Introduction
Oral cancer is one of most common types of
malignancy worldwide, and ~42,440 new cases were diagnosed in the
United States in 2014 (1). Oral
squamous cell carcinoma (OSCC) accounts for ~90% of oral cancer
cases. Despite significant advances in therapeutic strategies in
the last few years, the overall 5-year survival rates of patients
with OSCC is ~60% at the age of 62 years (2). Funk et al (3) reported that the 5-year survival rate
is almost 80% in the early stages, however, this rate decreases to
between 20 and 40% in advanced-stage OSCC. This indicates that
early detection is essential for improving the survival rates and
prognosis in OSCC. Therefore, an improved understanding of the
molecular biology and pathogenesis of OSCC is essential for the
development of novel biomarkers and therapies.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs of ~21–23 nucleotides in length, which regulate target genes
through the 3′-untranslated regions (3′-UTRs) to induce mRNA
degradation and inhibit mRNA translation (4). Accumulating evidence has reported
that miRNAs are important in tumorigenesis due to their aberrant
expression (5), and can function
either as oncogenes or tumor suppressors in various types of
cancer, depending on their downstream target genes (5,6). In
addition, several miRNAs have been identified to be dysregulated,
regulating the initiation and progression of OSCC (7). miR-216a has been observed to be
downregulated and function as a tumor suppressor in several types
of cancer (8–10). However, the role of miR-216a in the
regulation of key genes and signaling pathways associated with OSCC
remains to be elucidated.
In the present study, the expression levels of
miR-216a in OSCC specimens and cell lines, and the effects on the
growth and metastasis of OSCC cells were investigated. In addition,
the present study aimed to identify whether eukaryotic translation
initiation factor 4B (EIF4B) is a potential target of miR-216a and
identify correlations with miR-216a in the OSCC tissues.
The current study may not only provide a novel
understanding of the regulatory mechanism of miR-216a but
additionally offer a novel target for the treatment of OSCC.
Materials and methods
OSCC specimens, cell lines and
transfection
A total of 23 paired OSCC tissue samples and
adjacent non-tumor tissue samples were collected from patients
undergoing resection of OSCC at the No.1 People’s Hospital (Jining,
China) between June 2011 and December 2013. Of the patients, 15
were men and 8 were women, with a median age of 51 years (range,
22–83). No patients received chemotherapy or radiotherapy prior to
surgery. The tissue samples were immediately snap frozen in liquid
nitrogen and stored at −80°C. Histopathology was confirmed by two
independent pathologists. The study was approved by the Hospital
Ethical Committee and informed consent was obtained from each
patient prior to commencement of the investigation.
The SCC-4 and CAL 27 cell lines were obtained from
American Type Culture Collection (Manassas, VA, USA) and cultured
in RPMI-1640 medium (GE Healthcare, Logan UT, USA). The HEK293
cells were obtained from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China) and were maintained in
Dulbecco’s modified Eagle’s medium. All the media were supplemented
with 10% fetal bovine serum (FBS) at 37°C under 5% CO2.
For transfection, cells were transfected using Lipofectamine 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions. Following 4 h incubation at 37°C in
FBS-free medium, cells were incubated for 24 h at 37°C in 10% FBS
medium.
Plasmid construction
The pre-miR-216a plasmid was constructed using the
following synthetic oligonucleotides that were obtained from
SangonBiotech, Shanghai, China: Sense, 5′-AAT TCG ATG GCT GTG AGT
TGG CTT AAT CTC AGC TG G CAA CTG TGA GAT GTT CAT ACA ATC CCT CAC
AGT GGT CTC TGG GAT TAT GCT AAA CAG AGC AAT TTC CTA GCC C TC ACG
AA-3′; anti-sense, 5′-AGC TTT CGT GAG GGC TAG G AA ATT GCT CTG TTT
AGC ATA ATC CCA GAG ACCACT GT G AGG GAT TGT ATG AAC ATC TCA CAG TTG
CCA GCT GAG ACC AAG CCA ACT CAC AGC CAT CG-3′. These
oligonucleotides were then cloned into the pcDNA6.2-GW vector at
the EcoRIand HindIII sites (Promega Corporation,
Madison, WI, USA) using TargetScan, version 6.2 (www.targetscan.org/) to predict the target of
miR-216a. Furthermore, the complimentary sites in the 3′-UTRs of
the wild-type EIF4B (EIF4B-WT) and mutant EIF4B (EIF4B-MT) of
miR-216a were synthesized (SangonBiotech) and cloned into the
pmirGLO dual-luciferase reporter vector at the Sac1 and
Xho 1 sites (Promega Corporation). The EIF4B expression
plasmid was obtained from GeneCopoeia, Inc. (Rockville, MD,
USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was isolated from the OSCC tissues and
cells using TRIzol reagent (Invitrogen Life Technologies). All the
reagents used for RT-qPCR were obtained from Tiangen Biotech Co.,
Ltd. (Beijing, China). For mRNA analyses, first-strand cDNA was
synthesized using a FastQuant RT kit (Tiangen Biotech Co., Ltd.).
