Introduction
Biliary atresia (BA) is a rare hepatobiliary
disorder in infants with poor prognosis caused by an obstruction of
the bile ducts, leading to a disruption in bile flow, which can
rapidly progress to biliary cirrhosis (1,2). The
incidence of BA in Asia is higher than in western countries, with
reports indicating a rate of 1/9,600 to 1/16,700 of live births
(3). BA is reported to occur in
half of all cases of neonatal cholestasis with a greater incidence
in females, and associated congenital malformations occur in 3–10%
of cases of BA (4). Without
treatment, BA progresses into liver cirrhosis, hepatic failure and
mortality in <2 years following birth (5,6).
Natural mortality of BA is 100%, therefore liver transplantation is
a necessary requirement for late stage BA (7); however, current medical management of
patients with BA is unsatisfactory, placing an economic burden on
society and family members (5,7).
The etiology of BA remains to be fully elucidated;
however, previous studies have identified three factors suggested
to contribute to the formation of BA. One is congenital genetic
abnormalities, for example, the mutation of INVS gene, which leads
to abnormal functioning of inversin, and may terminate the
development of biliary ducts during the embryonic period (8). Another factor identified by various
studies is viral infection (9,10),
with cytomegalovirus, Epstein-Barr virus and human papilloma virus
having all been isolated from the livers of infants with BA
(11–13). The third factor identified was
immune disorders, which appeared following the viral infection
(14). Previous studies have
supported the hypothesis that the viral infection triggering host
immune responses may be the basis for the pathogenesis of BA
(15). Leonhardt et al
(16) and Schreiber and Kleinman
(17) additionally agreed that BA
is a virus-induced, immune-mediated disease that occurs in
genetically susceptible individuals. Previous studies have
demonstrated that BA was associated with T helper cell response
type I-mediated portal tract inflammation and that interleukin
(IL)-2, interferon γ, IL-12, tumor necrosis factor α and macrophage
activation all served important roles (18,19).
The current study aimed to elucidate the pathogenesis of BA using
biological information analysis in relation to immunology.
Microarray analysis has been reported as an
effective method to monitor alterations in gene expression and
identify genes that are associated with biological processes
induced by BA. Leonhardt et al (16) identified several targeted genes
that were implicated in the development of BA, including CCL2,
CCL5, CCR5, CXCL9, CXCL10 and IL1F5 (20). The majority of these genes were
associated with the immune system or with modulating
inflammation.
Previous studies have demonstrated that the
expression levels of cell signaling and transcription regulatory
genes, such as CIT or ActRII were upregulated during BA progression
(21), and BA was observed to be a
necrotizing process caused by viral cholangitis at the embryonic
stage and the expression level of the type II major
histocompatibility complex was increased in the livers of patients
(22). However, there is little
evidence indicating the relevance between human leukocyte
expression and BA. Thus, the cause and pathogenetic mechanism of BA
remain largely unknown.
In the present study, microarray analysis was used
to monitor differentially expressed genes (DEGs) in BA samples at
different time points (3, 7 and 14 days) following induction with
rhesus rotavirus (RRV) compared with sham-RRV-control.
Comprehensive bioinformatics analysis was used, which combined gene
ontology (GO) and Kyoto Enrichment of Genes and Genome (KEGG)
pathway information, in order to provide a more thorough
understanding of the biological mechanisms of BA. The current study
aimed to predict the hub genes that are most likely to be
associated with BA, and to identify their molecular mechanisms in
BA. This may aid in the development of novel therapeutic strategies
for BA.
Materials and methods
Affymetrix microarray data
The microarray data were obtained from the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database from the
National Center for Biotechnology Information (Bethesda, MD, USA)
(23), which is the largest public
database of its type. The gene expression profiles were extracted
from the GEO database (ID: GSE46967) based on the GPL6246
Affymetrix Mouse Gene 1.0 ST array (Affymetrix, Inc., Santa Clara,
CA, USA). The experimental mice (18 tissue samples) were injected
intraperitoneally with 1.5×106 units of rhesus rotavirus
(RRV) or 0.9% normal saline (NS; control) within 24 h. The
microarray data were collected from the experimental group and the
control group at 3, 7 and 14 days after RRV or saline injection.
