HIF-2α specifically
binds to the sequence between −251 and −228 in the mHIF-3α
promoter, which is essential in response to the activation of
HIF-2α (37). In human umbilical
venous endothelial cells, the expression of HIF-3α is driven by
HIF-1 and HIF-2 (18).
2-Deoxy-D-glucose (2-DG) and insulin
2-DG and insulin are able to cause a widespread
increase in the mRNA expression levels of HIF-3α. Following
treatment with 2-DG in rats, the expression of HIF-3α was markedly
increased in the lung, heart and kidney by 9.6-, 9.0- and 4.1-fold,
respectively (38). Following
treatment with insulin, the mRNA expression of HIF-3α is
significantly increased in every major tissue and organ; however,
the induction is not as high as that following treatment with 2-DG
(38).
Regulation of HIF-3α expression at the
post-transcriptional level
Hypoxia
The mRNA expression of HIF-3α is increased
significantly in A549 cells exposed to a low oxygen environment for
2 h (30). On one hand, hypoxia
markedly increases the protein synthesis of HIF-3α within 30 min
exposure. Conversely, hypoxia reduces the degradation of HIF-3α
protein and therefore, relatively increases its protein level
(30).
Von Hippel-Lindau (VHL)
VHL is a tumor suppressor gene involved in VHL
syndrome and renal cell carcinoma (39–41).
Maynard et al (22)
demonstrated that hHIF-3α1-3 splice variants share a common ODD
domain, which can be degraded by the pVHL ubiquitin-proteasome
under normal oxygen partial pressure. The ability of VHL to degrade
HIF3α is dependent on the proline 490 residue on HIF3α and this is
increased in the presence of prolyl hydroxylase (PHD).
PHD
PHD is a cellular sensor for low-oxygen. Under
normal oxygen partial pressure and in the presence of
Fe2+ and acetone dicarboxylic acid, PHD catalyzes the
hydroxylation of key amino acid residues in the HIF-α ODD domain
(42,43). This is followed by VHL binding to
HIF-α and inducing degradation via the ubiquitin-proteasome pathway
(44,45). Chen et al (46) demonstrated that the in vivo
protein level of HIF-3α under hypoxic conditions is negatively
correlated with the protein expression levels of PHD2 and PHD3,
whereas the content of HIF-3α mRNA is positively correlated with
the mRNA expression levels of PHD2 and PHD3. It is possible that
PHD increases the protein degradation of HIF-3α, which is followed
by negative feedback upregulation of the mRNA expression of HIF-3α
in order to compensate for the loss of the HIF-3α protein.
Deferoxamine (DFX) and
CoCl2
DFX and CoCl2 increase the protein
expression levels of HIF-3α (30).
DFX binds to iron and interrupts the hydroxylation of proline in
the ODD domain of HIFs, by preventing the binding of the VHL
ubiquitin-proteasome complex to HIF-3α and eventually leads to the
accumulation of intracellular HIF-3α protein (30,47,48).
Similarly, CoCl2 reduces the degradation of HIF-3α by
occupying its binding site for the VHL ubiquitin-proteasome
complex, resulting in an increased protein expression of HIF-3α
(30,49,50).
5. Biological functions of HIF-3
Following protein stabilization during hypoxia,
HIF-1α and HIF-2α dimerize with HIF-β, bind to co-activators,
including p300, and interact with the HRE of target genes (51,52).