The mature miRNA was reverse transcribed using specific primers for
miR-216a. Subsequently, qPCR was performed using SuperReal PreMix
Plus (SYBR Green) on an ABI7500 PCR machine (Applied Biosystems
Life Technologies, Foster City, CA, USA). The relative expression
levels of mRNA and miRNA were calculated based on the
−ΔΔCt method. β-actin and U6 were used as controls for
mRNA l and miRNA, respectively. The following primers that were
synthesized by SangonBiotech were used in the qPCR analysis: RT
5′-GTCGTATCCAGTGCGTGTCGT GGAGTCGGCAATTGCACTGGATACGACTCACAGT-3′ for
miR-216a; miR216a, forward 5′-ATCCAGTGCGTGTCGTG-3′ and reverse
5′-TGCTTAATCTCAGCTGGCA-3′; and EIF4B, forward
5′-AGCGTCAGCTGGATGAGCCAA-3′ and reverse
5′-TGTCCTCGACCGTTCCCGTT-3′.
Cell proliferation assay
The transfected cells were seeded into 96-well
plates at a density of 3,000 cells/well. At 24, 48 and 72 h
following transfection, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl
tetrazolium bromide (MTT; Sigma-Aldrich, St. Louis, MO, USA) assay
was used to determine the cell proliferation. The MTT solution (5
mg/ml; 200 μl) was added to the cells and incubated for 4 h
at 37°C. Following the removal of the culture medium, the remaining
crystals were dissolved in 150 μl dimethyl sulfoxide, and
the absorbance at 492 nm was measured (FluoStar Optima; BD
Biosciences, Franklin Lakes, NJ, USA).
Colony formation assay
The cells were cultured in 12-well plates at a
density of 2,000 cells/well between 24 h and 15 days. The clones
were washed with phosphate-buffered saline (PBS) and stained using
0.1% crystal violet (Beyotime Institute of Biotechnology, Haimen,
China) for 20 min. Images of the colonies were captured and the
numbers of cells were counted under a microscope (CK2; Olympus,
Tokyo, Japan).
Migration and invasion assays
The present study then performed cell migration and
invasion assays using uncoated or coated Matrigel, respectively.
Briefly, the cells (5×104) were added to the upper
chamber of Transwell inserts in serum-free medium containing 0.1%
FBS, and a medium supplemented with 10% FBS was added to the lower
chamber. The cells were cultured for 24 h at 37°C in 5%
CO2. The Matrigel and non-invading cells were then
gently removed, and the migrated and invaded cells in the lower
membrane were fixed with 4% paraformaldehyde (Nanjing Chemical
Material Corporation, Nanjing, China), stained using 0.1% crystal
violet and counted under a light microscope (CK2; Olympus).
Dual luciferase reporter assays
The HEK293 cells were seeded in 96-well plates
(3,000 cells/well) and co-transfected with 0.1 mg pmirGLO reporter
plasmid (EIF4B-WT/EIF4B-MT) and 0.1 mg pre-miR-216a plasmid or
control miRNA. Cells were incubated at 37°C for 24 h and then lysed
with a mixture of Dual-Glo Luciferase reagent and buffer (Promega
Corporation). The luciferase activities were measured using the
FluoStar Optima.
Western blot analysis
The total protein was extracted from the transfected
cells using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China), according to the
manufacturer’s instructions. The total proteins (50 μg) were
separated using 10% SDS-PAGE and transferred onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA), which
were then blocked in 5% non-fat milk in Tris-buffered saline with
0.05% Tween-20 for 2 h at 37°C, followed by incubation with rabbit
monoclonal anti-EIF4B antibody (1:1,000; 17917-1-AP; Protein Tech
Group, Inc., Wuhan, China). β-actin was used as a loading
control.
