Introduction
Osteosarcoma is the most common malignant tumor of
bone, which mainly affects children and adolescents and accounts
for ~60% of malignant bone tumors diagnosed in the first two
decades of life (1,2). Current treatment of osteosarcoma
consists of multiagent chemotherapy and surgical resection.
However, further therapy with additional chemotherapy is palliative
and too often toxic (3,4). Thus, safe and more effective
anti-cancer treatments are required for patients with
osteosarcoma.
Paxilitaxel, which was isolated from the bark of the
yew tree, inhibits the division of actively growing tumor cells and
has become increasingly important in the treatment of a number of
major cancers, including ovarian (5), pancreatic (6) and breast (7) cancer. However, the impact of
paxilitaxel on osteoblasts has remained to be elucidated.
Autophagy is an evolutionarily conserved lysosomal
degradation process by which cells recycle macromolecules and
organelles (8). Studies have
demonstrated that anti-cancer therapies (such as chemotherapy)
induce autophagy in cancer cells, while the association between
autophagy and therapeutic effects has remained controversial
(9,10). However, an increasing number of
studies have reported that autophagy is elevated in certain tumors
and contributes to poor outcome by promoting resistance to
chemotherapy (11). It has
remained elusive whether paxilitaxel is able to induce autophagy in
osteosarcoma cells and, if so, what the significance of this
response is.
Hypoxia-inducible factor 1 (HIF-1) is the most
critical nuclear transcription factor under hypoxic conditions in
normal cells and tumor cells. Studies have reported that HIF-1α
protein expression is upregulated in certain tumor types and
contributes to poor disease outcome by promoting tumor progression,
metastasis and resistance to chemotherapy (12,13).
Previous studies also showed that HIF and autophagy are closely
linked (14,15). Whether paxilitaxel is able to
affect the expression of HIF-1 and autophagy, as well as the
complex association between them, was the subject of the present
study.
The present study investigated whether paxilitaxel
induces apoptosis in human osteosarcoma cells. Furthermore, the
ability of paxilitaxel to induce autophagy and HIF-1α expression in
osteosarcoma and its possible association with resistance were
investigated.
Materials and methods
Cell cultures
The human osteosarcoma cell line MG-63 was obtained
from the Cell Collection of the Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) (Gibco-BRL, Invitrogen Life Technologies,
Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco-BRL) at
37°C in a 5% CO2 incubator. The cells were routinely
sub-cultured every 2–3 days. All experiments were performed on
cells harvested at the mid-logarithmic growth phase. Briefly, the
cells were treated with autophagy inhibitors: 50 µmol/l
3-(5′-hydroxy-methyl-2′-furyl)-1-benzyl indazole (YC-1) or 5 mM
3-methyladenine (3-MA) (both Sigma-Aldrich, St. Louis, MO, USA), or
with an autophagy inducer: 2 µM rapamycin (RAPA;
Sigma-Aldrich) in DMEM with 10% FBS.
Reagents and antibodies
Paxilitaxel was purchased from Sigma-Aldrich. Mouse
monoclonal anti-B-cell lymphoma 2-associated X protein (Bax; cat.
no. sc-20067), mouse monoclonal anti-apoptosis-inducing factor
(AIF; cat. no. sc-13116), and mouse monoclonal anti-β-actin (cat.
no. sc-8432) antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Rabbit monoclonal
anti-caspase 3 (cat. no. 9664), rabbit polyclonal anti-caspase 9
(cat. no. 9502), rabbit monoclonal anti-Beclin1 (cat. no. 3495),
rabbit monoclonal anti-microtubule-associated protein light chain 3
(LC3; cat. no. 12741) and rabbit monoclonal anti-HIF-1α (cat. no.
14179) were purchased from Cell Signaling Technology (Danvers, MA,
USA).
Analysis of apoptosis
Cellular apoptosis was determined using the Annexin
V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit I
(Clontech Laboratories Inc., Takara Bio Inc., Mountain View, CA,
USA). Brieflly, MG-63 cells were cultured at 4×106
cells/ml and seeded in six-well plates. Cells were harvested by
trypsinization (Sigma-Aldrich), then washed twice with cold
phosphate-buffered saline (PBS) and centrifuged at 700 × g.
1×105–1×106 cells were re-suspended in 300
µl 1X binding buffer (Clontech Laboratories Inc.),
centrifuged again at 700 × g for 5 min and then the supernatant was
removed. Cells were re-suspended in 300 µl 1X binding buffer
and transferred to a sterile flow cytometry glass tube. 10
µl Annexin V-FITC was added and cells were incubated in the
dark for 30 min at room temperature. Cells were then incubated in
the dark with 5 µl propidium iodide (PI) and analyzed by
flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ,
USA).
Hoechst 33342 staining
The cells were washed three times with PBS and fixed
with 4% paraformaldehyde (Sigma-Aldrich) for 60 min at 4°C. After
washing with PBS for three times, the cells were incubated with
0.2% Triton X-100 (Sigma-Aldrich) for 15 min. Cells were then
blocked with 5% bovine serum albumin (BSA; Sigma-Aldrich) for 60
min at room temperature and Hoechst 33342 (Sigma-Aldrich) was added
to the cells for 20 min. After washing three times with PBS, cells
were visualized under a fluorescence microscope (SZ51; Olympus
Corporation, Tokyo, Japan).
Immunofluorescence staining
MG-63 cells were seeded in six-well plates for 24 h.
Cells were then washed once with ice-cold PBS and fixed with 4%
paraformaldehyde for 30 min at 4°C. After washing with PBS three
times, the cells were incubated with 1% Triton X-100 for 10 min.