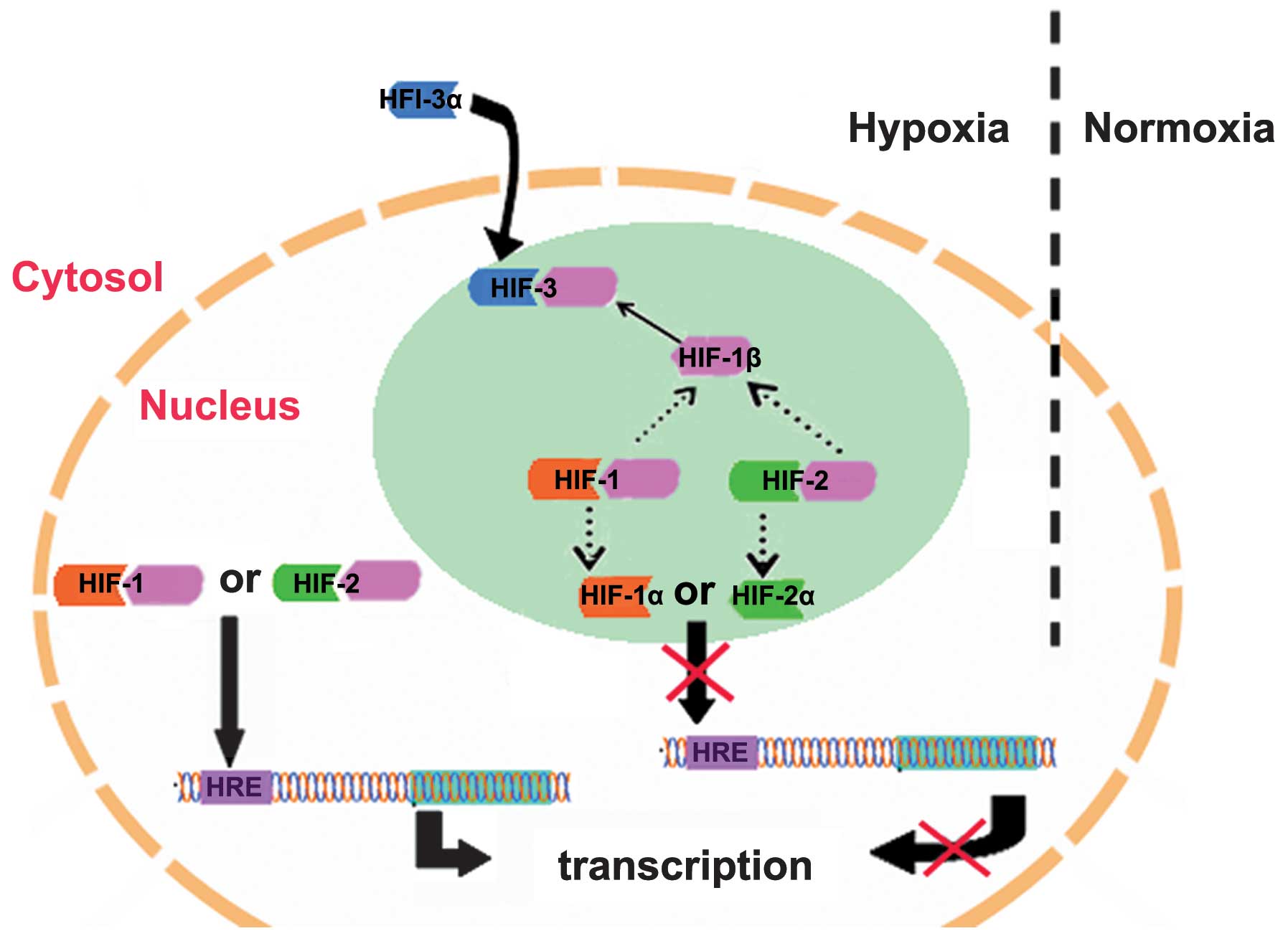
Compared with HIF-1α and HIF-2α, HIF-3α has dual functions:
Inhibition of the activities of HIF-1α and HIF-2α, and regulation
of its own target genes (19).