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analyses. All data are presented as the
mean ± standard error of the mean of at least three independent
samples. The differences between the two groups were analyzed using
Student’s t-test. The linear correlation coefficient was calculated
to estimate the correlation between miR-216a and EIF4B in the OSCC
specimens. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-216a is downregulated in OSCC tissues
and cell lines
The present study first determined the expression of
miR-216a using RT-qPCR in 23 OSCC tissues and in the SCC-4 and CAL
27 OSCC cell lines. As shown in Fig.
1A, miR-216a was significantly downregulated in the OSCC
tissues compared with the adjacent normal tissues (P= 0.034).
Similarly, miR-216a was also decreased in the two OSCC cells,
compared with the normal oral epithelial cells (P=0.002 and
P<0.001, respectively; Fig.
1B).
miR-216a suppresses the growth of OSCC
cells
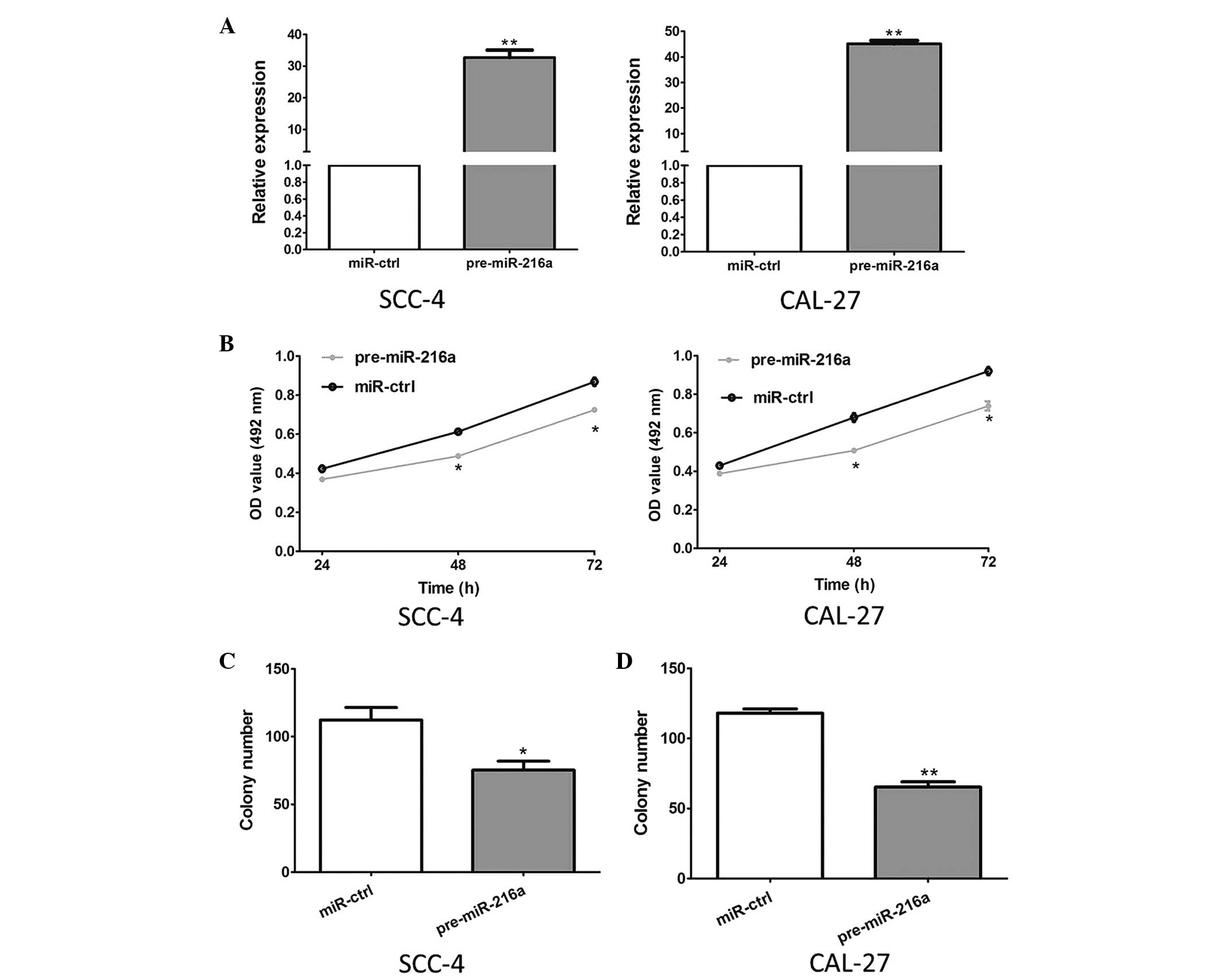
The effect of miR-216a on the growth of OSCC cells
was evaluated using MTT and colony formation assays. The results of
the RT-qPCR analysis revealed that the expression levels of miR-26a
in the SCC-4 and CAL 27 cells transfected with pre-miR-216a were
significantly increased, compared with the control cells
(P<0.001; Fig. 2A).
