Introduction
Colorectal cancer (CRC) is a major cause of cancer
mortality and morbidity worldwide. It is the third most commonly
diagnosed cancer in males and the second in females, with >1.2
million new cases and 608,700 mortalities estimated to have
occurred in 2008 (1). At present,
the identification of novel strategies for therapy is urgently
required.
The colorectal cancer stem cell (Co-CSC) hypothesis
provides a novel understanding of tumorigenesis. Increasing
evidence shows that Co-CSC subpopulations are capable of
self-renewal, driving tumor growth and differentiating to form all
of the lineages observed within a tumor (2–5).
Therefore, Co-CSCs may represent a novel target for the treatment
of tumors (6). Current strategies
for cancer treatment require modification to ensure that the
Co-CSCs which are driving tumor growth are specifically targeted.
However, there is increasing evidence that Co-CSCs are more
resistant to current chemotherapy (7) than other subpopulations of cells
within the tumor. This may be one of the reasons why the
effectiveness of standard chemotherapy is limited. Standard
chemotherapeutics are incapable of eradicating Co-CSCs, potentially
due to drug efflux, autocrine survival signaling and alterations in
DNA damage repair mechanisms in Co-CSCs (8,9).
However, the specific mechanism underlying Co-CSC chemoresistance
has yet to be elucidated.
5-fluorouracil (5FU) and oxaliplatin are the
predominant chemotherapeutic agents used for treating advanced CRC.
5FU and oxaliplatin have different mechanisms of action. 5FU
inhibits the activity of the thymidylate synthase enzyme during DNA
replication (10), while
oxaliplatin causes prolonged G2-phase arrest and
inhibits tumor cell growth through covalent DNA binding (11). Although the mechanisms of tumor
cell resistance to 5FU and oxaliplatin have been extensively
investigated, the specific mechanism remain to be fully elucidated.
Previous studies have reported that these chemoresistant cells
overexpress markers of CSCs (12).
This shows that chemoresistant tumor cells represent a
subpopulation of cells from the original tumor which are
molecularly and phenotypically distinct. Therefore, the association
between Co-CSCs and chemoresistant cells is of interest. Co-CSCs
exhibit characteristics of cells which are resistant to standard
chemotherapeutics and chemoresistant cells also express markers of
Co-CSCs. Therefore, Co-CSCs and chemoresistant cells may be
interrelated. Thus, the identification of a common target of
Co-CSCs and chemoresistant cells may be of significance for
clinical treatment.
A number of important signaling pathways, including
Wnt, Notch and Hedgehog, have been found to have a role in
regulating the self-renewal of CSCs in the hematopoietic system,
skin, nervous system and breast (13–15).
However, the mechanism by which pathways, including the Notch
pathway, regulate Co-CSCs and chemoresistant cells, as well as the
role that Notch has in the interrelation between these two types of
cells, has yet to be elucidated.
Based on the clinical significance of
chemoresistance and the ineffectiveness of chemotherapy in
eliminating Co-CSCs, the present study aimed to investigate the
interrelation between Co-CSCs and chemoresistant cells. Molecular
and phenotypic alterations were investigated in Co-CSCs and
chemoresistant cells in vitro, as well as using a mouse
model system in vivo. Notch pathway activation was also
detected in the Co-CSCs and chemoresistant cell lines and was
targeted using xenograft models. The present study has two clinical
implications associated with the interrelation between Co-CSCs and
chemoresistant cells. The first is that it translates the theory of
CSCs into clinical practice and shows that similar mechanisms may
act in Co-CSCs and chemoresistant cells. The second is that it
shows that Notch signaling simultaneously regulates Co-CSCs and
chemoresistant cells and identifies a novel mechanism of targeting
the Notch signaling pathway in CRC. Of note, the altered Notch
activity observed in the present study may partially explain the
chemoresistance in Co-CSCs and chemoresistant cells.
Materials and methods
Cell lines and culture
The HCT116 human CRC cell line was obtained from the
Colorectal Cancer Institute of Harbin Medical University (Harbin,
China). Colonospheres were cultured as described previously
(16). In brief, using a limited
dilution method, 1–3 cells were seeded on a 96-well
ultralow-attachment plate (Corning Life Sciences, Corning, NY, USA)
with Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium
containing B27 supplement (Invitrogen Life Technologies, Carlsbad,
CA, USA), 20 ng/ml basic fibroblast growth factor and 20 ng/ml
epidermal growth factor, which served as the stem cell medium (SCM)
for the experiments. Under these culture conditions, Co-CSCs, but
not differentiated cancer cells, are able to survive and
proliferate (17–19). The number of surviving cells in
each well of the 96-well plate was then observed and one cell well
was selected and marked. After seven days, the cells were observed
and the death cell well was removed and SCM was added to the
survival cell well. After 10–14 days, the well in which
colonospheres grew was marked and colonospheres were supplemented
with SCM every three days until the colonospheres were able to be
passaged.
An oxaliplatin-resistant cell line (HCT116/OxR) and
5FU-resistant cell line (HCT116/5FU-R) were developed as previously
described (20,21). The resistant cells and parental
cells were grown in DMEM containing 10% fetal bovine serum (FBS;
Sigma-Aldrich, St. Louis, MO, USA) and 1% antibiotic solution
(Mediatech Inc., Herndon, VA, USA) at 37°C in a humidified
incubator containing 5% CO2.
Drugs and antibodies
Oxaliplatin and 5FU were purchased from the
Colorectal Cancer Institute of Harbin Medical University. The
γ-secretase inhibitor N-[N-(3,5-difluoroph
enacetyl)-l-alanyl]-S-phenylglycine
t-butyl ester (DAPT) was purchased from Sigma-Aldrich and
was used to inhibit Notch signaling in vitro and in
vivo. The antibodies used for flow cytometry,
immunohistochemistry (IHC), immunofluorescence and western blot
analysis were as follows: Rabbit anti-cluster of differentiation
(CD) 133, rabbit anti-Notch1 (Cell Signaling Technology, Inc.,
Beverly, MA, USA), rabbit anti-β-actin (Sigma-Aldrich), mouse
anti-CD44 (Abcam PLC, Cambridge, UK), allophycocyanin
(APC)-conjugated anti-CD133, APC-conjugated mouse-immunoglobulin G
(IgG) 1 (Miltenyi Biotec, Bergisch Gladbach, Germany),
phycoerythrin (PE)-conjugated anti-CD44, PE-conjugated mouse-IgG2b
(Becton, Dickinson and Company, Franklin Lakes, NJ, USA), rabbit
anti-hairy and enhancer of split-1 (HES-1; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and mouse anti-Ki67
(Dako, Ely, UK).