Each experimental condition contained three sets of samples (three
treated with RRV three with normal saline).
A total of 28,816 probes were mapped with the gene
names labelled in the GPL6246 annotation platform, and a log2
transformation was performed on 25,299 genes (24). The limma package (http://bioinf.wehi.edu.au/limma) in R language
was used to select the DEGs of the case-group compared with the
control-group (25). The P-values
were adjusted by the Benjamin and Hochberg method, and the
FDR-value based on the multtest package of 0.05 was used as the
cut-off criterion (26).
Hiarchical clustering of DEGs
The expression values of DEGs in each group were
selected according to the probe information from the downloaded
file. The R language pheatmap package (http://cran.r-project.org/web/packages/pheatmap/index.html)
was used for hierarchical clustering analysis (27) of DEGs based on Euclidean distance
(28), and the results were
presented in heat maps. Genes with similar expression levels could
be collected together by hierarchical clustering for a convenient
search. Samples clustered based on the gene expression value can
aid in determining whether the screening DEGs have sample
specificity or not, which means the selected sample types (case or
control) can be identified based on the differential gene
expression information.
Common DEGs of three time points
The expression levels of selected genes from samples
at 3, 7 and 14 days were compared, in order to identify the common
DEGs (29).
GO analysis and pathway enrichment
analysis
GO analysis has become a commonly used approach for
functional studies of large-scale genomic or transcription data
(30). The KEGG pathway database
contains information of how molecules or genes are networked, which
is complementary to the majority of the existing molecular biology
databases containing the information of individual genes (31). The Database for Annotation,
Visualization and Integrated Discovery (DAVID) bioinformatics
resources consist of an integrated biological knowledge base and
analytical tools aimed at systematically extracting biological
meaning from large gene or protein lists (32). DAVID was used to analyze the
enriched functions and pathways of the common DEGs at the three
time points with P<0.05.