Furthermore, as shown in Fig. 2B,
the overexpression of pre-miR-216a markedly inhibited the
proliferation of the SCC-4 and CAL 27 cells (P<0.05). Consistent
with these results, the overexpression of pre-miR-216a
significantly suppressed the colony formation of the SCC-4 and CAL
27 cell lines (P=0.031 and P<0.001, respectively; Fig. 2C).
miR-216a suppresses the metastasis of
OSCC cells
The present study subsequently investigated the
effect of miR-216a on the metastasis of OSCC cells, including
migration and invasion. As shown in Fig. 3A, the overexpression of
pre-miR-216a significantly suppressed tumor cell migration in the
CAL 27 cells (P<0.001). In addition, the overexpression of
pre-miR-216a markedly inhibited the invasive capacity of the CAL 27
cells (P<0.001; Fig. 3B).
EIF4B is a direct target of miR-216a
The present study predicted that EIF4B may be a
target of miR-216a using TargetScan 6.2. To confirm whether
miR-216a directly targets EIF4B, as shown in Fig. 4A, EIF4B-WT and EIF4B-MT plasmids
were constructed and cloned into the region downstream of the
pmiRGLO dual-luciferase reporter vector. The subsequent luciferase
activity assay demonstrated that miR-216a significantly decreased
luciferase activity of EIF4B-WT, but not EIF4B-MT in the HEK293
cells (P<0.001; Fig. 4B). In