Proliferation and chemosensitivity
Cell proliferation and drug sensitivity were
assessed using a Cell Counting Kit-8 (CCK-8) assay as described
previously (22). In brief, the
cell lines were seeded at a density of 5,000 cells/well in 96-well
plates in 100 µl media with or without 5FU, oxaliplatin or
DAPT. At each time-point (0, 24, 48 and 72 h), 10 µl CCK-8
solution was added to each well and incubated for a further 2 h.
The absorbance was read at 450 nm using a standard microplate
reader.
Colonosphere assay
Each cell line was trypsinized and quantified by
plating a single cell in each well of a low-attachment 96-well
plate and counted under a microscope, assessing the rate of
colonospheres. The colonospheres were cultured using the
aformentioned process.
Clonogenic assay
Clonogenic assays were performed to determine the
proliferative capacity of the colonospheres and the chemoresistant
cells. A total of 500 cells/well were seeded on a six-well plate
and incubated for 14 days at 37°C in 5% CO2. Following
incubation, the colonies were formalin-fixed and stained with
hematoxylin. Colonies which were >50 µm were counted
under a light microscope (CKX31; Olympus, Tokyo, Japan) and
compared with the parental cells.
Western blot analysis
Cell lysates were subjected to SDS-PAGE and blotted
onto Immobilon®-P polyvinylidene difluoride membranes
(Millipore Corporation, Billerica, MA, USA). Specific proteins were
detected using an enhanced chemiluminescence system (GE Healthcare,
Little Chalfont, UK). Membranes were probed with the aforementioned
antibodies.
Flow cytometry
Single cells were prepared for the analysis of cell
surface markers by digesting with pancreatin. Cells were then
detached from the plates through incubation with enzyme-free cell
dissociation buffer (Invitrogen Life Technologies). Cells were then
washed with 10 mmol/l cold phosphate-buffered saline (PBS) and
resuspended in 1X binding buffer (BD Biosciences, San Jose, CA,
USA) at a concentration of 1×106 cells/ml. Cells were
subjected to direct immunofluorescence staining using
APC-conjugated anti-CD133 and PE-conjugated anti-CD44 antibodies
followed by flow cytometric analysis. Samples were analyzed using a
NucleoCounter® NC-3000 analyzer (ChemoMetec, Lillerød,
Denmark). The experiments were repeated at least three times.
In vivo assay and Notch pathway
inhibition
Male nude mice, aged four weeks, were purchased from
the Shanghai Laboratory Animal Center (Shanghai, China). All animal
experiments were approved by the Institutional Animal Care and Use
Committee of Harbin Medical University. Equal numbers of cells
(1×106) from colonospheres and chemoresistant cell lines
were suspended in 100 µl PBS and injected subcutaneously
into the flank of each mouse (six mice per group). When the tumors
reached ~100 mm3, mice were subjected to intraperitoneal
injection with 200 µg DAPT twice per week. Tumor growth was
observed and recorded over 10 weeks. When the tumors in the control
group exceeded 1.5 cm in diameter, the animals were euthanized and
the tumors were weighed and measured. Tumor volume was calculated
using the following formula: (length)/2×(width)2. Tumors
were then paraffin-embedded, sectioned and stained with hematoxylin
and eosin (H&E) for histological analysis. IHC and apoptotic
analysis were also performed. Data are shown from representative
experiments.
Immunohistochemistry and
immunofluorescent analysis
IHC was performed on 4-µm sections which were
prepared using the paraffin-embedded tissues. Following standard
procedures, tissues were deparaffinized using xylene, hydrated in
graded alcohol and pretreated for antigen retrieval in
Tris/ethylene diaminetetracetic acid buffer for 5 min in a 100°C
steamer. Slides were then H&E stained in order to assess
morphology or incubated with anti-Ki67 antibodies to visualize the
proliferative nuclei. All sections were developed using
3,3′-diaminobenzidine and counterstained with hematoxylin
(Invitrogen Life Technologies). Apoptotic cells within the tissue
were detected using the terminal
deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL)
assay. An Apoptag in situ apoptosis detection kit (Roche
Diagnostics, Mannheim, Germany) was used for TUNEL staining
according to the manufacturer’s instructions for paraffin-embedded
tissues. Immunohistochemical staining and fluorescence were
analyzed using a Zeiss Axioskop microscope (Carl Zeiss AG,
Oberkochen, Germany) and apoptosis was expressed as the percentage
of TUNEL positive cells.
Statistical analysis
All data are presented as the mean ± standard error
of three independent experiments, each performed in triplicate.
Data were analyzed using the Student’s t-test. Analysis of variance
was performed for multiple comparisons. P<0.05 was considered to
indicate a statistically significant difference. SPSS 17.0
statistical software (SPSS, Inc., Chicago, IL, USA) was used for
the analyses.
Results
Expression of CSC markers in the
colonospheres and chemoresistant cells
CRC has been proposed to arise specifically in stem
cell populations at the base of colonic crypts. Markers used for
the identification of Co-CSCs include CD44, CD133, CD24, CD29,
leucine-rich repeat-containing G-protein coupled receptor 5 and
doublecortin-like kinase 1 (23).
Among these markers, CD44 and CD133 have been widely used for the
identification of CSCs in CRC. The CSC population has been reported
to be capable of self-renewal and generating tumors resembling the
primary tumor. Moreover, CSCs have been found to be capable of
growth in serum-free medium and the formation colonospheres. In the
present study, the expression profiles of HCT116 human CRC
colonospheres and cells resistant to 5FU or oxaliplatin
(HCT116/5FU-R or HCT116/OxR, respectively) were assessed using
western blot analysis and flow cytometry. Compared with the
parental HCT116 cells, CD133 and CD44 expression were observed to
be significantly higher in the colonospheres, HCT116/5FU-R and
HCT116/OxR cells (Fig. 1A). The
number of cells expressing CD133 and CD44 was also found to be
significantly higher in the colonospheres and chemoresistant cells
compared with the parental cells (Fig.
1B), with only 2% of the parental cells expressing CD133 and
48% expressing CD44, while between 33 and 65% of the three cell
types expressed CD133, and between 84 and 93% of the three cell
types expressed CD44. Following CD133 and CD44 labeling, flow
cytometric analysis revealed a 4.8-fold enrichment of
CD133+/CD44+ cells in the HCT116/5FU-R cell
line, a 22-fold enrichment of CD133+/CD44+
cells in the oxaliplatin-resistant cell line and a 24.7-fold
enrichment of CD133+/CD44+ cells in the
colonospheres compared with the parental HCT116 cells (Fig. 1C).