In addition, the expression values of DEGs that
participated in the most significant GO functions and KEGG pathways
were compared.
Results
Screening and hierarchical clustering
analysis
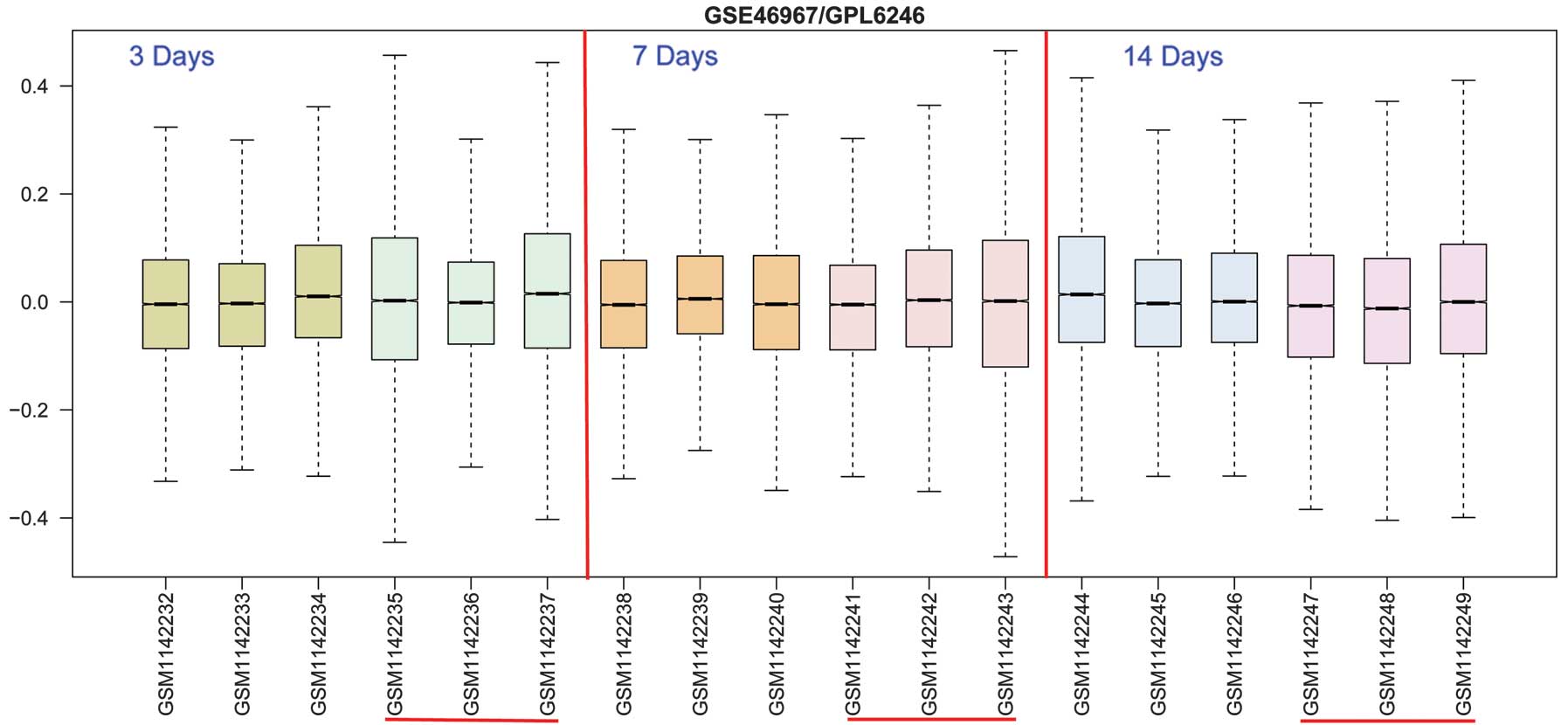
In order to investigate DEGs in mice with BA at 3, 7
and 14 days following viral induction, the publicly available
microarray dataset GSE46967 from GEO was used. A total of 306 DEGs
were selected in the 3 day samples, 721 DEGs in the 7 day samples
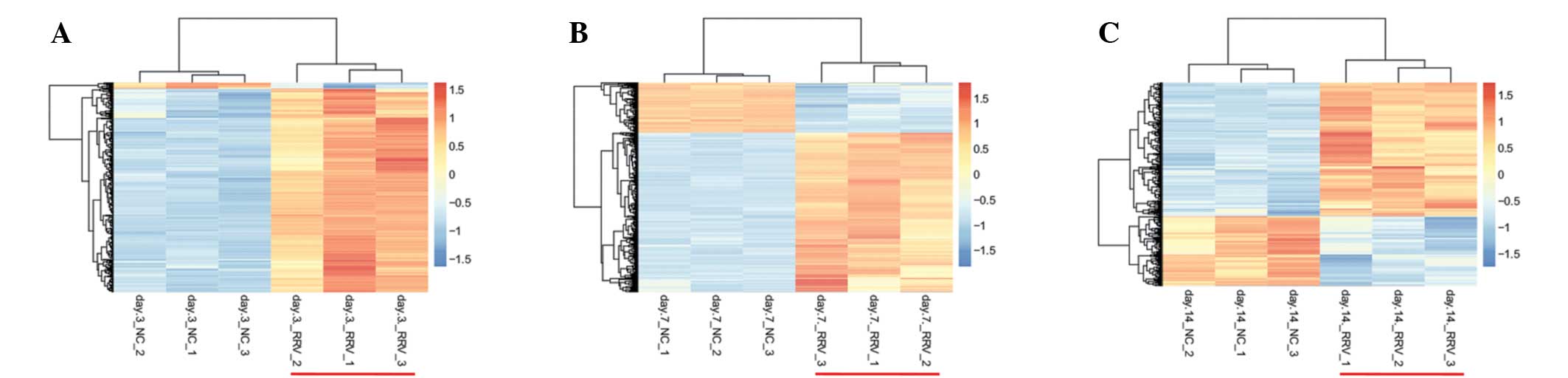
and 370 DEGs in the 14 day samples (Fig. 1). From the heat maps (Fig. 2), the results of hierarchical
clustering analysis revealed that the case- and control-groups can
be separated by the selected DEGs in each group, which means the
expression patterns of DEGs screened were significant, and they can
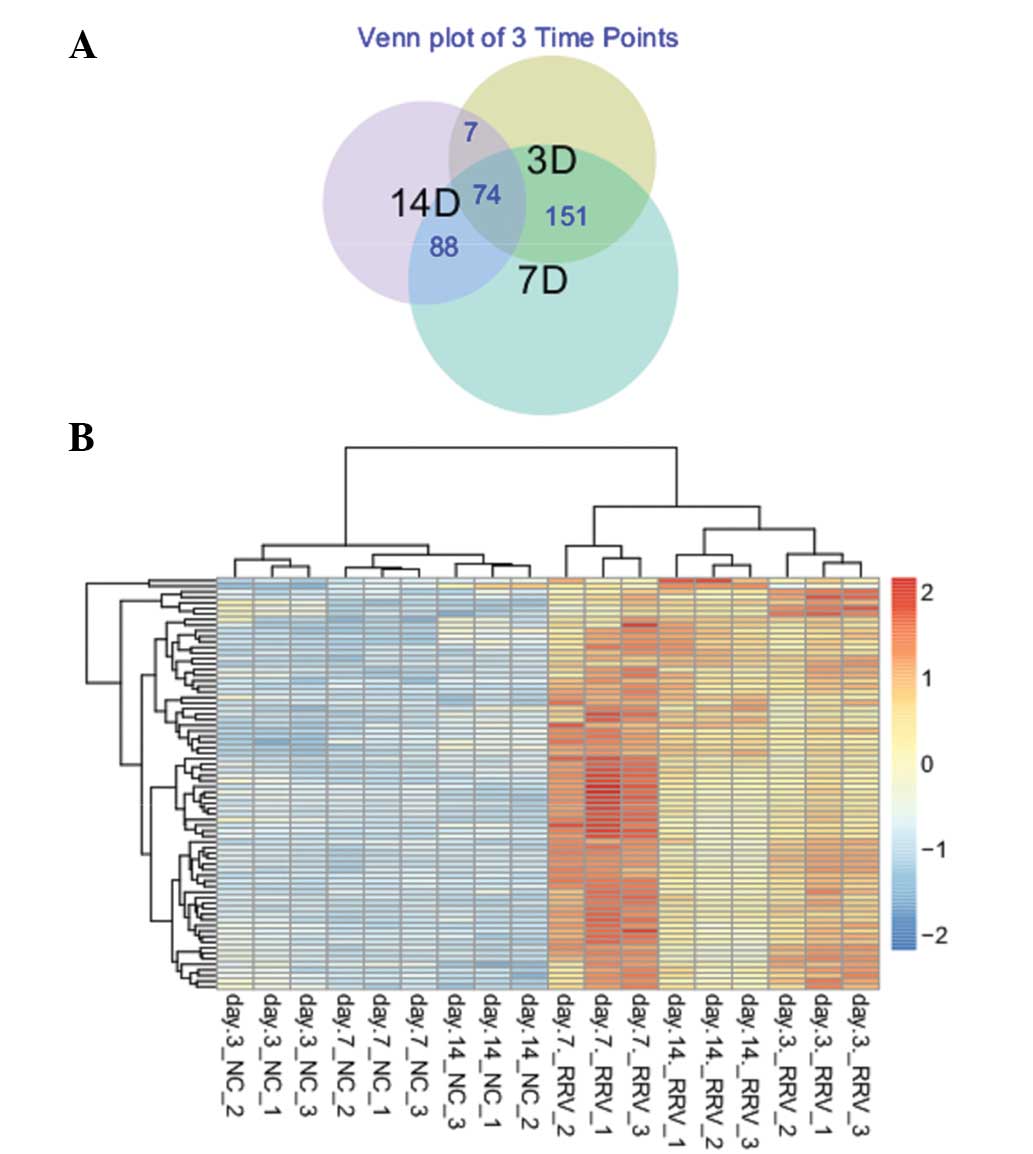
be used to distinguish case and control samples. A total of 74
common DEGs were identified between the samples from the different
time points (Fig. 3A), and the
expression values of all of these common DEGs were upregulated, as
compared with in the control samples(Fig. 3B).