addition, the overexpression of pre-miR-216a in the CAL 27 cells
significantly suppressed the mRNA and protein expression levels of
EIF4B (P<0.05; Fig. 4C and
D,).
miR-216a inhibits the progression of OSCC
cells by targeting EIF4B
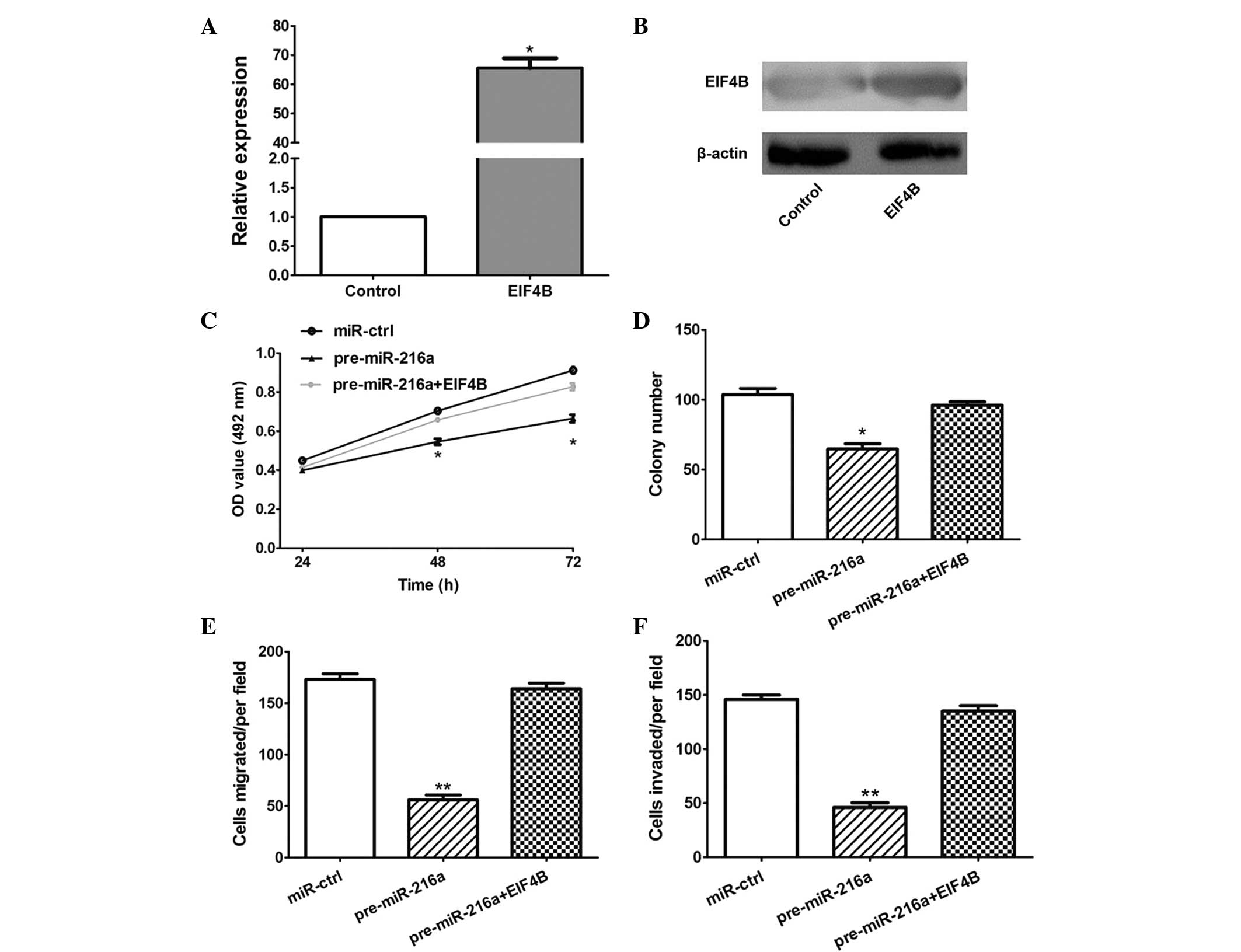
In order to establish whether the overexpression of
EIF4B attenuated the antitumor effects of miR-216a, an EIF4B
overexpression plasmid was constructed, which significantly
increased the expression of EIF4B at the mRNA and protein levels in
the CAL 27 cells (P<0.05; Fig. 5A
and B). Furthermore, the MTT, colony formation, migration and
invasion assays demonstrated that the overexpression of EIF4B
significantly attenuated the antitumor effects of miR-216a
(P<0.05; Fig. 5C–F).
Expression of miR-216a is inversely
correlated with that of EIF4B in OSCC tissues
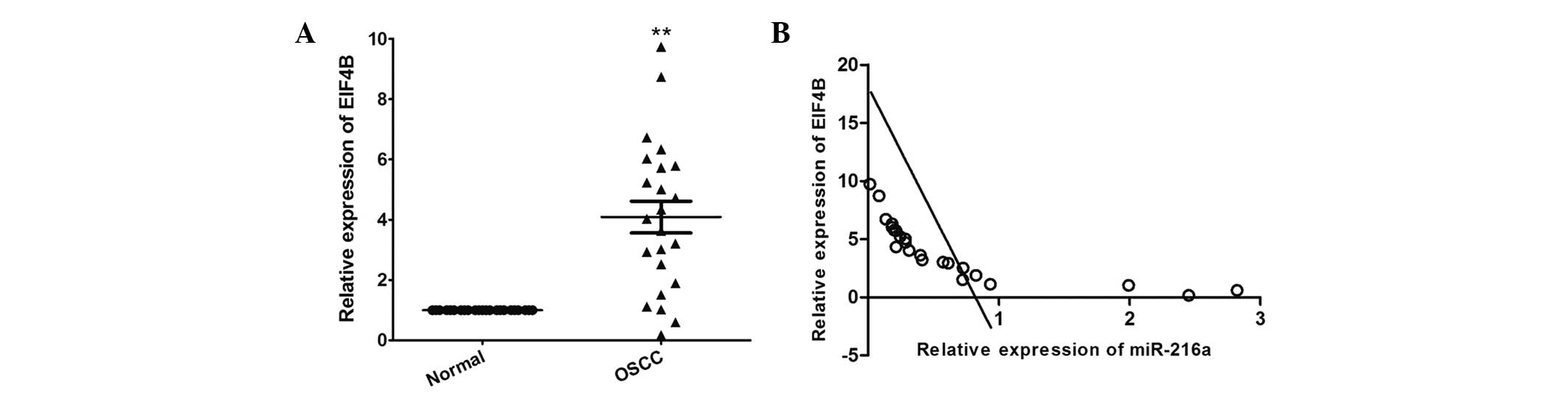
RT-qPCR was performed to determine the expression
levels of EIF4B in OSCC and in matched adjacent normal tissues. As
shown in Fig. 6A, the mRNA
expression level of EIF4B was significantly increased in The OSCC
tissues, compared with the adjacent normal tissues (P<0.001). In
addition, the mRNA expression level of EIF4B was inversely
correlated with that of miR-216a in the OSCC tissues (r=−0.761;
P<0.001; Fig. 6B).