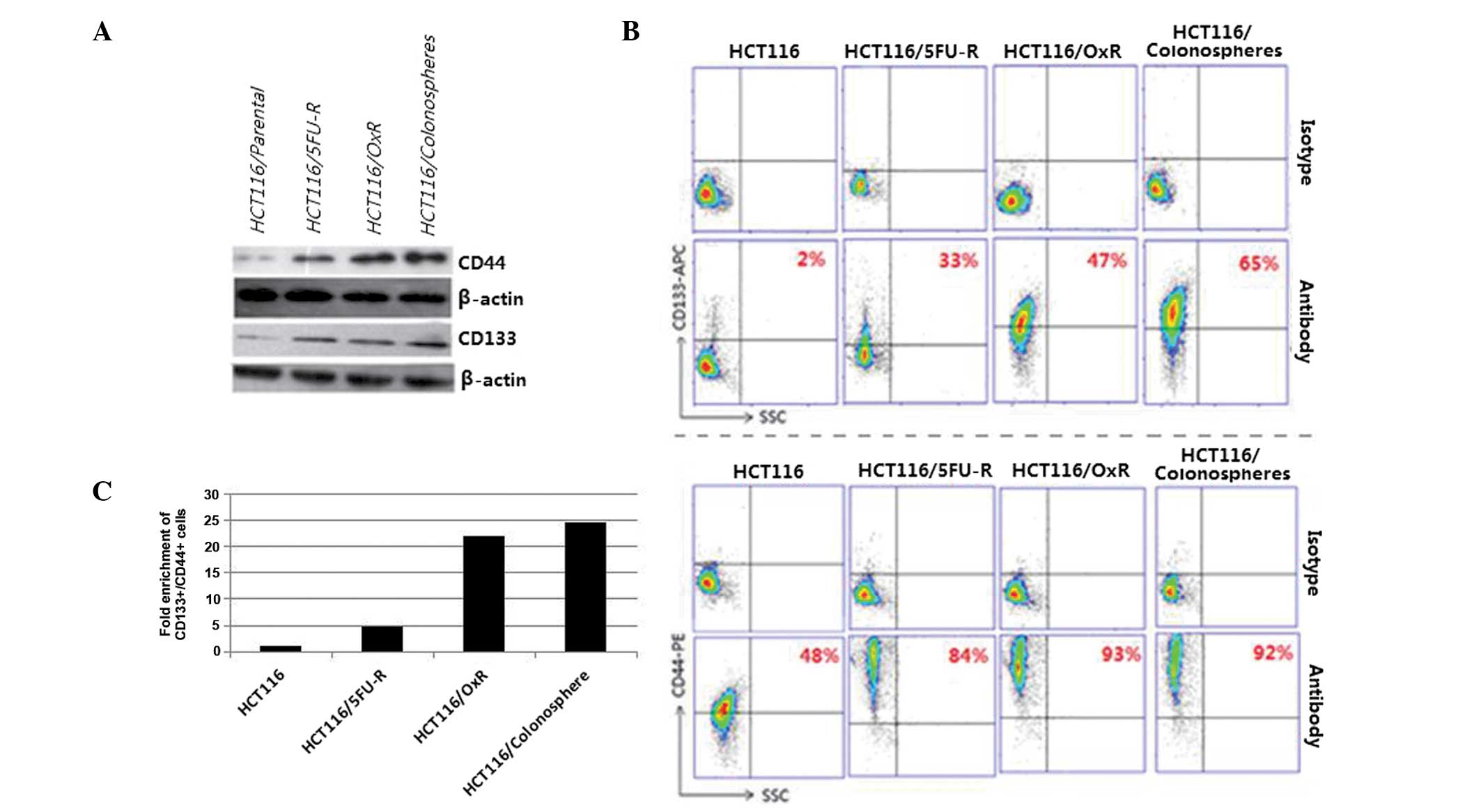 | Figure 1Colonospheres and chemoresistant cell
lines are enriched with Co-CSC markers. (A) Western blot analysis
revealed that expression of the Co-CSC markers CD133 and CD44 was
higher in the colonospheres and HCT116/5FU-R and HCT116/OxR
chemoresistant cells compared with the parental HCT116 human CRC
cells. β-actin was used as a loading control. (B) Flow cytometric
analysis revealed that the colonospheres and chemoresistant cell
lines were enriched with cells expressing CD133 and CD44 compared
with the parental cell line. A total of 33% of the HCT116/5FU-R
cells, 47% of the HCT116/OxR cells and 65% of the
HCT116/colonosphere cells expressed CD133 compared with 2% of the
parental HCT116 cells. Similarly, 84% of the HCT116/5FU-R cells,
93% of the chemoresistant cells and 92% of the HCT116/colonosphere
cells expressed CD44 compared with 48% of the parental cells.
Cytometric analysis plots using isotype control antibodies were
used as staining controls. (C) CD44 and CD133 labelling and flow
cytometric analysis revealed a 4.8-, 22- and 24.7-fold enrichment
of double-positive cells in the HCT116/5FU-R, HCT116/OxR and
colonosphere cells compared with the parental HCT116 cell line.
SCC, side scatter; Co-CSC, colorectal cancer stem cell; CD, cluster
of differentiation; 5-FU, 5-fluorouracil; R, resistant; Ox,
oxaliplatin. |
Cell phenotype in the colonospheres and
chemoresistant cells
In vitro proliferation was assessed through
plating an equal number of cells from each cell line and using a
CCK-8 assay as an index of cell number. The proliferation rates of
the colonospheres, 5FU- and oxaliplatin-resistant cells were found
to be significantly lower than those of the parental cells (52–72%;
P<0.05; Fig. 2A). The CCK-8
assay was also used to analyze cell sensitivity to chemotherapeutic
agents. Colonospheres, 5FU- and oxaliplatin-resistant cells were
exposed to clinically relevant doses of 5FU and oxaliplatin. The
number of cells remaining after 72 h was then assessed. Parental
cells were found to be sensitive to oxaliplatin and 5FU, with only
34 and 21% of the cells remaining viable following exposure to
oxaliplatin and 5FU, respectively (Fig. 2B). 5FU-resistant cells were
observed to be resistant to 5FU; however, these cells were also
resistant to oxaliplatin, with 77% of the cells remaining after 72
h of exposure. Similarly, oxaliplatin-resistant cells were found to
be resistant to oxaliplatin, but also exhibited cross-resistance to
5FU. Colonospheres were resistant to oxaliplatin and 5FU, with
79–87% of the cells remaining viable after 72 h of exposure.