The cells were blocked at non-specific antibody binding sites by
incubating with 10% goat serum in PBS containing 0.3% Triton X-100
and 0.5% BSA for 30 min at room temperature, followed by incubation
with a mouse antibody against Beclin1 (1:400 in PBS; Cell Signaling
Technology) or LC3 (1:200 in PBS; Cell Signaling Technology)
overnight. The samples were then incubated with either
tetramethylrhodamine (TRITC)- or FITC-conjugated goat anti-rabbit
immunoglobulin (Ig)G secondary antibodies (1:100 in PBS) for 0.5 h
at room temperature. Cells initially incubated with anti-Beclin1
were incubated with the TRITC-conjugated secondary antibody,
whereas those initially incubated with anti-LC3 were incubated with
the FITC-conjugated secondary antibody. Hoechst 33342 was added to
the cells for 15 min. After washing three times with PBS, cells
were visualized under a fluorescence microscope (SZ51; Olympus
Corporation).
Assessment of the mitochondrial membrane
potential (Δψm)
The Δψm was analyzed using the
tetraethylbenzimidazolylcar-bocyanine iodide (JC-1) assay
(Beyotime, Nantong, China). JC-1 is a cationic dye that indicates
the Δψm by reversibly shifting its fluorescence emission between
green (JC-1 monomers indicating loss of Δψm) and red (JC-1
aggregates indicating intact Δψm). In brief, a staining mixture
(300 nM JC-1) was prepared according to the manufacturer's
instructions of the JC-1 kit. Cells were incubated in the staining
mixture for 30 min at 37°C. Thereafter, cells were washed twice
with medium and re-suspended in fresh medium. Δψm was monitored
using a fluorescence microscope.
Isolation of proteins from mitochondria
and cytosol and western blot analysis
The preparation of proteins from the mitochondrial
and cytosolic fractions and western blot were performed as
described previously (16). The
cells were washed twice in ice-cold PBS and re-suspended in five
volumes of ice-cold extraction buffer [20 mM western blotting
analysis (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-KOH,
1.5 mM MgCl2, 1 mM EDTA, 1 mM ethylene glycol
tetraacetic acid, 1 mM dithiothreitol and 0.1 mM
phenylmethanesulfonyl fluoride, pH 7.5; Sigma-Aldrich]. The
re-suspended cells were homogenized with ten strokes of a Teflon
homogenizer (2–16P; Sigma-Aldrich). The homogenates were
centrifuged twice at 750 ×g for 10 min at 4°C. The supernatants
were centrifuged at 10,000 ×g for 15 min at 4°C to obtain the
mitochondrial pellets. Cytosolic fractions were obtained after
further centrifugation at 100,000 ×g for 1 h at 4°C. The protein
concentrations of the resulting supernatants and mitochondrial
fractions were measured. The samples (10 µg protein) were
separated by 10% SDS-PAGE (Invitrogen Life Technologies). The
proteins separated by SDS-PAGE were electrotransferred onto a
Hybond-polyvinylidenefluoride (PVDF) membrane (Invitrogen Life
Technologies). The individual SDS gels were distinguished by
placing the protein molecular weight marker (Invitrogen Life
Technologies) in different but consistent positions. The PVDF
membrane was then soaked in a blocking solution [5% nonfat milk in
TBST buffer (20 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 0.1% Tween 20;
Sigma-Aldrich)] for 2 h at room temperature. The soaked PVDF
membrane was then incubated in TBST containing primary antibodies
overnight at 4°C. The following primary antibodies were used:
Anti-Bax, anti-AIF, and anti-β-actin (all Santa Cruz
Biotechnology); Anti-caspase 3, anti-caspase 9, anti-LC3,
anti-Beclin1 and anti-HIF-1α (1:400 dilution; all Cell Signaling
Technology). Following incubation, the blot was washed with TBST
buffer three times for 5 min each and incubated at room temperature
for 2 h in TBST containing horseradish peroxidase (HRP)-conjugated
goat anti-mouse and goat anti-rabbit IgG (Santa Cruz Biotechnology,
Inc.). The membrane was washed with TBST buffer three times for 10
min each. The membranes were incubated in enhanced
chemiluminescence reagent (Pierce Biotechnology, Thermo Fisher
Scientific, Waltham, MA, USA) for HRP (30 sec) and exposure to
autoradiography film for visualization of the bands using a
LAS-1000 system (Fujifilm, Tokyo, Japan). The relative amounts of
various proteins were analyzed. The results were quantified by
Quantity One version 5.2.1 software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Statistical analysis
Values are expressed as the mean ± standard error of
the mean (n=10). Statistical analysis was performed using SPSS 18
(International Business Machines, Armonk, NY, USA). Data were
analyzed using the one-way analysis of variance first. Individual
comparisons were made using Tukey's multiple comparisons test.
P<0.05 was considered to indicate a statistically significant
difference between values.
Results
Paxilitaxel induces apoptosis of MG-63
cells via a mitochondria-mediated pathway
First, the effects of paxilitaxel on human
osteosarcoma cancer cells were tested and the mechanisms by which
paxilitaxel induces MG-63 apoptosis at different concentrations
were investigated. Cells were treated with 0, 0.4 and 1
µg/ml paxilitaxel for 24 h and the apoptotic rate was
determined by Annexin V/PI double staining followed by flow
cytometric analysis. The results showed a substantial increase in
the apoptotic population among cells treated with paxilitaxel (0.4
or 1 µg/ml) at 24 h (Fig.
1A). Fluorescence microscopy (Hoechst 33342 staining)
demonstrated that the paxilitaxel-treated cells showed obvious
nuclear damage in the form of chromatin condensation, which
distinguished them from the cells in the control group (paxilitaxel
0 µg/ml) (Fig. 1B).
