Introduction
Renal cell carcinoma (RCC) is the most common
neoplasm of the adult kidney with the highest rate of recurrence
and mortality among malignances in urologic systems (1). Nearly 30% of RCC patients present at
advanced stages, and ~40% of patients that undergo surgical
resection experience recurrence during subsequent follow-up
(2,3). Metastatic RCC has an extremely poor
prognosis and remains an incurable disease despite the great
improvement in surgery and personalized treatments (4,5). The
high rates of recurrence and mortality of RCC create an urgent
requirement for personalized care and reliable biomarkers for early
detection and prognosis prediction (6). Therefore, exploring the molecular
mechanisms underlying the disease and identifying novel molecular
biomarkers is important (7).
In recent years, miRNAs have emerged as important
molecules in the complex networks of gene regulation (8). These small, endogenous non-coding RNA
molecules that regulate the expression of protein coding genes at a
post-transcriptional level have been implicated in a variety of
human disorders, such as infectious diseases, metabolic diseases
and cancer (9,10). Aberrantly expressed miRNAs are
prevalent in a number of types of human cancer, and are important
in cancer initiation, development and metastasis (11,12).
Certain highly expressed miRNAs may function as oncogenes by
repressing tumor suppressors, whereas downregulated miRNAs may
function as tumor suppressors by negatively regulating oncogenes
(13). Their stable expression in
the blood render them reliable biomarkers for early detection,
diagnosis and prognosis prediction in various diseases, including
cancer (14,15).
miR-510 has been demonstrated to be involved in lung
cancer, breast cancer, gastric cancer and ovarian serous carcinoma
(16–19); however, the expression and function
of miR-510-5p in renal cancer remains unclear. The present study
aimed to determine the expression of miR-510-5p in RCC tissues and
paired normal adjacent tissues, and analyzed the impact of
miR-510-5p on renal cancer by
3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT), wound scratch and apoptosis assays.
Materials and methods
Clinical specimens
The present study was approved by the Ethics
Committee of Peking University Shenzhen Hospital (Shenzhen, China)
and Anhui Medical University First Affiliated Hospital (Hefei,
China). Written informed consent was obtained from every patient
prior to sample collection. A total of 48 paired renal cell
carcinoma (RCC) and adjacent normal specimens were collected from
patients receiving radical nephrectomies at Peking University
Shenzhen Hospital or Anhui Medical University First Affiliated
Hospital. All samples were processed and stored at -80°C in
RNAlater (Qiagen, Valencia, CA, USA) until RNA isolation. The
clinical and pathological information of all the patients is
summarized in Table I. These
samples were staged according to the American Joint Committee on
Cancer (AJCC) staging system (20).
 | Table IClinical and pathological features of
48 patients. |
Table I
Clinical and pathological features of
48 patients.
| Variable | Number of
cases |
|---|
| Total | 48 |
| Age (years) |
| ≥52 | 29 |
| <52 | 19 |
| Gender |
| Male | 30 |
| Female | 18 |
| Histological
type |
| Clear cell | 39 |
| Papillary | 9 |
| Primary tumor
stage |
| T1 | 27 |
| T2 | 19 |
| T3 and T4 | 2 |
| AJCC clinical
stage |
| I | 27 |
| II | 18 |
| III+IV | 3 |
Cell culture and RNA extraction
Two human RCC cell lines, ACHN and 786-O (Guangdong
and Shenzhen Key Laboratory of Male Reproductive Medicine and
Genetics, Shenzhen, China) were used in this study. They were
incubated in Dulbecco’s modified Eagle’s medium (Invitrogen Life
Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum (Invitrogen Life Technologies) and maintained in a humidified
incubator containing 5% CO2 at 37°C. Total-RNA of each
sample was isolated with TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) and purified with an RNeasy Maxi
kit (Qiagen) according to the manufacturer’s instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To obtain the cDNA templates, 1 μg total RNA
of each sample was isolated for reverse transcription with the
miScript Reverse Transcription reagent (Qiagen).