hHIF-3α1 has been demonstrated to suppress HIF-1 and
HIF-2-mediated gene expression (13). Hara et al (13) transfected expression vectors of
HIF-1α, HIF-2α or HIF-3α into COS-7 cells and demonstrated that
HIF-1α and HIF-2α promote the transcription of HREs, whereas
HIF-3α1 inhibits HRE transcription. A previous study revealed that
HIF-3α1 inhibits the expression levels of HIF-1α and HIF-2α
(13). It is suggested that HIF-3α
competes with HIF-1α and HIF-2α in binding to HIF-1β subunits,
reduces the levels of HIF-1 and HIF-2, and ultimately inhibits the
upregulation of the target genes of HIF-1 and HIF-2 (Fig. 2) (13). Additionally, HIF-3α lacks a
transcriptional activation domain, and its bHLH and PAS domains
suppress the expression of target genes, which are typically
inducible by HIF-1α and HIF-2α (13). Splice variant IPAS dimerizes with
HIF-1α protein and disrupts the interaction between HIF-1α and the
HRE of its target genes (36).
Makino et al (36)
demonstrated that there is a negative feedback mechanism between
HIF-1α and IPAS. At first, HIF-1α binds to the HRE in the IPAS
promoter and induces the expression of IPAS (36). Increased levels of IPAS dimerize
with HIF-1α protein and inhibits further induction of IPAS
(36). In hepatoma cells, ectopic
expression of IPAS decreases the expression of vascular endothelial
growth factor (VEGF), resulting in reduced tumor growth and
decreased tumor vascular density in vivo (27). The splice variant, HIF-3α4, is
different from IPAS in terms of its structure and gene regulation.
For example, IPAS only binds to HIF-1α, whereas HIF-3α4 binds to
HIF-1α and ARNT (53). The
HIF-3α4/HIF-β complex binds to HREs, which inhibits the binding of
the HIF-1α/HIF-β complex to the HRE (53). The HIF-3α4/HIF-β complex is not
transcriptionally active, however, it significantly reduces
HIF-1-mediated promoter activation by acting as a dominant negative
regulator of HIF-1 (53). Similar
to IPAS, ectopic expression of HIF-3α4 inhibits the endogenous
expression of hypoxia-responsive genes, including glucose
transporter-1 (GLUT-1), and knocking down the endogenous expression
of HIF-3α4 using siRNA increases the transcription of HIF target
genes (53). Besides HIF-1α and
HIF-β, HIF-3α4 also binds to HIF-2α and inhibits HIF-2-mediated
transactivation of HRE-driven genes (54). In addition, overexpression of
HIF-3α4 in clear-cell renal cell carcinoma (CCRCC) cells reduces
endogenous expression of HIF-2 target genes and inhibits the growth
of CCRCC xenografts in severe combined immunodeficiency mice
(54). These findings suggest that
HIF-3α4 has a dominant negative role in suppressing CCRCC growth
and has a potential therapeutic role in the treatment of CCRCC
(54). A previous study revealed
that overexpression of HIF-3α4 impairs angiogenesis, proliferation,
and metabolism/oxidation of hypervascular meningioma (55). Therefore, HIF-3α4 is a potential
molecular target for the treatment of meningioma (55).
It is reported that siRNA-mediated knockdown of
HIF-3α induces the expression of certain HIF-1α-mediated genes and
decreases the expression of ANGPTL4 in response to hypoxia
(31). This indicates that HIF-3α
also possesses transcriptional activity. All the hHIF-3α variants
were demonstrated to be able to bind to HIF-β and overexpression of
certain HIF-3α variants, together with HIF-β, induces the mRNA
expression levels of several HIF-1 and HIF-2 target genes,
including EPO (56,57), ANGPTL4 (58,59)
and GLUT1 (60,61). However, the overexpression of
HIF-3α variants reveals no significant stimulation of the
expression of HRE-driven reporter genes (62), suggesting that the target genes
induced by HIF-3α variants may contain specific response elements,
which are not canonical HREs (31,62).