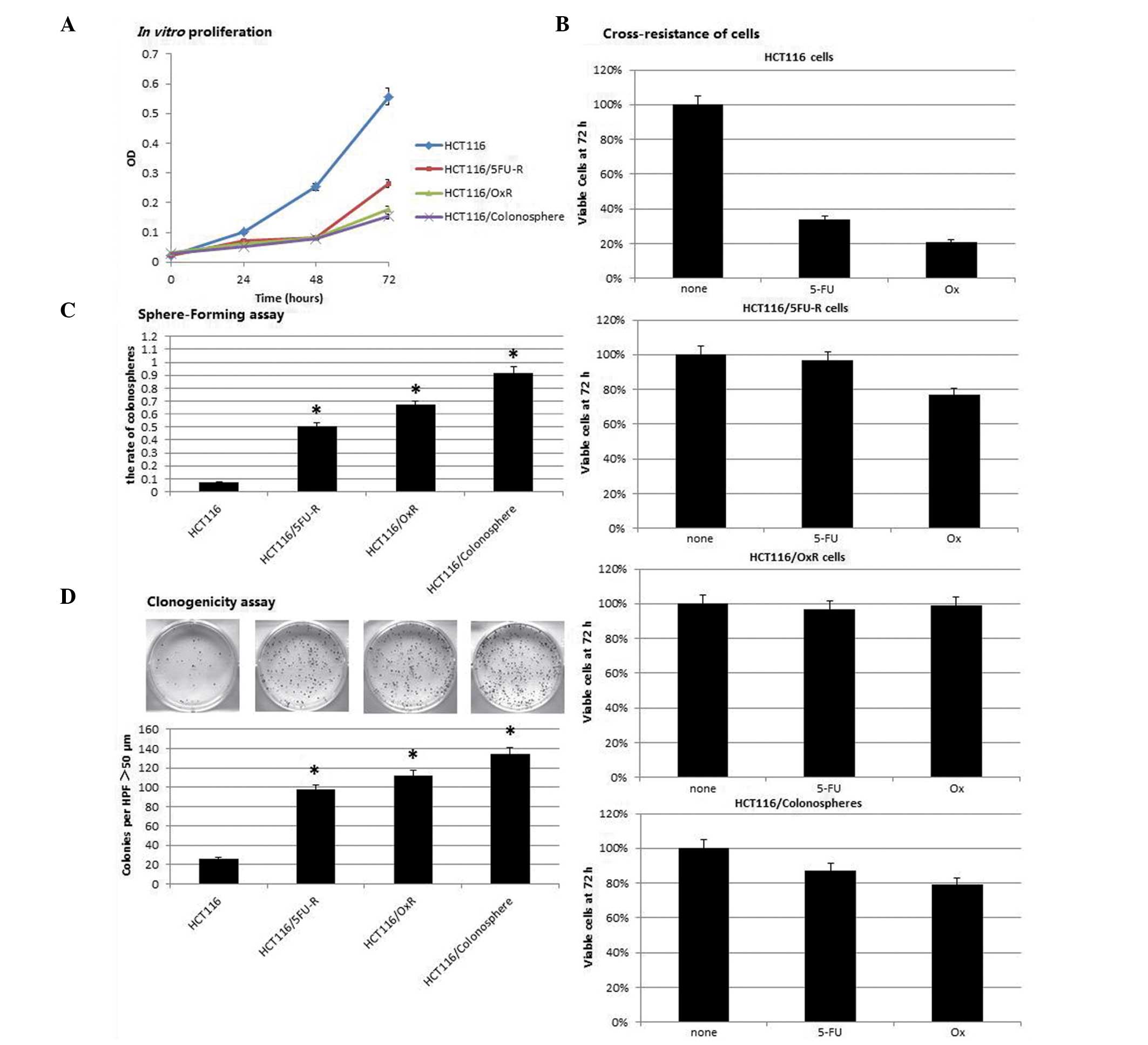 | Figure 2Colonospheres and chemoresistant cell
exhibit a cancer stem cell phenotype. (A) Colonospheres and
chemoresistant cells proliferated at a significantly slower rate
than parental cells, detected using the Cell Counting Kit-8 assay.
(B) Parental cells were sensitive to 5FU and Ox following exposure
for 72 h, with only 34 and 21% of cells remaining, respectively,
compared with the untreated cells. By contrast, HCT116/5FU-R cells
were resistant to 5FU; however, they also exhibited increased
resistance to Ox compared with the parental cells. Similarly,
HCT116/Ox-R cells were resistant to Ox, as well as 5FU.
Colonosphere cells were also resistant to Ox and 5-FU. (C) Cells
were plated in an ultralow-attachment 96-well plate in the absence
of serum and after 14 days, the rate of viable sphere-forming cells
was assessed. The colonosphere formation rate was significantly
increased in the colonospheres and chemoresistant cells compared
with the parental cells. (D) Clonogenic assay revealed that the
number of colonies larger than 50 µm in diameter which were
formed under standard growth conditions was significantly higher in
the colonospheres and chemoresistant compared with the parental
HCT116 cells. Data are presented as the mean ± standard error.
*P<0.05 vs. HCT116 cells. OD, optical density, 5-FU,
5-fluorouracil; Ox, oxaliplatin; R, resistant. |
CSCs have the capacity to form colonies, also known
as spheres, in the absence of serum and without attachment to
culture plates. In the present study, the capacity of colonospheres
and chemoresistant cell lines to grow colonospheres under
serum-free conditions was analyzed. Cell lines were trypsinized and
quantified by plating a single cell in each well of a
low-attachment 96-well plate and assessing the capacity of the
cells to form colonospheres. In the colonosphere cells, the rate of
secondary sphere generation was higher than that in the
HCT116/5FU-R and HCT116/OxR cells. However, compared with the
parental cells, an increased number of colonospheres was found in
the three cell types (P<0.05; Fig.
2C). The clonogenic assay revealed that the three types of
cells exhibited an increased colonosphere formation capacity after
14 days of culture (Fig. 2D).