Discussion
The importance of miRNAs in tumor development have
been identified, and miRNAs dysregulation can drive tumorigenesis
as tumor suppressors or oncogenes in several types of tumor
(11–14). The results of the present study
provide important evidence, which supports miR-216a acting as a
tumor suppressor in OSCC. In addition, the present study further
identified EIF4B as a direct target of miR-216a, and the
overexpression of EIF4B was observed to significantly attenuate the
effects of miR-216a on suppressing the progression of OSCC.
Accumulating evidence has demonstrated that the
dysfunction of miR-126a is involved in human malignancies and
carcinogenesis, however, the detail mechanisms of miR-216a remain
to be fully elucidated. In human hepatocellular carcinoma (HCC),
miR-216a is significantly increased and contributes to early
hepatocarcinogenesis through suppressing the gene expression of
tumor suppressor in lung cancer-1 (15). Similarly, Xia et al also
indicated that the expression of miR-216a was significantly
upregulated in HCC tissue samples and cell lines, and was
associated with early tumor recurrence and poor rates of
disease-free survival. The overexpression of miR-216a-induced
epithelial-mesenchymal transition activated the
phosphatidylinositol 3-kinase/Akt and transforming growth factor-β
pathways and increased the stem-like cell population and migration
and metastatic abilities of epithelial HCC cells by targeting
phosphatase and tensin homolog and mothers against decapentaplegic
homolog 7 (9). However, Wang et
al demonstrated that the expression of miR-216a is
downregulated in non-small cell lung cancer specimens, and the
overexpression of miR-216a suppresses the growth and metastasis,
and enhances cisplatin-induced cell growth inhibition and apoptosis
in NSCLC cells (8). The results of
the present study were consistent with those of Wang et al
(8), and demonstrated the tumor
suppressive role of miR-216a in the proliferation, colony
formation, migration and invasion of OSCC cells. Therefore, it was
suggested that miR-216a may have a unique pattern of expression in
various types of cancer and function as an oncogene or suppressor
gene.
EIF4B is comprised of eIF4A, eIF4B, eIF4E and eIF4G;
and potentiates ribosome recruitment to the mRNA in the positioning
of the ribosome over the start codon (16,17).
Deregulated translational control of the EIF4B complex members is
considered to be important in oncogenic transformation (18–20).
Horvilleur et al (18)
demonstrated that the expression of EIF4B was increased in diffuse
large B-cell lymphoma (BCL) patient samples. Reducing the
expression of EIF4B alone was sufficient to decrease the synthesis
of proteins associated with enhanced tumor cell survival, including
excision repair cross-complementation group 5, death-associated
protein 6 and BCL2; and the translational dysregulation of EIF4B
may have resulted from aberrant signaling via the mammalian target
of rapamycin pathway. In addition, Ren et al (19) demonstrated that the level of
proviral integrations of moloney virus 2 (Pim-2) determined the
phosphorylation level of EIF4B and the apoptotic rate of prostatic
cells, suggesting that the overexpression of Pim-2 may inhibit the
apoptosis of prostatic cells by phosphorylating EIF4B. In addition
to the antiapoptotic effect of EIF4B on cancer cells,
overexpression of EIF4B may also contribute to NSCLC cell growth
and metastasis (8). In the present
study, the 3′-UTR sequences of EIF4B were directly targeted by
miR-216a, and EIF4B significantly attenuated the tumor suppressive
effects of miR-216a on the OSCC cells. In addition, the expression
of EIF4B was inversely correlated with miR-216a in OSCC
tissues.