Zhang et al (19) revealed that HIF-3α exhibits
significant transactivation activity in zebrafish. The authors
performed transcriptomic analyses and identified a large number of
HIF-3 target genes, which can be divided to three categories: i)
Genes that are upregulated by HIF-3a only (e.g. sqrdl, mclb and
zp3v2); ii) genes that are regulated by HIF-1α and HIF-3α with
similar potencies (e.g. redd1 and mlp3c); and iii) genes that are
regulated by HIF-1a and HIF-3a, however, with different potencies
(e.g. igfbp1a) (19). Notably, the
authors demonstrated that the transcriptional activity is conserved
across species and hHIF-3a-9 isoforms stimulate similar target
genes in different human cell types, including LC3C, REDD1 and
SQRDL cells (19). These findings
suggest that HIF-3 is an oxygen-dependent transcription factor,
which activates a distinctive set of genes in response to
hypoxia.
6. Association between HIF-3 and
diseases
Tissue hypoxia is a pathological feature of several
human diseases, including myocardial infarction, stroke and kidney
disease (63–65). The expression of HIF-3α is often
altered in these diseases and may contribute to their development
(32,66). It has been reported that the mRNA
expression of HIF-3a is increased as an early response to acute
hypoxia and acute myocardial ischemia in humans and experimental
animal models (32,66). Zolk et al (67) demonstrated that the mRNA expression
level of HIF-1α is 51% lower in cardiac tissue from a patient with
heart failure compared with that of a healthy control. By contrast,
the expression of HIF-2α remains unchanged and the mRNA expression
of HIF-3α is 72% higher in cardiac tissue from a patient with heart
failure compared with healthy control (67).
In addition, HIF-3 exerts abnormal expression
patterns in liver and kidney disease (68). Hypoxia-associated molecules are
upregulated during cystic alteration into a heterogeneous
appearance (68). In polycystic
liver, VEGF is markedly and widely expressed in the cytoplasm of
hepatocytes (68), and the
expression of HIF-3α, however not HIF-1α, is observed in a few
nuclei of hepatocytes adjacent to the biliary areas (68). By contrast, VEGF, HIF-1α and HIF-3α
proteins are not present in the cytoplasm or nuclei of hepatocytes
in the control livers (68).
Therefore, it is hypothesized that the presence of HIF-3α in
periportal hepatocytes is associated with the induction of VEGF
(68). Fang et al (69) demonstrated that HIF-3α is one of
the mediators, which contribute to the development of primary
spontaneous pneumothorax.
7. Conclusion
Based on the current knowledge, HIF-3α has a dual
role in response to hypoxia: It suppresses HIF-1 and HIF-2-mediated
gene expression and induces the expression of their own target
genes by binding to the HRE or specific response elements of
varying lengths, which are distinct from the canonical HRE. The
function of HIF-3α remains to be fully elucidated, however, it is
an important factor for the fine-tuning of the hypoxic response in
humans in physiological and pathological conditions (62,70).
A previous study identified certain target genes of HIF-3α and
confirmed its role as a transcription factor (19). Understanding the biological roles
of HIF-3α is important for identifying a potential therapeutic
target for the treatment of diseases.
Acknowledgments
The authors would like to thank Dr Shu-Cai Yang from
the Chinese University of Hong Kong for providing the figures.
References
|
1
|
Rankin EB, Giaccia AJ and Schipani E: A
central role for hypoxic signaling in cartilage, bone and
hematopoiesis. Curr Osteoporos Rep. 9:46–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Loenarz C, Coleman ML, Boleininger A, et
al: The hypoxia-inducible transcription factor pathway regulates
oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO
Rep. 12:63–70. 2011. View Article : Google Scholar
|
|
3
|
Semenza GL: Hypoxia-inducible factors in
physiology and medicine. Cell. 148:399–408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greer SN, Metcalf JL, Wang Y and Ohh M:
The updated biology of hypoxia-inducible factor. EMBO J.
31:2448–2460. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goda N and Kanai M: Hypoxia-inducible
factors and their roles in energy metabolism. Int J Hematol.