PCR amplification was performed using 1 μl
cDNA in a 20 μl reaction system, containing 10 μl
QuantiTect SYBR Green PCR Master mix, 2 μl miScript
Universal Primer, 0.5 μl specific microRNA primer and 6.5
μl RNase-free water. The sequence of the miR-510-5p forward
primer was 5′-TAGCAGCACGTAAATATTGGCG-3′ and the reverse primer was
provided by the miScript SYBR Green PCR kit. The sequence of the U6
forward primer was 5′-CTCGCTTCGGCAGCACA-3′ and reverse primer was
5′-ACGCTTCACGAATTTGCGT-3′. PCR amplification conditions were set
as: 95°C for 2 min, then 40 cycles of 95°C for 15 sec, 58°C for 30
sec and 72°C for 30 sec. The relative expression levels of
miR-510-5p were calculated using the 2−ΔΔCt method
(21).
Mature miRNA and negative control
transfection
For the restoration of miR-510-5p, ACHN and 786-O
cells were trans-fected with synthetic mature molecules (miR-510-5p
mimics; Shanghai GenePharma Co., Ltd., Shanghai, China) with
Lipofectamine 2000 (Invitrogen Life Technologies) according to the
manufacturer’s instructions. Mature miR-510-5p mimics and negative
control were used in the gain-of-function experiments. The cancer
cells were harvested and RNA was isolated for RT-qPCR to analyze
the fold changes of miR-510-5p 24 h after transfection.
MTT assay
The capacity for cellular proliferation was
determined using an MTT assay, according to the manufacturer’s
instructions. Approximately 5×103 cells were seeded into
96-well culture plates and transfected with 5 pmol miR-510-5p
mimics or negative control. At 0, 24, 48 or 72 h after
transfection, the cells were incubated with 20 μl MTT (5
μg/ml) for 4 h, followed by the addition of 150 μl
dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA) and shaking
for 15 min at room temperature to solubilize the crystals. The
optical density (OD) was determined using a microplate reader
(Model 680; Bio-Rad) at dual wavelength of 490/630 nm.
Flow cytometric analysis of
apoptosis
Approximately 300,000 renal cancer cells were
cultured in 6-well plates at 37°C and transfected with miR-510-5p
mimics or negative control within 24 h. Cancer cells, including
floating cells, were harvested 48 h after transfection, washed
twice with cold phosphate-buffered saline and resuspended in 100
μl of 1X binding buffer (Invitrogen Life Technologies),
followed by the addition of 5 μl Annexin V-fluorescein
isothiocyanate (Invitrogen Life Technologies) and 3 μl
propidium iodide (PI). The fluorescence of stained cells was then
analyzed by flow cytometry (Beckman Coulter, Miami, FL, USA) using
488 nm excitation within 30 min of staining, according to the
manufacturer’s instructions.
Migration scratch assay
Wound scratch assay was used to assess the migration
ability of 786-O and ACHN renal cancer cells in vitro.
Approximately 350,000 cells were seeded per 12-well dish and
transfected with 80 pmol miR-510-5p mimics or 80 pmol negative
control 24 h later using Lipofectamine 2000.
After 5 h of transfection, the cell monolayer was
scraped using a P-20 micropipette tip. The initial gap length (0 h)
and the residual gap length 24 h after wound-healing were
calculated using the software program MIAS-2000 (Leica Microsystems
GmbH, Wetzlar, Germany). The experiments were performed in
triplicate, repeated at least three times, and analyzed in a
double-blind manner by at least two observers.
Bioinformatics
The potential targets of miR-510-5p were predicted
by combining four public algorithms, miRanda (http://www.targetscan.org/), TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
Putative genes predicted by all four algorithms were accepted and
candidates were selected based on the gene function.
Statistical analysis
All statistical analysis was conducted with SPSS
17.0 statistical software package (SPSS Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference. The different expression of miR-510-5p in RCC and
paired normal samples was analyzed by a paired t-test.