Following treatment with paxilitaxel, the Δψm in the cells was
examined. Compared with control group, MG-63 cells in paxilitaxel
groups exhibited green JC-1 fluorescence, indicating a reduction of
the Δψm (Fig. 1C). As shown in
Fig. 1D and E, the expression of
Bax in the mitochondria was significantly increased in the
paxilitaxel-treated groups compared to that in the control group
(P<0.01), suggesting that the translocation of Bax into the
mitochondria was involved in the induction of cell death by
paxilitaxel. Mitochondria-mediated apoptosis comprises
caspase-dependent and caspase-independent processes, where AIF is
involved in the caspase-independent response. As shown in Fig. 1E, the expression of AIF in the
cytosol was significantly increased in the paxilitaxel-treated
groups compared to that in the control group (P<0.01). This
result indicated that the release of AIF from the mitochondria to
the cytosol was involved in cell death. Similarly, caspase-3 and
caspase-9 were examined by western blot analysis. Levels of cleaved
caspase-3 and caspase-9 in the paxilitaxel-treated groups were
upregulated compared with those in the control group. These results
indicated that paxilitaxel induced apoptosis in MG-63 cells was via
a mitochondrial pathway involving the caspase-dependent pathway
(caspase-3 and -9) as well as the caspase-independent pathway
(AIF).
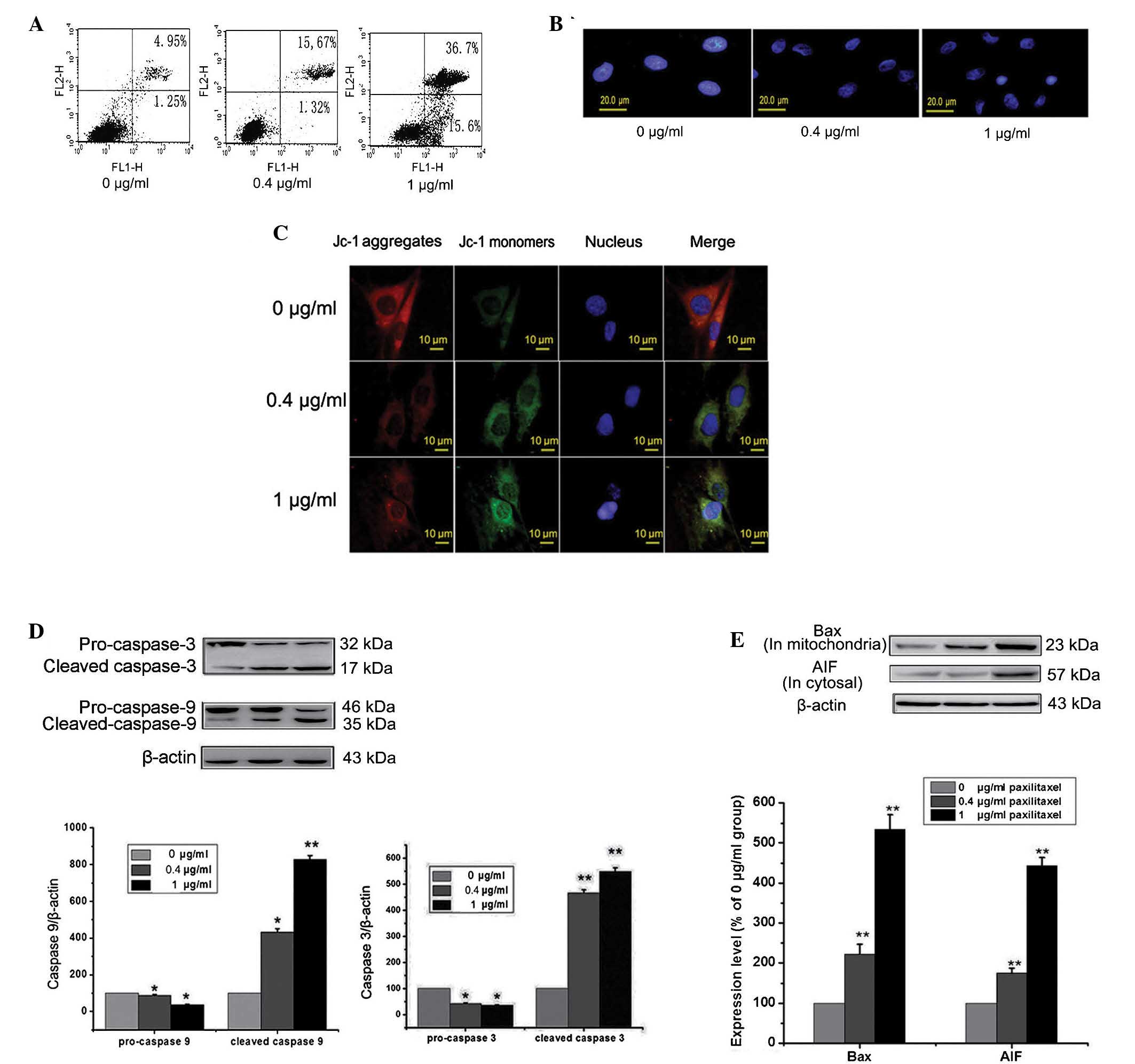 | Figure 1Effects of paxilitaxel on the
apoptosis of MG-63 cells. (A) Cells were treated with 0, 0.4 or 1
µg/ml paxilitaxel for 24 h and apoptosis was determined by
flow cytometry followed by Annexin V-propidium iodide double
staining. (B) Cells were treated as above, and the nuclei were
stained with Hoechst 33342 and observed under a fluorescence