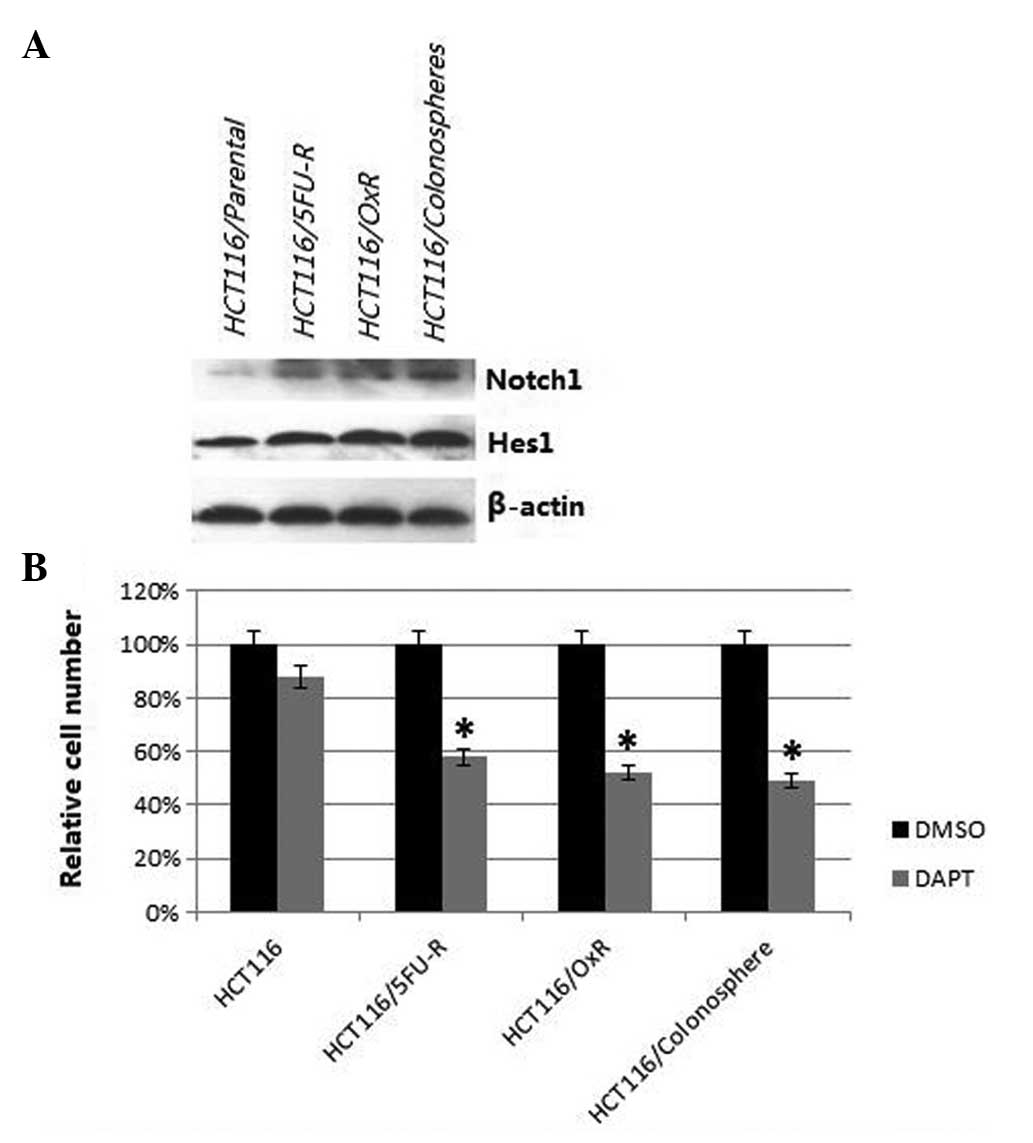
Notch pathway in colonospheres and
chemoresistant cells
Western blot analysis was used to assess
constitutive signaling in the parental, colonosphere and
chemoresistant cell lines, with a focus on targets for which agents
that inhibited target function were readily available. Several
signaling pathways were investigated, but the most marked
alterations were observed in the Notch signaling pathway, thus the
present study focused on Notch signaling. Notch1 levels were found
to be higher in the colonospheres and chemoresistant cell lines
compared with the parental cells (Fig.
3A). Furthermore, the levels of hairy and enhancer of split 1
(Hes1) were also observed to be increased in the colonospheres and
chemoresistant cell lines compared with the parental cells.
DAPT, a γ-secretase inhibitor, was used to determine
the dependence of the cells on Notch signaling for survival. The
CCK-8 assay revealed that DAPT treatment caused a minor decrease
(12%) in cell number in the parental cells, but a significantly
greater reduction in cell number in the colonospheres and
chemoresistant cells compared with the parental cells (42% for
HCT116/5FU-R, 48% for HCT116/OxR and 51% for HCT116/colonospheres;
all P<0.05; Fig. 3B).
Effect of Notch pathway inhibition on in
vivo tumor growth
Colonospheres and chemoresistant cells were injected
subcutaneously in the flanks of the nude mice and tumor growth was
assessed during biweekly treatment with DAPT or dimethyl sulfoxide
(DMSO). After ~four weeks, at which point the maximum tumor size
was ~1.5 cm3, the tumors were harvested and analyzed.
The tumors derived from the colonospheres, parental and
chemoresistant cells which were treated with DAPT were found to be
significantly smaller than those treated with DMSO (control).
However, the tumors derived from the colonospheres and
chemoresistant cells exhibited significantly greater DAPT-induced
growth inhibition compared with the parental cells. The
5FU-resistant and oxaliplatin-resistant cells showed 52 and 67%
growth inhibition, respectively, while growth of the colonosphere
cells was inhibited by 71% compared with the parental cells (21%;
P<0.05; Fig. 4A).
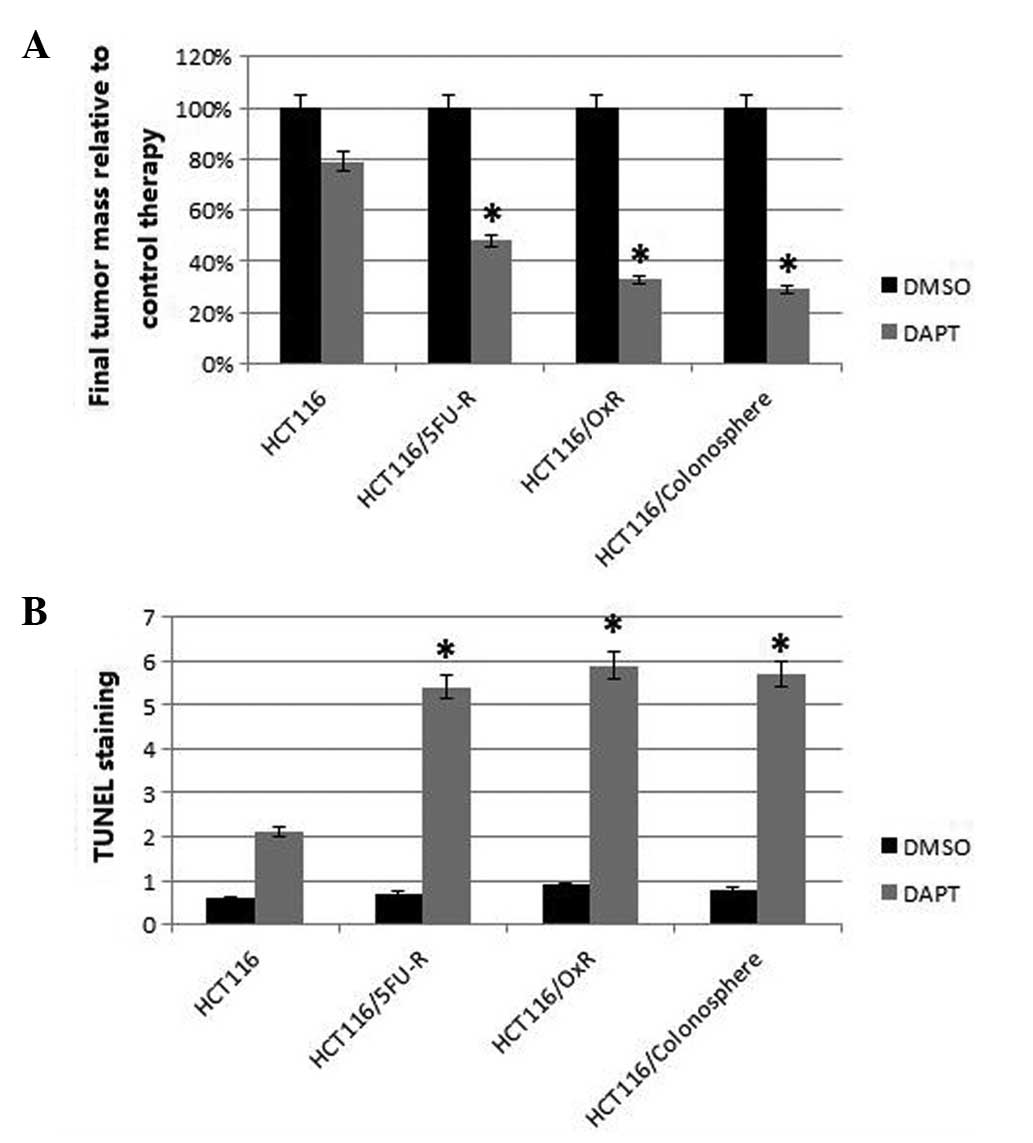 | Figure 4Effect of Notch pathway inhibition on
in vivo tumor growth, proliferation and apoptosis. Mice were
subcutaneously injected with 1×106 HCT116, HCT116/5FU-R,
HCT116/OxR or HCT116/colonosphere cells and treated with DMSO
(control) or DAPT twice weekly. Final tumor masses were measured
and compared between mice bearing tumors from each cell line. (A)
In the DAPT-treated mice, the HCT116/5FU-R-, HCT116/OxR- and
HCT116/colonosphere-derived tumors showed significantly greater
growth inhibition than the HCT116-derived tumors. (B) In the
DAPT-treated mice, TUNEL staining revealed significantly greater
apoptosis in the HCT116/5FU-R-, HCT116/OxR- and
HCT116/colonospheres-derived tumors than in tumors derived from the
HCT116 cells. Data are presented as the mean ± standard error.