Results
miR-510-5p is downregulated in RCC
tissues quantified by RT-qPCR
Previous miRNA expression profiles of RCC indicated
that miR-510-5p was downregulated (22,23).
In order to confirm the results of former studies, RT-qPCR was used
to quantify the expression of miR-510-5p in RCC and paired adjacent
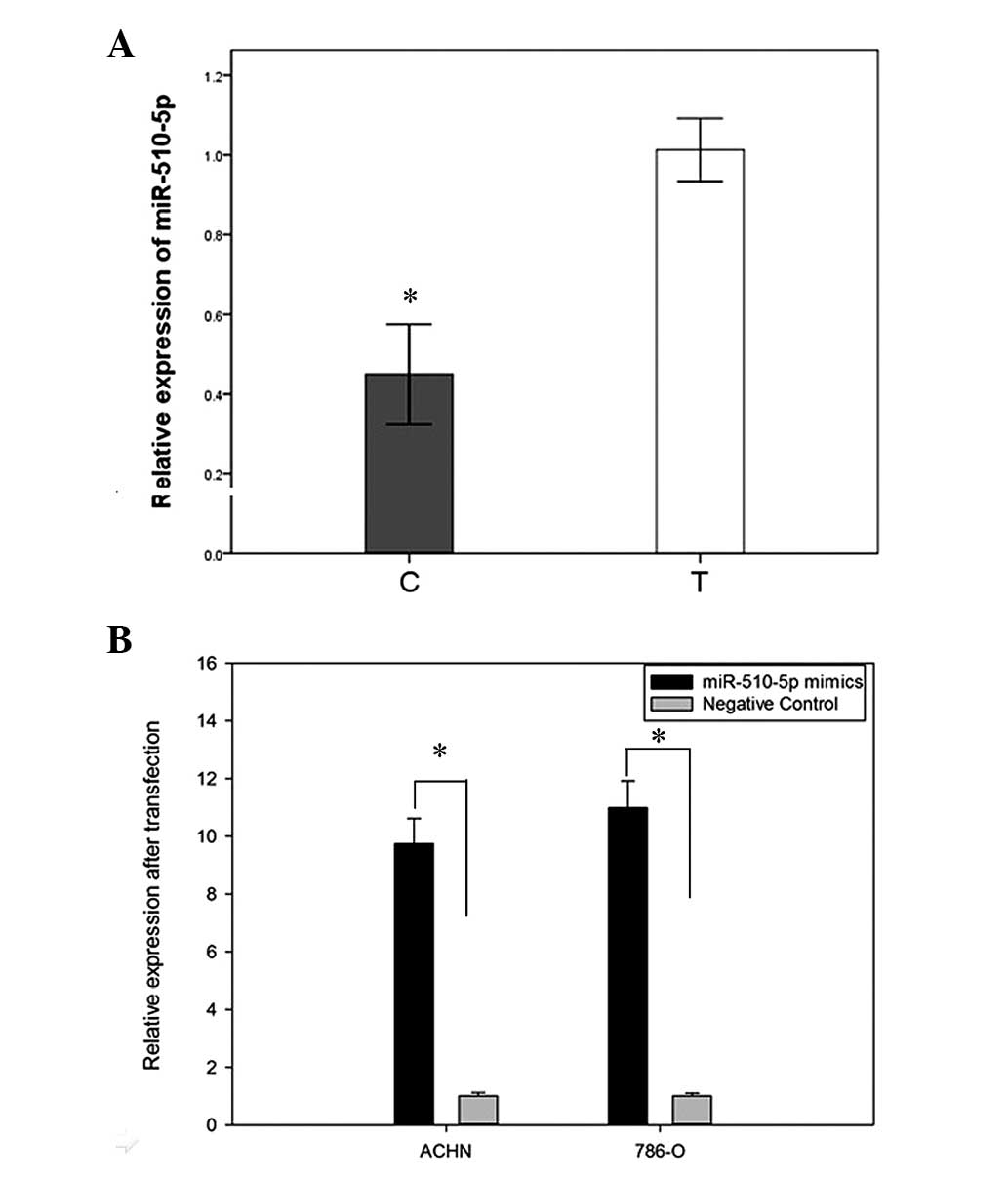
normal tissues from 48 patients. The results showed the expression
of miR-510-5p decreased in 81.25% (39/48) of RCC tissues, compared
with paired normal tissues, with an average reduction in expression
of 0.4283-fold (Fig. 1A). To
investigate the effects of miR-510-5p on renal cancer cells,
synthetic miR-510-5p mature mimics and negative controls were
transfected into ACHN and 786-O cell lines. As shown in Fig. 1B, RT-qPCR analysis indicated the
fold changes of miR-510-5p in ACHN and 786-O cells after
transfection were 9.58 and 11.32, respectively.