microscope (scale bar, 20 µm). (C) Cells were treated as
above and subjected to JC-1 staining for assessment of the
mitochondrial membrane potential. JC-1 monomers (green
fluorescence) indicate mitochondrial membrane potential loss, while
JC-1 aggregates (red fluorescence) indicate a retained
mitochondrial membrane potential (scale bar, 10 µm). (D)
Cells were treated as described above, and levels of caspase 3,
caspase 9 were detected by western blot analysis. β-Actin was used
as loading control. (E) Cells were treated as described above, and
the expression of Bax (in the mitochondria) and AIF (in the
cytosol) were detected by western blot analysis. β-Actin was used
as loading control. Values are expressed as the mean ± standard
error (n=10). (**P<0.01 vs. control;
*P<0.05 vs. control). JC-1,
tetraethylbenzimidazolylcarbocyanine iodide; Bax, B-cell lymphoma
2-associated X protein; AIF, apoptosis-inducing factor. |
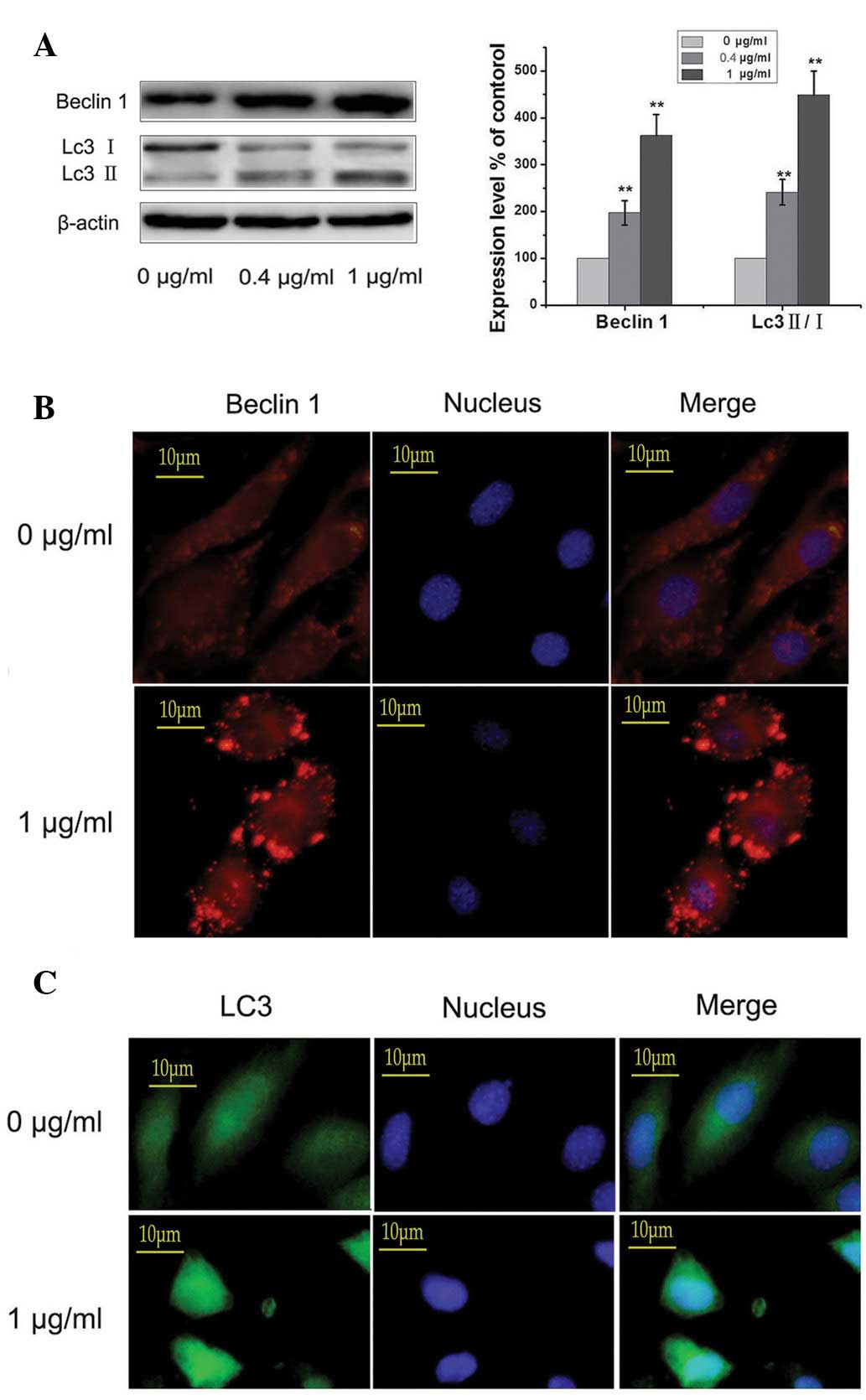
Paxilitaxel induces autophagy of MG-63
cells
Cells were treated with 0, 0.4 and 1 µg/ml
paxilitaxel for 24 h and the occurrence of autophagy was
determined. As shown in Fig. 2A,
the expression levels of Beclin1 and LC3 II/I were significantly
increased in the paxilitaxel-treated groups compared with those in
the control group (paxilitaxel 0 µg/ml; P<0.01),
suggesting that paxilitaxel, in addition to triggering apoptosis,
induced autophagy in MG-63 cells. Immunofluorescence microscopy
revealed that in the control group, low levels of autophagy were
sustained, while paxilitaxel markedly enhanced the levels of LC3
and Beclin1 (Fig. 2B and C). These
findings indicated that paxilitaxel did not block autophagic flux,
but induced the autophagic activity.