95:457–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye J, Wu D, Wu P, Chen Z and Huang J: The
cancer stem cell niche: Cross talk between cancer stem cells and
their microenvironment. Tumour Biol. 35:3945–3951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ
and Wu C: The correlation of expression levels of HIF-1α and HIF-2α
in hepatocellular carcinoma with capsular invasion, portal vein
tumor thrombi and patients’ clinical outcome. Jpn J Clin Oncol.
44:159–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao S, Yang S, Wu C, Wang Y, Jiang J and
Lu Z: Protein expression of hypoxia-inducible factor-1 alpha and
hepatocellular carcinoma: A systematic review with meta-analysis.
Clin Res Hepatol Gastroenterol. 38:598–603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai YP and Wu KJ: Hypoxia-regulated
target genes implicated in tumor metastasis. J Biomed Sci.
19:1022012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Semenza GL and Wang GL: A nuclear factor
induced by hypoxia via de novo protein synthesis binds to the human
erythropoietin gene enhancer at a site required for transcriptional
activation. Mol Cell Biol. 12:5447–5454. 1992.PubMed/NCBI
|
|
11
|
Tian H, McKnight SL and Russell DW:
Endothelial PAS domain protein 1 (EPAS1), a transcription factor
selectively expressed in endothelial cells. Genes Dev. 11:72–82.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu YZ, Moran SM, Hogenesch JB, Wartman L
and Bradfield CA: Molecular characterization and chromosomal
localization of a third alpha-class hypoxia inducible factor
subunit, HIF3alpha. Gene Expr. 7:205–213. 1998.PubMed/NCBI
|
|
13
|
Hara S, Hamada J, Kobayashi C, Kondo Y and
Imura N: Expression and characterization of hypoxia-inducible
factor (HIF)-3alpha in human kidney: Suppression of HIF-mediated
gene expression by HIF-3alpha. Biochem Biophys Res Commun.
287:808–813. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ku JH, Park YH, Myung JK, Moon KC, Kwak C
and Kim HH: Expression of hypoxia inducible factor-1α and 2α in
conventional renal cell carcinoma with or without sarcomatoid
differentiation. Urol Oncol. 29:731–737. 2011. View Article : Google Scholar
|
|
15
|
Luan Y, Gao C, Miao Y, Li Y, Wang Z and
Qiu X: Clinicopathological and prognostic significance of HIF-1α
and HIF-2α expression in small cell lung cancer. Pathol Res Pract.
209:184–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kroeger N, Seligson DB, Signoretti S, et
al: Poor prognosis and advanced clinicopathological features of
clear cell renal cell carcinoma (ccRCC) are associated with
cytoplasmic subcellular localisation of Hypoxia inducible
factor-2α. Eur J Cancer. 50:1531–1540. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong L, Zhang W, Zhou J, et al: Prognostic
value of HIFs expression in head and neck cancer: A systematic
review. PLoS One. 8:e750942013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Augstein A, Poitz DM, Braun-Dullaeus RC,
Strasser RH and Schmeisser A: Cell-specific and hypoxia-dependent
regulation of human HIF-3α: Inhibition of the expression of HIF
target genes in vascular cells. Cell Mol Life Sci. 68:2627–2642.
2011. View Article : Google Scholar
|
|
19
|
Zhang P, Yao Q, Lu L, Li Y, Chen PJ and
Duan C: Hypoxia-inducible factor 3 is an oxygen-dependent
transcription activator and regulates a distinct transcriptional
response to hypoxia. Cell Reports. 6:1110–1121. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Semenza GL: Hypoxia-inducible factor 1:
Master regulator of O2 homeostasis. Curr Opin Genet Dev.
8:588–594. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pasanen A, Heikkila M, Rautavuoma K,
Hirsila M, Kivirikko KI and Myllyharju J: Hypoxia-inducible factor
(HIF)-3alpha is subject to extensive alternative splicing in human
tissues and cancer cells and is regulated by HIF-1 but not HIF-2.