Overexpression of miR-510-5p inhibits RCC
cell proliferation
The impact on cell proliferation was analyzed by an
MTT assay, OD values of two groups (miR-510-5p mimics and negative
control) were measured at 0, 24, 48 and 72 h after transfection.
The present results showed the proliferation of ACHN cells
decreased by 5.16% (P>0.05), 15.26% (P<0.05) and 29.06%
(P<0.05) while the proliferation of 786-O cells decreased by
5.06 (P>0.05), 12.42 (P<0.05) and 21.78% (P<0.05)
(Fig. 2), suggesting that
miR-510-5p can reduce the proliferation of cancer cells.
Restoration of miR-510-5p induces RCC
cell apoptosis
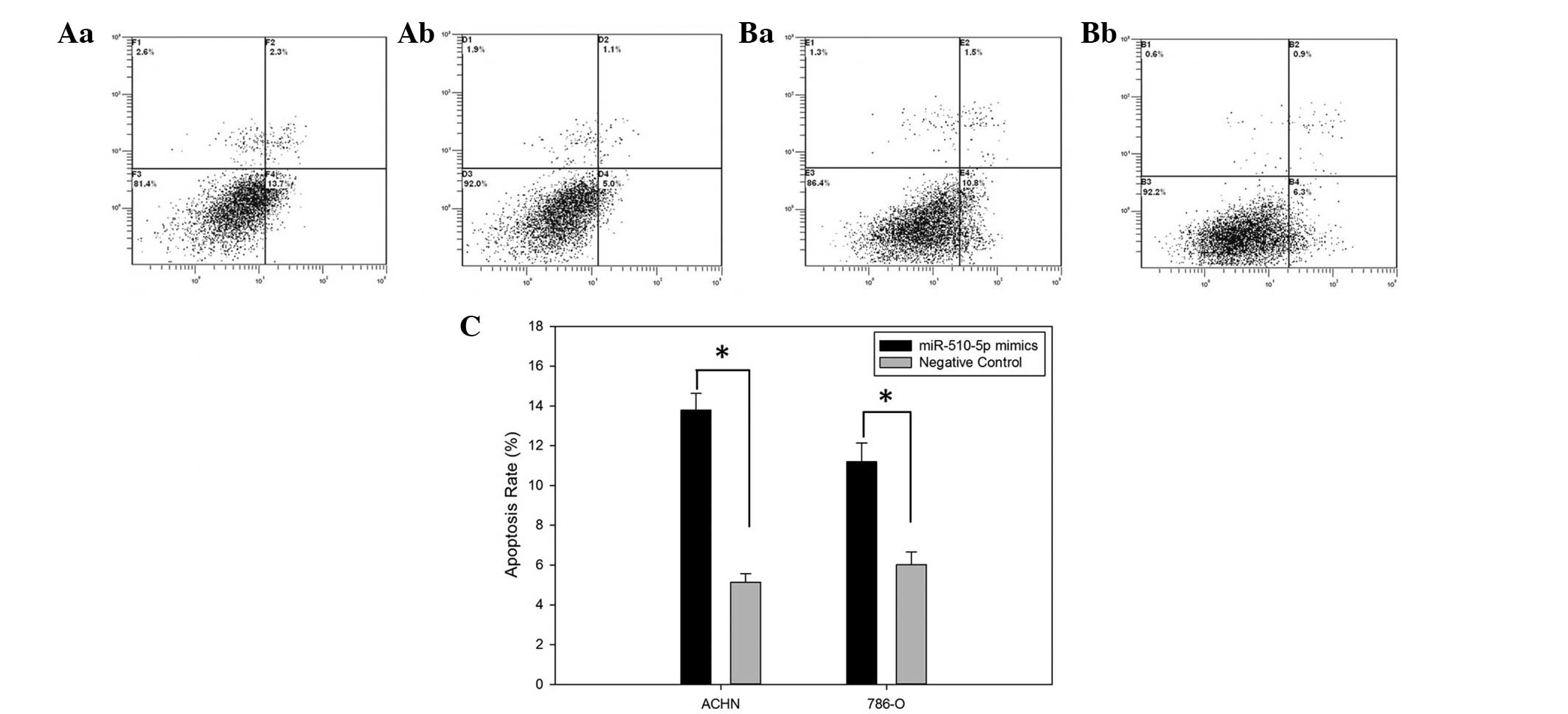
To demonstrate the effect of miR-510-5p on cell
apoptosis, a flow cytometry assay was performed to detect the
apoptosis rates of ACHN and 786-O cells after transfection. The
results revealed that apoptosis rates of ACHN transfected with
miR-510-5p mimics and negative control were 13.7 versus 5.0 while
the apoptosis rates of 786-O cells were 10.8 versus 6.1
(P<0.05), suggesting that restoration of miR-510-5p induces
apoptosis of renal cancer cells (Fig.
3).
miR-510-5p mimics inhibited cell
migration
The influence of miR-510-5p on cell migration was
observed by a wound scratch assay. As presented in Fig. 4, wound width of the group
transfected with miR-510-5p mimics was greater than that of the
negative control group (P<0.05), suggesting that overexpressed
miR-510-5p inhibited the ability of migration of renal cancer cells
(Fig. 4).
Target gene prediction
To investigate the downstream target genes of