In conclusion, the present study demonstrated for
the first time, to the best of our knowledge, that miR-216a, as a
tumor suppressor, negatively suppressed the growth and metastasis
of OSCC by targeting the 3′-UTR of EIF4B, suggesting that miR-216a
may have important therapeutic potential for patients with
OSCC.
Acknowledgments
The current study was supported by the Science
Foundation of Shandong Province, China (grant no. ZR2014HL053).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, et al:
Cancer treatment and survivorship statistics, 2014. CA Cancer J
Clin. 64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Funk GF, Karnell LH, Robinson RA, Zhen WK,
Trask DK and Hoffman HT: Presentation, treatment, and outcome of
oral cavity cancer: a National Cancer Data Base report. Head Neck.
24:165–180. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alsaleh G and Gottenberg JE:
Characterization of microRNAs and their targets. Methods Mol Biol.
1142:55–63. 2014.PubMed/NCBI
|
|
5
|
Di Leva G and Croce CM: miRNA profiling of
cancer. Curr Opin Genet Dev. 23:3–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bajan S and Hutvagner G: Regulation of
miRNA Processing and miRNA Mediated Gene Repression in Cancer.
Microrna. 3:10–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu BH, Xiong XP, Jia J and Zhang WF:
MicroRNAs: new actors in the oral cancer scene. Oral Oncol.
47:314–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang RT, Xu M, Song ZG, Xu CX and Jin H:
Decreased Expression of miR-216a Contributes to Non-small Cell Lung
Cancer Progression. Clin Cancer Res. 20:4705–4716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou B, Jian Z, Chen S, Ou Y, Li S and Ou
J: Expression of miR-216a in pancreatic cancer and its clinical
significance. Nan Fang Yi Ke Da Xue Xue Bao. 32:1628–1631. 2012.In
Chinese. PubMed/NCBI
|
|
11
|
Jia AY, Castillo-Martin M, Bonal DM,
Sánchez-Carbayo M, Silva JM and Cordon-Cardo C: MicroRNA-126
inhibits invasion in bladder cancer via regulation of ADAM9. Br J
Cancer. 110:2945–2954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Kim K, Li X, et al: MicroRNA-26b
Represses Colon Cancer Cell Proliferation by Inhibiting Lymphoid
Enhancer Factor 1 Expression. Mol Cancer Ther. 13:1942–1951. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song Q, Xu Y, Yang C, et al: miR-483-5p
promotes invasion and metastasis of lung adenocarcinoma by
targeting RhoGDI1 and ALCAM. Cancer Res. 74:3031–3042. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pan W, Wang H, Jianwei R and Ye Z:
MicroRNA-27a promotes proliferation, migration and invasion by
targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem.
33:402–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen PJ, Yeh SH, Liu WH, et al: Androgen
pathway stimulates microRNA-216a transcription to suppress the
tumor suppressor in lung cancer-1 gene in early
hepatocarcinogenesis. Hepatology. 56:632–643. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benne R and Hershey JW: The mechanism of
action of protein synthesis initiation factors from rabbit
reticulocytes. J Biol Chem. 253:3078–3087. 1978.PubMed/NCBI
|
|
17
|
Schreier MH, Erni B and Staehelin T:
Initiation of mammalian protein synthesis. I Purification and
characterization of seven initiation factors. J Mol Biol.
116:727–753. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Horvilleur E, Sbarrato T, Hill K, et al: A
role for eukaryotic initiation factor 4B overexpression in the
pathogenesis of diffuse large B-cell lymphoma. Leukemia.
28:1092–1102. 2014. View Article : Google Scholar :
|
|
19
|
Ren K, Gou X, Xiao M, et al: The
over-expression of Pim-2 promote the tumorigenesis of prostatic
carcinoma through phosphorylating eIF4B. Prostate. 73:1462–1469.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Wang J, Chen K, et al: eIF4B
phosphorylation by pim kinases plays a critical role in cellular
transformation by Abl oncogenes. Cancer Res. 73:4898–4908. 2013.
View Article : Google Scholar : PubMed/NCBI
|