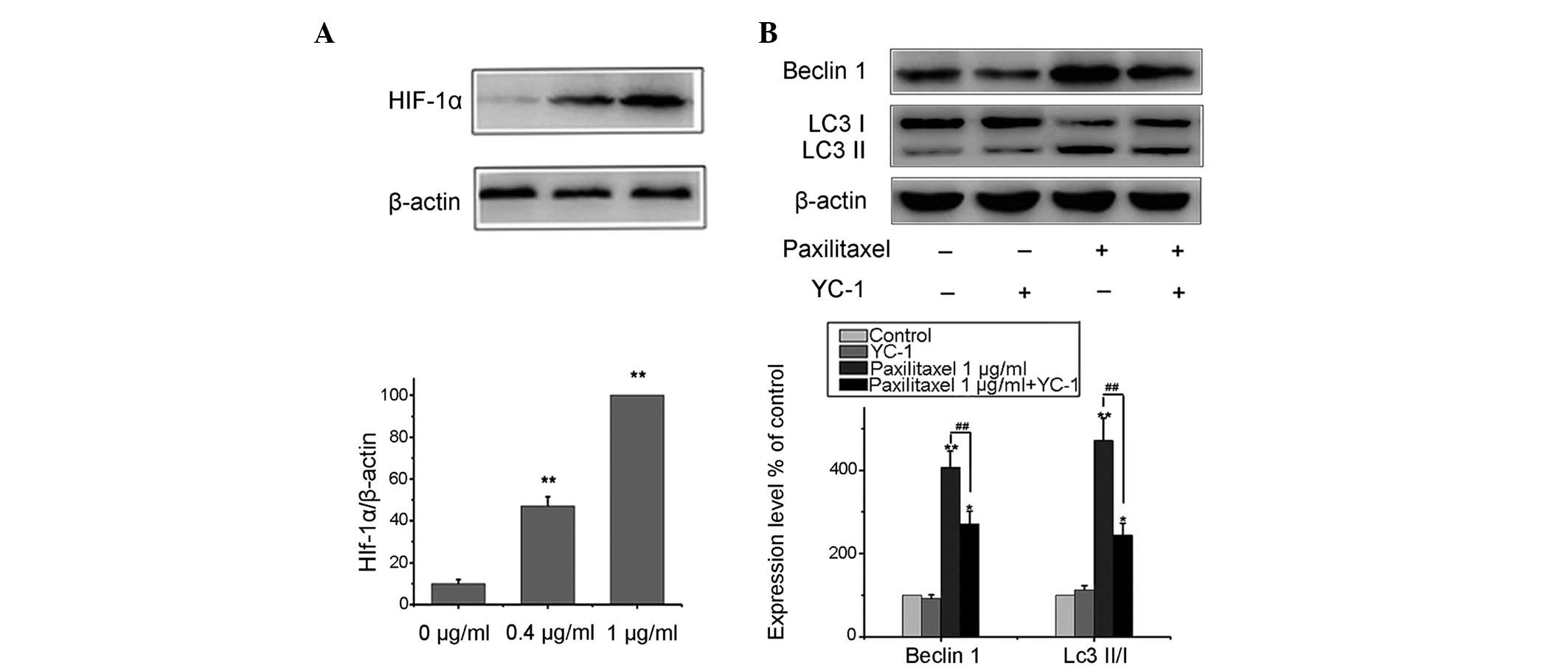
Paxilitaxel induces autophagy of MG-63
cells through the HIF-1α pathway
It is well known that HIF and autophagy are closely
linked (13,14). Therefore, the present study
investigated whether paxilitaxel treatment of MG-63 cells had any
effect on the expression of HIF-1α. MG-63 cells were treated with
0.4 or 1 µg/ml paxilitaxel for 24 h, and the expression
levels of HIF-1α were assessed by western blot analysis. As shown
in Fig. 3A, paxilitaxel enhanced
the levels of HIF-1α compared to those in the control group. These
results indicated that paxilitaxel treatment not only resulted in
apoptosis, but also in the induction of autophagy and the
expression of HIF-1α. Following treatment with 0.4 and 1
µg/ml paxilitaxel, autophagy-associated protein as well as
and HIF-1α protein levels were increased in a dose-dependent manner
(Figs. 2A and 3A). However, it appeared unlikely that
paxilitaxel induced autophagy by upregulating the expression of
HIF-1α alone. Therefore, MG-63 cells were exposed to 1 µg/ml
paxilitaxel in the presence of the HIF-1α inhibitor YC-1 [50
µmol/l in accordance with reference (17)] for 24 h and the expression of
Beclin1 and LC3 II/I was detected by western blot analysis. The
results indicated that paxilitaxel-mediated upregulation of
autophagy-associated protein expression was attenuated, but not
fully abolished by HIF-1α inhibitor YC-1 (Fig. 3B). Therefore, these results
indicated that HIF-1α generation by paxilitaxel has an important
role in autophagy induction; however, there are likely to be
additional signaling proteins via which autophagy is induced by
paxilitaxel.