miR-510-5p, miRanda, TargetScan, PicTar and miRWalk were used in
combination to predict the putative targets. AKT2, AKT3 and FAS
were three of the potential targets predicted by all four
algorithms simultaneously, of which the 3′ untranslated region
(UTR) of the mRNA contained a complementary site for the seed
sequences of miR-510-5p (Fig.
5).
Discussion
Carcinogenesis is a complicated process that
involves numerous genetic aberrations and signaling pathways. The
recent identification of miRNAs and their capability of
simultaneously regulating multiple downstream genes may be
important in explaining the complex mechanisms underlying cancer
formation (24). These short RNAs
of 19–25 nucleotides are key in a wide variety of biological
processes, including cell fate specification, proliferation,
migration, apoptosis and tumorigenesis (25,26).
A number of studies have validated that miRNAs contribute to the
development of various types of malignances, as well as to their
invasive and metastatic capacities, including RCC (27). For example, miR-204 was confirmed
to be a Von hippel-Lindau-regulated tumor suppressor acting by
inhibiting macroautophagy, with MAP1LC3B (LC3B) as a direct and
functional target (28). In
addition, miR-21 regulates PTEN to force the canonical oncogenic
Akt/TORC1 signaling conduit to drive renal cancer cell
proliferation and invasion (29).
Further research into the function and interaction with the target
genes of deregulated miRNAs may reveal the molecular mechanisms
underlying the tumorigenesis of RCC.