Int J Biochem Cell Biol. 42:1189–1200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maynard MA, Qi H, Chung J, et al: Multiple
splice variants of the human HIF-3 alpha locus are targets of the
von Hippel-Lindau E3 ubiquitin ligase complex. J Biol Chem.
278:11032–11040. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang LE, Arany Z, Livingston DM and Bunn
HF: Activation of hypoxia-inducible transcription factor depends
primarily upon redox-sensitive stabilization of its alpha subunit.
J Biol Chem. 271:32253–32259. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Whitelaw ML, Gustafsson JA and Poellinger
L: Identification of transactivation and repression functions of
the dioxin receptor and its basic helix-loop-helix/PAS partner
factor Arnt: inducible versus constitutive modes of regulation. Mol
Cell Biol. 14:8343–8355. 1994.PubMed/NCBI
|
|
25
|
Reyes H, Reisz-Porszasz S and Hankinson O:
Identification of the Ah receptor nuclear translocator protein
(Arnt) as a component of the DNA binding form of the Ah receptor.
Science. 256:1193–1195. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Makino Y, Kanopka A, Wilson WJ, Tanaka H
and Poellinger L: Inhibitory PAS domain protein (IPAS) is a
hypoxia-inducible splicing variant of the hypoxia-inducible
factor-3alpha locus. J Biol Chem. 277:32405–32408. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Makino Y, Cao R, Svensson K, et al:
Inhibitory PAS domain protein is a negative regulator of
hypoxia-inducible gene expression. Nature. 414:550–554. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamashita T, Ohneda O, Nagano M, et al:
Abnormal heart development and lung remodeling in mice lacking the
hypoxia-inducible factor-related basic helix-loop-helix PAS protein
NEPAS. Mol Cell Biol. 28:1285–1297. 2008. View Article : Google Scholar :
|
|
29
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li QF, Wang XR, Yang YW and Lin H: Hypoxia
upregulates hypoxia inducible factor (HIF)-3alpha expression in
lung epithelial cells: Characterization and comparison with
HIF-1alpha. Cell Res. 16:548–558. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tanaka T, Wiesener M, Bernhardt W, Eckardt
KU and Warnecke C: The human HIF (hypoxia-inducible factor)-3alpha
gene is a HIF-1 target gene and may modulate hypoxic gene
induction. Biochem J. 424:143–151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heidbreder M, Frohlich F, Johren O,
Dendorfer A, Qadri F and Dominiak P: Hypoxia rapidly activates
HIF-3alpha mRNA expression. FASEB J. 17:1541–1543. 2003.PubMed/NCBI
|
|
33
|
Rajatapiti P, de Rooij JD, Beurskens LW,
et al: Effect of oxygen on the expression of hypoxia-inducible
factors in human fetal lung explants. Neonatology. 97:346–354.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li QF and Dai AG: Differential expression
of three hypoxia-inducible factor-alpha subunits in pulmonary
arteries of rat with hypoxia-induced hypertension. Acta Biochim
Biophys Sin (Shanghai). 37:665–672. 2005. View Article : Google Scholar
|
|
35
|
Zhang P, Lu L, Yao Q, et al: Molecular,
functional and gene expression analysis of zebrafish
hypoxia-inducible factor-3alpha. Am J Physiol Regul Integr Comp
Physiol. 303:R1165–R1174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Makino Y, Uenishi R, Okamoto K, et al:
Transcriptional up-regulation of inhibitory PAS domain protein gene
expression by hypoxia-inducible factor 1 (HIF-1): a negative
feedback regulatory circuit in HIF-1-mediated signaling in hypoxic
cells. J Biol Chem. 282:14073–14082. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hatanaka M, Shimba S, Sakaue M, et al:
Hypoxia-inducible factor-3alpha functions as an accelerator of
3T3-L1 adipose differentiation. Biol Pharm Bull. 32:1166–1172.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heidbreder M, Qadri F, Johren O, et al:
Non-hypoxic induction of HIF-3alpha by 2-deoxy-D-glucose and
insulin. Biochem Biophys Res Commun. 352:437–443. 2007. View Article : Google Scholar
|
|
39
|
Choueiri TK, Fay AP, Gagnon R, et al: The
role of aberrant VHL/HIF pathway elements in predicting clinical
outcome to pazopanib therapy in patients with metastatic clear-cell
renal cell carcinoma. Clin Cancer Res. 19:5218–5226. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kennedy BK: A new connection between VHL
and cancer threads through progerin. Cell Cycle. 12:2721–2722.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bausch B, Jilg C, Glasker S, et al: Renal
cancer in von Hippel-Lindau disease and related syndromes. Nat Rev
Nephrol. 9:529–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pientka FK, Hu J, Schindler SG, et al:
Oxygen sensing by the prolyl-4-hydroxylase PHD2 within the nuclear
compartment and the influence of compartmentalisation on HIF-1
signalling. J Cell Sci. 125:5168–5176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Niecknig H, Tug S, Reyes BD, Kirsch M,
Fandrey J and Berchner-Pfannschmidt U: Role of reactive oxygen
species in the regulation of HIF-1 by prolyl hydroxylase 2 under
mild hypoxia. Free Radic Res. 46:705–717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Groulx I and Lee S: Oxygen-dependent
ubiquitination and degradation of hypoxia-inducible factor requires
nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor
suppressor protein. Mol Cell Biol. 22:5319–5336. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ivan M, Kondo K, Yang H, et al: HIFalpha
targeted for VHL-mediated destruction by proline hydroxylation:
Implications for O2 sensing. Science. 292:464–468. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen YR, Dai AG, Hu RC and Jiang YL:
Differential and reciprocal regulation between hypoxia-inducible
factor-alpha subunits and their prolyl hydroxylases in pulmonary
arteries of rat with hypoxia-induced hypertension. Acta Biochim
Biophys Sin (Shanghai). 38:423–434. 2006. View Article : Google Scholar
|
|
47
|
Harvey AJ, Kind KL and Thompson JG:
Regulation of gene expression in bovine blastocysts in response to
oxygen and the iron chelator desferrioxamine. Biol Reprod.
77:93–101. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Woo KJ, Lee TJ, Park JW and Kwon TK:
Desferrioxamine, an iron chelator, enhances HIF-1alpha accumulation
via cyclooxygenase-2 signaling pathway. Biochem Biophys Res Commun.
343:8–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Triantafyllou A, Liakos P, Tsakalof A,
Georgatsou E, Simos G and Bonanou S: Cobalt induces
hypoxia-inducible factor-1alpha (HIF-1alpha) in HeLa cells by an
iron-independent, but ROS-, PI-3K- and MAPK-dependent mechanism.
Free Radic Res. 40:847–856. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuan Y, Hilliard G, Ferguson T and
Millhorn DE: Cobalt inhibits the interaction between
hypoxia-inducible factor-alpha and von Hippel-Lindau protein by
direct binding to hypoxia-inducible factor-alpha. J Biol Chem.
278:15911–15916. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu D, Zhang R, Zhao R, Chen G, Cai Y and
Jin J: A novel function of novobiocin: Disrupting the interaction
of HIF 1alpha and p300/CBP through direct binding to the HIF1alpha
C-terminal activation domain. PLoS One. 8:e620142013. View Article : Google Scholar
|
|
52
|
Mendonca DB, Mendonca G, Aragao FJ and
Cooper LF: NF-kappaB suppresses HIF-1alpha response by competing
for P300 binding. Biochem Biophys Res Commun. 404:997–1003. 2011.
View Article : Google Scholar
|
|
53
|
Maynard MA, Evans AJ, Hosomi T, Hara S,
Jewett MA and Ohh M: Human HIF-3alpha4 is a dominant-negative
regulator of HIF-1 and is down-regulated in renal cell carcinoma.