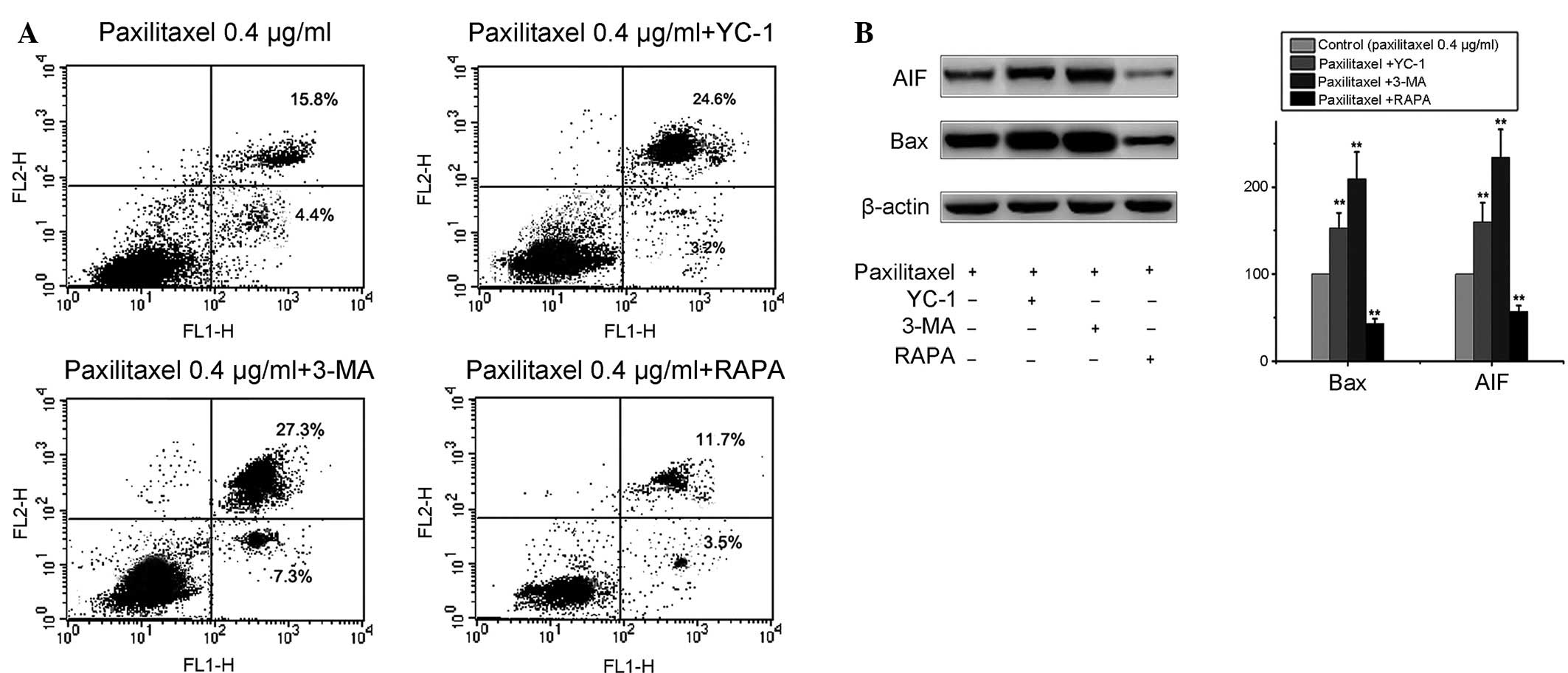
Autophagy induced by paxilitaxel protects
MG-63 cells from apoptosis
To investigate the effect of autophagy on the
apoptosis of MG-63 cells, 3-MA, a potent pharmacological inhibitor
of autophagy, was used to suppress the autophagy induced by
paxilitaxel. The results demonstrated that pre-treatment with 3-MA
[5 mM according to reference (18)] was able to block autophagy in MG-63
cells without significant cytotoxicity. 3-MA itself scarcely
induced cell apoptosis and cell death, but significantly increased
apoptosis at 24 h after paxilitaxel exposure (paxilitaxel group,
20.2%; paxilitaxel plus 3-MA, 34.6%). In addition, in the group of
combined treatment of 0.4 µg/ml paxilitaxel and 50
µmol/l YC-1 for 24 h, the apoptotic rate was higher than
that in the group treated with paxilitaxel only (paxilitaxel group,
20.2%; paxilitaxel plus YC-1, 27.8%) (Fig. 4A). These results suggested that
suppression of autophagy by 3-MA or YC-1 increased
paxilitaxel-induced injury in MG-63 cells. By contrast,
paxilitaxel-induced apoptosis decreased following co-treatment with
autophagy inducer RAPA [2 µM according to reference
(19)] compared to that of cells
treated with paxilitaxel alone (Fig.
4A). Treatment of MG-63 cells with RAPA or 3-MA alone did not
affect cell viability (data not shown). The results were further
confirmed by western blot analysis. The upregulation of the
expression of apoptotic proteins AIF and Bax was more marked when
cells were co-treated with 3-MA, while it was attenuated by
co-treatment with RAPA (Fig. 4B).
These results indicated that autophagy induced by paxilitaxel had a
protective effect on MG-63 cells, and blockage of autophagy
enhanced the anti-tumor effect of paxilitaxel.
Discussion
Paxilitaxel is an active component of the herbal
medicine Curcuma wenyujin with reported anti-tumor activity
and has been approved for the treatment of malignant effusion and
certain types of solid tumor in China (20,21).
However, to the best of our knowledge, the effects of paxilitaxel
on osteosarcoma have not been documented. In the present study,
paxilitaxel was shown to inhibit the proliferation and induce
apoptosis of human osteosarcoma cells. Mitochondria has an
important role in the regulation of cell death. Mitochondrial
dysfunction is considered an early event in apoptosis and this
process accompanied with a conspicuous reduction of the
mitochondrial membrane potential (22,23).
The present study found that paxilitaxel-treated cells exhibited
green JC-1 fluorescence, indicating a reduction of the Δψm.
Paxilitaxel treatment led to a marked upreguration of
cleaved-caspase 3, cleaved-caspase 9, Bax (in the mitochondria) and
AIF (in the cytosol), while significantly downregulating the levels
of pro-caspase 3 and pro-caspase 9. These results suggested that
paxilitaxel is capable of inducing mitochondria-mediated apoptosis
involving the caspase-dependent (via caspase-3 and -9) as well as
the caspase-independent pathway (via AIF).
Autophagy, an evolutionarily conserved
lysosome-dependent cellular catabolic degradation process, is
characterized by the formation of autophagosomes (24). Autophagy has a housekeeping role in
clearing damaged organelles, including mitochondria and
peroxisomes, as well as eliminating intracellular pathogens. Thus,
autophagy is generally regarded as a survival mechanism (25,26).
Autophagy and apoptosis are closely associated. Apoptosis is often
accompanied by the occurrence of autophagy (27,28).