Aberrant expression of miR-510 has been observed in
several types of cancer, as described above. In breast cancer,
miR-510 directly binds to the 3′UTR of peroxiredoxin1 and prevents
its protein expression, thereby suppressing the migration of cancer
cells (17). While in ovarian
serous carcinoma (OSC), low miR-510 expression was significantly
associated with poorer overall survival, indicating that miR-510
may be considered a novel-candidate clinical biomarker for
predicting OSC outcome (18).
However, the expression and function of miR-510 in RCC remains
unclear.
In the present study, the expression of miR-510-5p
in 48 paired RCC and normal renal specimens was quantified by
RT-qPCR and found that miR-510-5p was downregulated in RCC. The
present results of decreased expression of miR-510-5p was in
accordance with the results of recent miRNA expression profiles of
RCC (30,31). To investigate whether miR-510-5p
was important for the tumorigenesis of RCC, MTT and wound scratch
assays, as well as flow cytometry were used to analyze the impact
of miR-510-5p on renal cancer by transfecting synthetic miR-510-5p
mimics. The results show that cancer cells transfected with
miR-510-5p mimics displayed less cellular proliferation and
migration and more cellular apoptosis compared with the negative
control groups, indicating that miR-510-5p may act as a tumor
suppressor by reducing cell proliferation and migration, and
inducing cell apoptosis in RCC.
It is generally acknowledged that miRNAs are
important in various biological processes by ‘imperfect’
complementary binding to the 3′UTR of the downstream genes. To
determine the target genes of miR-510-5p, several algorithms were
combined to predict putative target genes and AKT2, AKT3 and FAS
were identified as potential targets of miR-510-5p. AKT is a major
transducer of the phosphoinositide 3-kinase (PI3K) pathway and is
crucial in the regulation of cellular processes, such as growth,
metabolism and survival. Mammalian cells are characterized by the
expression of three different AKT isoforms (AKT1, AKT2 and AKT3),
encoded by distinct genes (32).
Emerging evidence has shown that AKT2 and AKT3 serve as significant
contributors to malignancy (33).
While FAS is a member of the TNF-receptor superfamily, which has
been shown to be central in the physiological regulation of
programmed cell death, and has been implicated in the pathogenesis
of various malignancies (34). It
has been reported that FAS expression is a surrogate biomarker of
active cancer cell proliferation and accurately predicts RCC
patient survival (35). Decreased
expression of miR-510-5p may regulate cellular proliferation,
migration and apoptosis by targeting oncogenes AKT2, AKT3 and FAS;
however, this requires further research.
In conclusion, the present study revealed that
downregu-lated miR-510-5p functioned as a tumor suppressor by
reducing cellular proliferation and migration and inducing
apoptosis in RCC. Further research is required to define target
genes of miR-510-5p to elucidate the cellular mechanism underlying
the effect of miR-510-5p in the carcinogenesis of RCC.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (no. 81101922), the Medical Scientific
Research Foundation of Guangdong Province of China (nos. A2012584
and A2013606), the Science and Technology Development Fund Project
of Shenzhen (grant no. JCYJ20130402114702124) and the fund of
Guandong Key medical subject.
References
|
1
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hofmann HS, Neef H, Krohe K, Andreev P and
Silber RE: Prognostic factors and survival after pulmonary
resection of metastatic renal cell carcinoma. Eur Urol. 48:77–81.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arsanious A, Bjarnason GA and Yousef GM:
From bench to bedside: current and future applications of molecular
profiling in renal cell carcinoma. Mol Cancer. 8:202009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wood CG: Molecular markers of prognosis in
renal cell carcinoma: Insight into tumor biology helps define risk
and provides targets for therapy. J Surg Oncol. 94:264–265. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Motzer RJ, Hutson TE, Tomczak P, et al:
Sunitinib versus interferon alfa in metastatic renal-cell
carcinoma. N Engl J Med. 356:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma: a reappraisal. Urol Nurs. 32:182–190.