*P<0.05 vs. HCT116 cells. 5-FU, 5-fluorouracil; R,
resistant; Ox, oxaliplatin; DAPT,
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine
t-butyl ester; DMSO, dimethyl sulfoxide; TUNEL, terminal
deoxynucleotidyltransferase-mediated dUTP nick-end labeling. |
Analysis of the proliferation marker Ki67 using
tumor section staining revealed that DAPT caused a decrease in the
number of proliferating cells in all of the tumors compared with
those treated with DMSO. Similar to tumor growth inhibition, the
inhibition of Notch signaling had a greater effect on the tumors
derived from the colonospheres and chemo-resistant cells than on
the tumors derived from the parental cells; however, this
difference was not found to be statistically significant. TUNEL
staining was used to analyze apoptosis in the xenografts.
Quantification of TUNEL staining showed that DAPT treatment caused
significantly more apoptosis in the tumors derived from the
colonospheres and chemoresistant cells compared with those derived
from the HCT116 cells (P<0.05). Specifically, upon Notch
signaling inhibition, HCT116 tumors showed a 2.1-fold increase in
apoptotic nuclei compared with 5.4-, 5.9- and 5.7-fold increases in
the HCT116/5FU-, HCT116/OxR- and colonosphere-derived tumors,
respectively (all P<0.05; Fig.
4B). Representative images from the analyzed tumor sections and
the subcutaneous tumors are shown in Fig. 5.
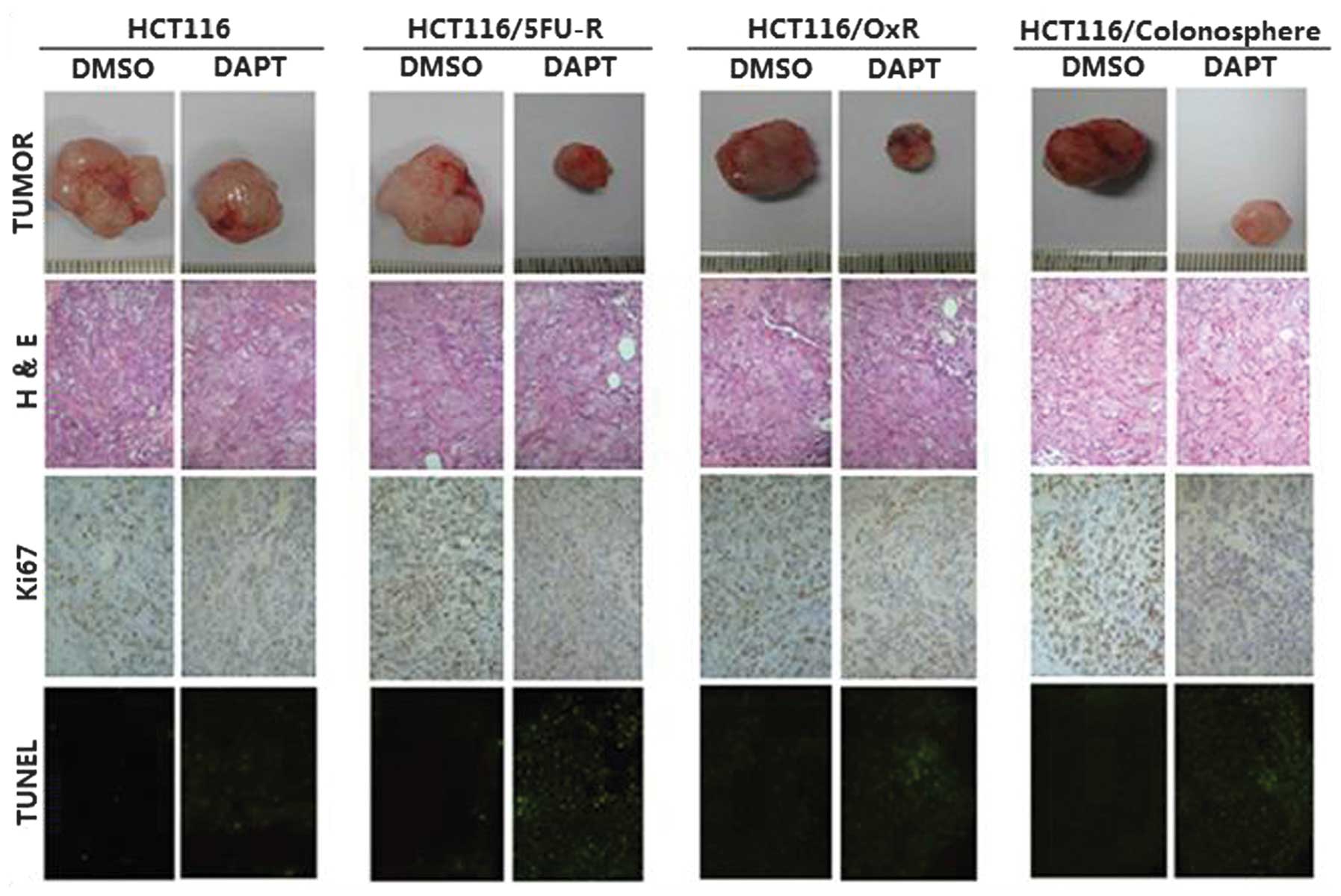 | Figure 5Effect of Notch pathway inhibition on
in vivo tumor characteristics. Immunohistochemical analysis
of tumors was performed and multiple tumor fields were analyzed per
group. Representative images of all groups and treatments are
presented (magnification, x100). H&E staining revealed similar
subcutaneous tumor morphology among all groups of tumors. Ki67
staining showed decreased cell proliferation in the tumors treated
with DAPT; however, no significant differences were observed in the
cell proliferation between the HCT116-, colonosphere-,
HCT116/5FU-R- and HCT116/Ox-R-derived tumor sections. TUNEL
staining revealed significantly increased apoptosis in response to
DAPT in colonosphere-, HCT116/5FU-R- and HCT116/Ox-R-derived tumors
compared with tumors derived from HCT116 cells (P<0.05). 5-FU,
5-fluorouracil; R, resistant; Ox, oxaliplatin; DAPT,
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine
t-butyl ester; DMSO, dimethyl sulfoxide; TUNEL, terminal
deoxynucleotidyltransferase-mediated dUTP nick-end labeling;
H&E, hematoxylin and eosin. |
Discussion
CRC is the second leading cause of cancer-associated
mortality worldwide. Despite recent therapeutic regimens which have
markedly increased survival in CRC, almost all CRC tumors become
chemoresistant (24). Thus, it is
necessary to understand the mechanisms of resistance in order to
improve current treatment protocols in CRC.