It was found that paxilitaxel not only induced apoptosis but also
induced autophagy in MG-63 cells. The autophagy proteins Beclin1
and LC3 were upregulated by paxilitaxel. Moreover, in accordance
with the results of previous studies (29,30),
the present study reported that paxilitaxel activated autophagy by
modulation of HIF-1α signaling. Paxilitaxel induced autophagy as
well as HIF-1α expression in MG-63 cells. This autophagic induction
by paxili-taxel was partly mediated through the activation of
HIF-1α, as inhibition of HIF-1α expression by YC-1 reduced
autophagy.
Autophagy has an important role in mediating the
effects of drugs in cells. Autophagy and HIF-1α are often
associated with resitance to chemotherapy (31–33).
The results of the present study suggested that autophagy is a
pro-survival mechanism of MG-63 cells following paxilitaxel
treatment and facilitates the development of acquired paxilitaxel
resistance. Paxilitaxel-induced apoptosis of MG-63 cells was
markedly decreased by autophagy. In addition, co-treatment with
clinically applicable inhibitors of autophagy or HIF-1α may be one
of the important strategies for human osteosarcoma therapy.
Suppression of autophagy by 3-MA or YC-1 was able to increase
paxilitaxel-induced apoptosis in MG-63 cells. By contrast,
paxilitaxel-induced apoptosis was decreased following co-treatment
with the autophagy inducer RAPA. However, the molecular mechanisms
of HIF-1α-mediated activation of autophagy and paxilitaxel
resistance, as well as the association between autophagy, apoptosis
and other factors of paxilitaxel-resistance require further study.
At present, the precise underlying mechanism of autophagy mediated
by anti-apoptotic proteins remains elusive and is under
investigation in our laboratory.
Abbreviations:
|
HIF-1
|
hypoxia-inducible factor 1
|
|
3-MA
|
3-methyladenine
|
|
RAPA
|
rapamycin
|
|
LC3I/II
|
microtubule-associated protein light
chain 3-I/II
|
|
AIF
|
apoptosis-inducing factor
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
PI
|
propidium iodide
|
|
ΔΨm
|
mitochondrial membrane potential
|
|
YC-1
|
3-(5′-hydroxymethyl-2′furyl)-1-benzyl
indazole
|
References
|
1
|
Markiewicz K, Zeman K, Kozar A and
Gołebiowska-Wawrzyniak M: Evaluation of selected cytokines in
children and adolescents with osteosarcoma at diagnosis-preliminary
report. Med Wieku Rozwoj. 15:25–31. 2011.In Polish. PubMed/NCBI
|
|
2
|
Daw NC, Neel MD, Rao BN, Billups CA, Wu J,
Jenkins JJ, Quintana J, Luchtman-Jones L, Villarroel M and Santana
VM: Frontline treatment of localized osteosarcoma without
methotrexate: results of the St. Jude Children's Research Hospital
OS99 trial. Cancer. 117:2770–2778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeRegis CJ, Moore AS, Rand WM and Berg J:
Cisplatin and doxorubicin toxicosis in dogs with osteosarcoma. J
Vet Intern Med. 17:668–673. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moore-Maxwell CA, Datto MB and Hulette CM:
Chemotherapy-induced toxic leukoencephalopathy causes a wide range
of symptoms: a series of four autopsies. Mod Pathol. 17:241–247.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoneyama K, Konishi H, Yahata T, Fujita K,
Aoki Y, Doi D, Matsushima T, Kodama S, Honma S, Kato H, Nakayama H,
et al: A Phase II study of paclitaxel and carboplatin with a
biweekly schedule in patients with epithelial ovarian cancer:
Gynecologic cancer network trial. J Nippon Med Sch. 81:28–34. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Von Hoff DD, Goldstein D and Renschler MF:
Albumin-bound paclitaxel plus gemcitabine in pancreatic cancer. N
Engl J Med. 370:479–480. 2014.PubMed/NCBI
|
|
7
|
Dank M, Budi L, Piko B, Mangel L, Erfan J,
Cseh J, Ruzsa A and Landherr L: First-line bevacizumab-paclitaxel
in 220 patients with metastatic breast cancer: Results from the
AVAREG study. Anticancer Res. 34:1275–1280. 2014.PubMed/NCBI
|
|
8
|
Orvedahl A, Sumpter R Jr, Xiao G, Ng A,
Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, et
al: Image-based genome-wide siRNA screen identifies selective
autophagy factors. Nature. 480:113–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maycotte P, Aryal S, Cummings CT, Thorburn
J, Morgan MJ and Thorburn A: Chloroquine sensitizes breast cancer
cells to chemotherapy independent of autophagy. Autophagy.
8:200–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakamura O, Hitora T, Akisue T, Kawamoto
T, Yamagami Y and Yamamoto T: Inhibition of induced autophagy
increases apoptosis of Nara-H cells. Int J Oncol. 39:1545–1452.
2011.PubMed/NCBI
|
|
11
|
Mani J, Vallo S, Rakel S, Antonietti P,
Gessler F, Blaheta R, Bartsch G, Michaelis M, Cinatl J, Haferkamp A
and Kögel D: Chemoresistance is associated with increased
cytoprotective autophagy and diminished apoptosis in bladder cancer
cells treated with BH3 mimetic (−)-Gossypol (AT-101). BMC Cancer.
15:2242015. View Article : Google Scholar
|
|
12
|
Monti E and Gariboldi MB: HIF-1 as a
target for cancer chemotherapy, chemosensitization and
chemoprevention. Curr Mol Pharmacol. 4:62–77. 2011. View Article : Google Scholar
|
|
13
|
Generali D, Buffa FM, Berruti A, Brizzi
MP, Campo L, Bonardi S, Bersiga A, Allevi G, Milani M, Aguggini S,
Papotti M, et al: Phosphorylated ERalpha, HIF-1alpha and MAPK
signaling as predictors of primary endocrine treatment response and
resistance in patients with breast cancer. J Clin Oncol.