2012.PubMed/NCBI
|
|
7
|
Maroto P and Rini B: Molecular biomarkers
in advanced renal cell carcinoma. Clin Cancer Res. 20:2060–2071.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16–1 in cancer: discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar
|
|
9
|
White NM, Khella HW, Grigull J, et al:
miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tiwari M: Microarrays and cancer
diagnosis. J Cancer Res Ther. 8:3–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Perez-Diez A, Morgun A and Shulzhenko N:
Microarrays for cancer diagnosis and classification. Adv Exp Med
Biol. 593:74–85. 2007.PubMed/NCBI
|
|
13
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koberle V, Kronenberger B, Pleli T, et al:
Serum microRNA-1 and microRNA-122 are prognostic markers in
patients with hepatocellular carcinoma. Eur J Cancer. 49:3442–3449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao A, Li G, Péoc’h M, Genin C and
Gigante M: Serum miR-210 as a novel biomarker for molecular
diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol.
94:115–120. 2013. View Article : Google Scholar
|
|
16
|
Patnaik SK, Kannisto E, Knudsen S and
Yendamuri S: Evaluation of microRNA expression profiles that may
predict recurrence of localized stage I non-small cell lung cancer
after surgical resection. Cancer Res. 70:36–45. 2010. View Article : Google Scholar
|
|
17
|
Guo QJ, Mills JN, Bandurraga SG, et al:
MicroRNA-510 promotes cell and tumor growth by targeting
peroxiredoxin1 in breast cancer. Breast Cancer Res. 15:R702013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu X, Zhang X, Bi T, et al: MiRNA
expression signature for potentially predicting the prognosis of
ovarian serous carcinoma. Tumour Biol. 34:3501–3508. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen W, Tang Z, Sun Y, et al: miRNA
expression profile in primary gastric cancers and paired lymph node
metastases indicates that miR-10a plays a role in metastasis from
primary gastric cancer to lymph nodes. Exp Ther Med. 3:351–356.
2012.PubMed/NCBI
|
|
20
|
Guinan P, Sobin LH, Algaba F, et al: TNM
staging of renal cell carcinoma: Workgroup No. 3 Union
International Contre le Cancer (UICC) and the American Joint
Committee on Cancer (AJCC). Cancer. 80:992–993. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
White NM, Bao TT, Grigull J, et al: miRNA
profiling for clear cell renal cell carcinoma: biomarker discovery
and identification of potential controls and consequences of miRNA
dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Brannon AR, Reddy AR, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: with
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koscianska E, Baev V, Skreka K, et al:
Prediction and preliminary validation of oncogene regulation by
miRNAs. BMC Mol Biol. 8:792007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Volinia S, Galasso M, Costinean S, et al:
Reprogramming of miRNA networks in cancer and leukemia. Genome Res.
20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kefas B, Godlewski J, Comeau L, et al:
microRNA-7 inhibits the epidermal growth factor receptor and the
Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mikhaylova O, Stratton Y, Hall D, et al:
VHL-regulated MiR-204 suppresses tumor growth through inhibition of
LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell.
21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dey N, Das F, Ghosh-Choudhury N, et al:
microRNA-21 governs TORC1 activation in renal cancer cell
proliferation and invasion. PLoS One. 7:e373662012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
White NM, Bao TT, Grigull J, et al: miRNA
profiling for clear cell renal cell carcinoma: Biomarker discovery
and identification of potential controls and consequences of miRNA
dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu H, Brannon AR, Reddy AR, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Krzeslak A: Akt kinase: a key regulator of
metabolism and progression of tumors. Postepy Hig Med Dosw
(Online). 64:490–503. 2010.In Polish.
|
|
33
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Macher-Goeppinger S, Bermejo JL, Wagener
N, et al: Expression and prognostic relevance of the death receptor
CD95 (Fas/APO1) in renal cell carcinomas. Cancer Lett. 301:203–211.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sejima T, Morizane S, Hinata N, et al: Fas
expression in renal cell carcinoma accurately predicts patient
survival after radical nephrectomy. Urol Int. 88:263–270. 2012.
View Article : Google Scholar : PubMed/NCBI
|