FASEB J. 19:1396–1406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Maynard MA, Evans AJ, Shi W, Kim WY, Liu
FF and Ohh M: Dominant-negative HIF-3 alpha 4 suppresses VHL-null
renal cell carcinoma progression. Cell Cycle. 6:2810–2816. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ando H, Natsume A, Iwami K, et al: A
hypoxia-inducible factor (HIF)-3alpha splicing variant, HIF-3alpha4
impairs angiogenesis in hypervascular malignant meningiomas with
epigenetically silenced HIF-3alpha4. Biochem Biophys Res Commun.
433:139–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shah YM and Xie L: Hypoxia-inducible
factors link iron homeostasis and erythropoiesis. Gastroenterology.
146:630–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Haase VH: Regulation of erythropoiesis by
hypoxia-inducible factors. Blood Rev. 27:41–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li H, Ge C, Zhao F, et al:
Hypoxia-inducible factor 1 alpha-activated angiopoietin-like
protein 4 contributes to tumor metastasis via vascular cell
adhesion molecule-1/integrin beta1 signaling in human
hepatocellular carcinoma. Hepatology. 54:910–919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Imamura T, Kikuchi H, Herraiz MT, et al:
HIF-1alpha and HIF-2alpha have divergent roles in colon cancer. Int
J Cancer. 124:763–771. 2009. View Article : Google Scholar :
|
|
60
|
Marin-Hernandez A, Gallardo-Perez JC,
Ralph SJ, Rodriguez-Enriquez S and Moreno-Sanchez R: HIF-1alpha
modulates energy metabolism in cancer cells by inducing
over-expression of specific glycolytic isoforms. Mini Rev Med Chem.
9:1084–1101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Airley RE and Mobasheri A: Hypoxic
regulation of glucose transport, anaerobic metabolism and
angiogenesis in cancer: Novel pathways and targets for anticancer
therapeutics. Chemotherapy. 53:233–256. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Heikkila M, Pasanen A, Kivirikko KI and
Myllyharju J: Roles of the human hypoxia-inducible factor
(HIF)-3alpha variants in the hypoxia response. Cell Mol Life Sci.
68:3885–3901. 2011. View Article : Google Scholar
|
|
63
|
Deshmukh AB, Patel JK, Prajapati AR and
Shah S: Perspective in chronic kidney disease: targeting
hypoxia-inducible factor (HIF) as potential therapeutic approach.
Ren Fail. 34:521–532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kones R: Oxygen therapy for acute
myocardial infarction-then and now. A century of uncertainty. Am J
Med. 124:1000–1005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shi H: Hypoxia inducible factor 1 as a
therapeutic target in ischemic stroke. Curr Med Chem. 16:4593–4600.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lee SH, Wolf PL, Escudero R, Deutsch R,
Jamieson SW and Thistlethwaite PA: Early expression of angiogenesis
factors in acute myocardial ischemia and infarction. N Engl J Med.
342:626–633. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zolk O, Solbach TF, Eschenhagen T,
Weidemann A and Fromm MF: Activation of negative regulators of the
hypoxia-inducible factor (HIF) pathway in human end-stage heart
failure. Biochem Biophys Res Commun. 376:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yoshida T, Kuwahara M, Maita K and Harada
T: Immunohistochemical study on hypoxia in spontaneous poly-cystic
liver and kidney disease in rats. Exp Toxicol Pathol. 53:123–128.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fang HY, Lin CY, Chow KC, Huang HC and Ko
WJ: Microarray detection of gene overexpression in primary
spontaneous pneumothorax. Exp Lung Res. 36:323–330. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Drevytska T, Gavenauskas B, Drozdovska S,
Nosar V, Dosenko V and Mankovska I: HIF-3alpha mRNA expression
changes in different tissues and their role in adaptation to
intermittent hypoxia and physical exercise. Pathophysiology.
19:205–214. 2012. View Article : Google Scholar : PubMed/NCBI
|