27:227–234. 2009. View Article : Google Scholar
|
|
14
|
Zhao Y, Chen G, Zhang W, Xu N, Zhu JY, Jia
J, Sun ZJ, Wang YN and Zhao YF: Autophagy regulates hypoxia-induced
osteoclastogenesis through the HIF-1α/BNIP3 signaling pathway. Cell
Physiol. 227:639–648. 2012. View Article : Google Scholar
|
|
15
|
Zhang H, Bosch-Marce M, Shimoda LA, Tan
YS, Baek JH, Wesley JB, Gonzalez FJ and Semenza GL: Mitochondrial
autophagy is an HIF-1-dependent adaptive metabolic response to
hypoxia. Biol Chem. 283:10892–10903. 2008. View Article : Google Scholar
|
|
16
|
Wessel D and Flügge UI: A method for the
quantitative recovery of protein in dilute solution in the presence
of detergents and lipids. Anal Biochem. 138:141–143. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adesida AB, Grady LM, Khan WS,
Millward-Sadler SJ, Salter DM and Hardingham TE: Human meniscus
cells express hypoxia inducible factor-1α and increased SOX9 in
response to low oxygen tension in cell aggregate culture. Arthritis
Res Ther. 9:R692007. View
Article : Google Scholar
|
|
18
|
Zeng Y, Yang X, Wang J, Fan J, Kong Q and
Yu X: Aristolochic acid I induced autophagy extenuates cell
apoptosis via ERK 1/2 pathway in renal tubular epithelial cells.
PLoS One. 7:e303122012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amarnath S, Flomerfelt FA, Costanzo CM,
Foley JE, Mariotti J, Konecki DM, Gangopadhyay A, Eckhaus M, Wong
S, Levine BL, June CH and Fowler DH: Rapamycin generates
anti-apoptotic human Th1/Tc1 cells via autophagy for induction of
xenogeneic GVHD. Autophagy. 6:523–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao YQ, Ding X, Jia YC, Huang CX, Wang YZ
and Xu YH: Anti-tumor effect of betapaxilitaxel in glioblastoma
cells depends on p38 MAPK activation. Cancer Lett. 264:127–134.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang G, Li X, Huang F, Zhao J, Ding H,
Cunningham C, Coad JE, Flynn DC, Reed E and Li QQ: Antitumor effect
of betapaxilitaxel in non-small-cell lung cancer cells is mediated
via induction of cell cycle arrest and apoptotic cell death. Cell
Mol Life Sci. 62:881–893. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo B, Yang M, Liang D, Yang L, Cao J and
Zhang L: Cell apoptosis induced by zinc deficiency in osteoblastic
MC3T3-E1 cells via a mitochondrial-mediated pathway. Mol Cell
Biochem. 361:209–216. 2012. View Article : Google Scholar
|
|
23
|
Rasul A, Yu B, Yang LF, Ali M, Khan M, Ma
T and Yang H: Induction of mitochondria-mediated apoptosis in human
gastric adenocarcinoma SGC-7901 cells by kuraridin and
Nor-kurarinone isolated from Sophora flavescens. Asian Pac J Cancer
Prev. 12:2499–2504. 2011.
|
|
24
|
Granell S and Baldini G: Inclusion bodies
and autophagosomes: are ER-derived protective organelles different
than classical autophagosomes? Autophagy. 4:375–377. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deretic V, Delgado M, Vergne I, Master S,
De Haro S, Ponpuak M and Singh S: Autophagy in immunity against
mycobacterium tuberculosis: a model system to dissect immunological
roles of autophagy. Curr Top Microbiol Immunol. 335:169–188.
2009.PubMed/NCBI
|
|
26
|
Hofius D, Munch D, Bressendorff S, Mundy J
and Petersen M: Role of autophagy in disease resistance and
hypersensitive response-associated cell death. Cell Death Differ.
18:1257–1262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang T, Li Y, Park KA, Byun HS, Won M,
Jeon J, Lee Y, Seok JH, Choi SW, Lee SH, Man Kim J, et al:
Cucurbitacin induces autophagy through mitochondrial ROS production
which counteracts to limit caspase-dependent apoptosis. Autophagy.
8:559–576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ait-Mohamed O, Battisti V, Joliot V,
Fritsch L, Pontis J, Medjkane S, Redeuilh C, Lamouri A, Fahy C,
Rholam M, Atmani D and Ait-Si-Ali S: Acetonic extract of Buxus
sempervirens induces cell cycle arrest, apoptosis and autophagy in
breast cancer cells. PLoS One. 6:e245372011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu HH, Kao SY, Liu TY, Liu ST, Huang WP,
Chang KW and Lin SC: Areca nut extract induced oxidative stress and
upregulated hypoxia inducing factor leading to autophagy in oral
cancer cells. Autophagy. 6:725–737. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bellot G, Garcia-Medina R, Gounon P,
Chiche J, Roux D, Pouysségur J and Mazure NM: Hypoxia-induced
autophagy is mediated through hypoxia-inducible factor induction of
BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol.
29:2570–2581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu
Y, Xie M, Yin X, Livesey KM, Loze MT, Tang D and Cao L:
DAMP-mediated autophagy contributes to drug resistance. Autophagy.
7:112–124. 2011. View Article : Google Scholar :
|
|
32
|
Carew JS, Nawrocki ST, Kahue CN, Zhang H,
Yang C, Chung L, Houghton JA, Huang P, Giles FJ and Cleveland JL:
Targeting autophagy augments the anticancer activity of the histone
deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug
resistance. Blood. 110:313–322. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Doublier S, Belisario DC, Polimeni M,
Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A and Sapino A:
HIF-1 activation induces doxorubicin resistance in MCF7 3-D
spheroids via P-glycoprotein expression: a potential model of the
chemo-resistance of invasive micropapillary carcinoma of the
breast. BMC Cancer. 12:42012. View Article : Google Scholar : PubMed/NCBI
|