The eradication of drug-resistant Co-CSCs is an
important area of investigation (25). Numerous studies have used
fluorescence-activated cell sorting (FACS) in order to identify and
isolate CSCs. In the present study, FACS was not performed on CSCs
as no reliable surface markers are available for identifying
Co-CSCs. However, CSCs have the capacity to form colonies, also
know as spheres, when cultured in the absence of serum. Therefore,
colonosphere formation was used to investigate the characteristics
of Co-CSCs (26). Chemoresistance
is an important feature of Co-CSCs, thus the present study aimed to
investigate the interrelation between Co-CSCs and chemoresistant
cells. The present study focused on colonospheres and two
chemoresistant cell lines and identified certain common features.
Colonospheres and chemoresistant cells were found to be
significantly enriched in the CSC markers CD133 and CD44 (27). This finding suggested that
colonospheres may be abundant in Co-CSCs. Furthermore, the
chemoresistance imparted in the two chemoresistant cell lines may
be due to the acquisition of CSC phenotypes in these cell lines.
Thus, in the present study, to further investigate the
characteristics of Co-CSCs, colonospheres were cultured in
serum-free media and chemoresistant cell lines were developed.
Colonospheres and chemoresistant cells were also
found to be more quiescent in vitro, with decreased cell
proliferation compared with the parental cells. However, a
clonogenic assay revealed that colonospheres and chemoresistant
cells had an increased capacity to form colonies and spheres in
specialized serum-free media, characteristics which are consistent
with the CSC phenotype (5).
Furthermore, lysates obtained from cell line-derived colonospheres
were observed to have increased resistance to 5FU and oxaliplatin
as compared with the adherent parental cells. Oxaliplatin-resistant
cells and 5FU-resistant cells also exhibited cross-resistance to
5FU and oxaliplatin, respectively. This suggested that
colonospheres and chemoresistant cells activated general resistance
pathways leading to multi-drug resistance. Colonospheres and
chemoresistant cells were also found to be enriched in CSC markers
and properties, consistent with the CSC phenotype. It is most
likely that the process of developing colonospheres and
chemoresistant cell lines involved increasing the expression of
such CSC markers, rather than enriching the population of cells
which already expressed these markers. Assessing this hypothesis
may be difficult; however, previous studies have shown that the
tumor microenvironment, including soluble factors and hypoxia, may
affect colonosphere characteristics (28,29).
However, little is known about the pathway involved in the
expression of CSC markers in Co-CSCs and chemoresistant cells and
further investigations into the mechanism of resistance and
increased marker expression are required.
Therefore, the present study aimed to investigate
the pathways involved in CSCs and chemoresistant cells and aimed to
identify potential targets in the colonospheres and chemoresistant
cells which would allow specific targeting of these cells with
optimal agents. One such pathway was the Notch signaling pathway,
which is involved in CRC progression and growth (30,31).
The Notch1 gene copy number has been reported to be increased in
colorectal adenocarcinomas and to be correlated with aggressive
tumor behavior and poor prognosis (32–34).
In the present study, constitutive Notch1 expression was observed
to be higher in the colonospheres and chemoresistant cell lines
compared with the parental cell lines, and most markedly increased
in the colonospheres. Furthermore, the increase in Hes1 was
associated with an increase in the levels of the targeted gene. In
the present study, the Notch signaling pathway, particularly
Notch1, was found to have a key role in colonospheres and
chemoresistant cell lines, and it was hypothesized that the
activation of Notch was involved in maintaining a phenotypic
characteristic in the colonospheres and chemoresistant cell lines.
However, the specific mechanism underlying the increase in the
Notch1 expression in these cells has yet to be elucidated. The
inhibition of Notch signaling in vitro was found to cause a
decrease in cell growth, as determined by CCK-8 assay, and these
effects were observed to be greater in the colonospheres and
chemoresistant cell lines than in the parental cells. These
findings showed that the inhibition of the Notch pathway using DAPT
significantly depleted the number of colonosphere cells and
chemoresistant cells, further validating the hypothesis that the
activation of Notch is necessary for the maintenance of phenotypic
characteristics in colonospheres and chemoresistant cell lines.
In vivo, following DAPT treatment, the inhibition of the
growth of colonosphere- and chemoresistant cell-derived tumors was
found to be significantly higher than that of the parental
cell-derived tumors. This finding suggested that the Notch
signaling pathway may be important for Co-CSC maintenance and tumor
resistance to standard chemotherapeutics. A previous study reported
that colonic cancer cells may upregulate Notch1 as a protective
mechanism in response to chemotherapy (35). Furthermore, in the present study,
Notch inhibition was found to have a greater effect on the growth
of colonosphere- and chemoresistant cell-derived tumors than
parental cell-derived tumors in vivo, which was largely due
to an increase in apoptosis. It is unlikely that colonospheres and
chemoresistant cells acquired common molecular alterations to
resist certain agents. However, the present study found that the
colonospheres and chemoresistant cells acquired similar molecular
and phenotypic alterations when cultured in serum-free media or
when chronically exposed to chemotherapeutic agents. Of note, the
colonospheres and chemoresistant cells also exhibited increased
Notch1 and Hes1 expression, which led to these cells becoming more
sensitive to the inhibition of the Notch pathway. Previous studies
have suggested that targeting the Notch signaling pathway may be an
effective method for targeting CSCs and chemoresistant cells
(36,37). The findings of the present study
suggest that inhibiting the Notch pathway using DAPT may be an
effective strategy for targeting Co-CSCs and overcoming the
chemoresistance of CRC cells in a clinical setting.
Acknowledgments
The authors would like to thank Qiang Li for his
helpful advice.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
O’Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
4
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
et al: Identification and expansion of human
colon-cancer-initiating cells. Nature. 445:111–115. 2007.
View Article : Google Scholar
|
|
5
|
Yeung TM and Mortensen NJ: Colorectal
cancer stem cells. Dis Colon Rectum. 52:1788–1796. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sugihara E and Saya H: Complexity of
cancer stem cells. Int J Cancer. 132:1249–1259. 2013. View Article : Google Scholar
|
|
7
|
Lou H and Dean M: Targeted therapy for
cancer stem cells: the patched pathway and ABC transporters.
Oncogene. 26:1357–1360. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sussman RT, Ricci MS, Hart LS, Sun SY and
El-Deiry WS: Chemotherapy-resistant side-population of colon cancer
cells has a higher sensitivity to TRAIL than the non-SP, a higher
expression of c-Myc and TRAIL-receptor DR4. Cancer Biol Ther.
6:1490–1495. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Todaro M, Alea MP, Di Stefano AB, et al:
Colon cancer stem cells dictate tumor growth and resist cell death
by production of interleukin-4. Cell Stem Cell. 1:389–402. 2007.
View Article : Google Scholar
|
|
10
|
Walko CM and Lindley C: Capecitabine: a
review. Clin Ther. 27:23–44. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kelland L: The resurgence of
platinum-based cancer chemotherapy. Nat Rev Cancer. 7:573–584.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wen K, Fu Z, Wu X, et al: Oct-4 is
required for an antiapoptotic behavior of chemoresistant colorectal
cancer cells enriched for cancer stem cells: effects associated
with STAT3/Survivin. Cancer Lett. 333:56–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Holland JD, Klaus A, Garratt AN and
Birchmeier W: Wnt signaling in stem and cancer stem cells. Curr
Opin Cell Biol. 25:254–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pannuti A, Foreman K, Rizzo P, et al:
Targeting Notch to target cancer stem cells. Clin Cancer Res.
16:3141–3152. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takebe N, Harris PJ, Warren RQ and Ivy SP:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar
|
|
16
|
Lee J, Kotliarova S, Kotliarov Y, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh SK, Clarke ID, Terasaki M, et al:
Identification of a cancer stem cell in human brain tumors. Cancer
Res. 63:5821–5828. 2003.PubMed/NCBI
|
|
18
|
Liu JC, Deng T, Lehal RS, Kim J and
Zacksenhaus E: Identification of tumorsphere- and tumor-initiating
cells in HER2/Neu-induced mammary tumors. Cancer Res. 67:8671–8681.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vlashi E, Kim K, Lagadec C, et al: In vivo
imaging, tracking, and targeting of cancer stem cells. J Natl
Cancer Inst. 101:350–359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang AD, Fan F, Camp ER, et al: Chronic
oxaliplatin resistance induces epithelial-to-mesenchymal transition
in colorectal cancer cell lines. Clin Cancer Res. 12:4147–4153.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dallas NA, Xia L, Fan F, et al:
Chemoresistant colorectal cancer cells, the cancer stem cell
phenotype, and increased sensitivity to insulin-like growth
factor-I receptor inhibition. Cancer Res. 69:1951–1957. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chu ZH, Liu L, Zheng CX, et al: Proteomic
analysis identifies translationally controlled tumor protein as a
mediator of phosphatase of regenerating liver-3-promoted
proliferation, migration and invasion in human colon cancer cells.
Chin Med J (Engl). 124:3778–3785. 2011.
|
|
23
|
Vaiopoulos AG, Kostakis ID, Koutsilieris M
and Papavassiliou AG: Colorectal cancer stem cells. Stem Cells.
30:363–371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marin JJ, Sanchez de Medina F, Castaño B,
et al: Chemoprevention, chemotherapy, and chemoresistance in
colorectal cancer. Drug Metab Rev. 44:148–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Puglisi MA, Tesori V, Lattanzi W,
Gasbarrini GB and Gasbarrini A: Colon cancer stem cells:
controversies and perspectives. World J Gastroenterol.
19:2997–3006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tirino V, Desiderio V, Paino F, et al:
Cancer stem cells in solid tumors: an overview and new approaches
for their isolation and characterization. FASEB J. 27:13–24. 2013.
View Article : Google Scholar
|
|
27
|
Yu Y, Kanwar SS, Patel BB, et al:
Elimination of colon cancer stem-like cells by the combination of
curcumin and FOLFOX. Transl Oncol. 2:321–328. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bose D, Zimmerman LJ, Pierobon M, et al:
Chemoresistant colorectal cancer cells and cancer stem cells
mediate growth and survival of bystander cells. Br J Cancer.
105:1759–1767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yeung TM, Gandhi SC and Bodmer WF: Hypoxia
and lineage specification of cell line-derived colorectal cancer
stem cells. Proc Natl Acad Sci USA. 108:4382–4387. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koch U and Radtke F: Notch and cancer: a
double-edged sword. Cell Mol Life Sci. 64:2746–2762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiao L and Wong BC: Role of notch
signaling in colorectal cancer. Carcinogenesis. 30:1979–1986. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arcaroli JJ, Powell RW, Varella-Garcia M,
et al: ALDH+ tumor-initiating cells exhibiting gain in NOTCH1 gene
copy number have enhanced regrowth sensitivity to a γ-secretase
inhibitor and irinotecan in colorectal cancer. Mol Oncol.
6:370–381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Li B, Ji ZZ and Zheng PS: Notch1
regulates the growth of human colon cancers. Cancer. 116:5207–5218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin HY, Zhang HY, Wang X, Xu J and Ding Y:
Expression and clinical significance of notch signaling genes in
colorectal cancer. Tumour Biol. 33:817–824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meng RD, Shelton CC, Li YM, et al:
gamma-Secretase inhibitors abrogate oxaliplatin-induced activation
of the Notch-1 signaling pathway in colon cancer cells resulting in
enhanced chemosensitivity. Cancer Res. 69:573–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McAuliffe SM, Morgan SL, Wyant GA, et al:
Targeting Notch, a key pathway for ovarian cancer stem cells,
sensitizes tumors to platinum therapy. Proc Natl Acad Sci USA.
109:E2939–E2948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ponnurangam S, Mammen JM, Ramalingam S, et
al: Honokiol in combination with radiation targets notch signaling
to inhibit colon cancer stem cells. Mol Cancer Ther. 11:963–972.
2012. View Article : Google Scholar : PubMed/NCBI
|