Functional annotation analysis of common
DEGs
To investigate the alterations in the function of
selected common DEGs, DAVID was used to identify the significant GO
categories in the biological process. A total of 12 GO terms were
observed to be enriched among the 74 DEGs (Table I), of which several significantly
enriched functions were associated with immune function, including
the defense response, leukocyte migration, cell chemotaxis and
leukocyte chemotaxis. C3AR1, CCL3, C5AR1, CXCL5, FPR1, CXCR2, CCL5,
ITGAM, CCL6, CXCL13, CSF3R, IL1B and FCER1G were the genes enriched
in the significant functions.
 | Table IFunction enrichment analysis of the
common DEGs. |
Table I
Function enrichment analysis of the
common DEGs.
| Term | Count | FDR |
|---|
|
GO:0042330~taxis | 13 |
1.67×10−13 |
|
GO:0006935~chemotaxis | 14 |
1.67×10−13 |
| GO:0006955~immune
response | 21 |
1.67×10−13 |
|
GO:0007626~locomotory behavior | 14 |
4.38×10−09 |
| GO:0006952~defense
response | 17 |
5.54×10−09 |
|
GO:0007610~behavior | 14 |
3.07×10−06 |
|
GO:0030595~leukocyte chemotaxis | 6 |
6.90×10−05 |
| GO:0060326~cell
chemotaxis | 6 |
6.90×10−05 |
|
GO:0006954~inflammatory response | 10 |
1.98×10−04 |
|
GO:0030593~neutrophil chemotaxis | 5 |
6.03×10−04 |
| GO:0009611~response
to wounding | 11 |
7.79×10−04 |
|
GO:0050900~leukocyte migration | 6 |
7.86×10−04 |
Pathway analysis of common DEGs
To further understand the pathways of the DEGs
screened in the current study, DAVID was used to identify the
significant pathways. Two pathways (Table II), were identified to be
enriched, the 'cytokine-cytokine receptor interaction' and
'chemokine signaling pathway'. The 'cytokine-cytokine receptor
interaction' pathway was the most significant pathway with ten
genes involved, as follows: IL1R2, CCL3, CXCL5, CXCL13, CCR1, IL1B,
CSF3R, CXCR2, CCL5 and CCL6.
 | Table IIPathways involved with common
DEGs. |
Table II
Pathways involved with common
DEGs.
| Kyoto enrichment of
genes and genomes | Gene number | FDR |
|---|
| mmu04060:
Cytokine-cytokine receptor interaction | 10 | 0.003256 |
| mmu04062: Chemokine
signaling pathway | 9 | 0.003469 |
Expression values of DEGs participating
in significant functions and pathways
A total of 13 DEGs were identified to be associated
with the significant functions, and 10 DEGs with the significant
pathways. Following the comparison of the two types of DEGs, the
following 6 common DEGs were identified: CCL3, CXCL5, CXCL13,
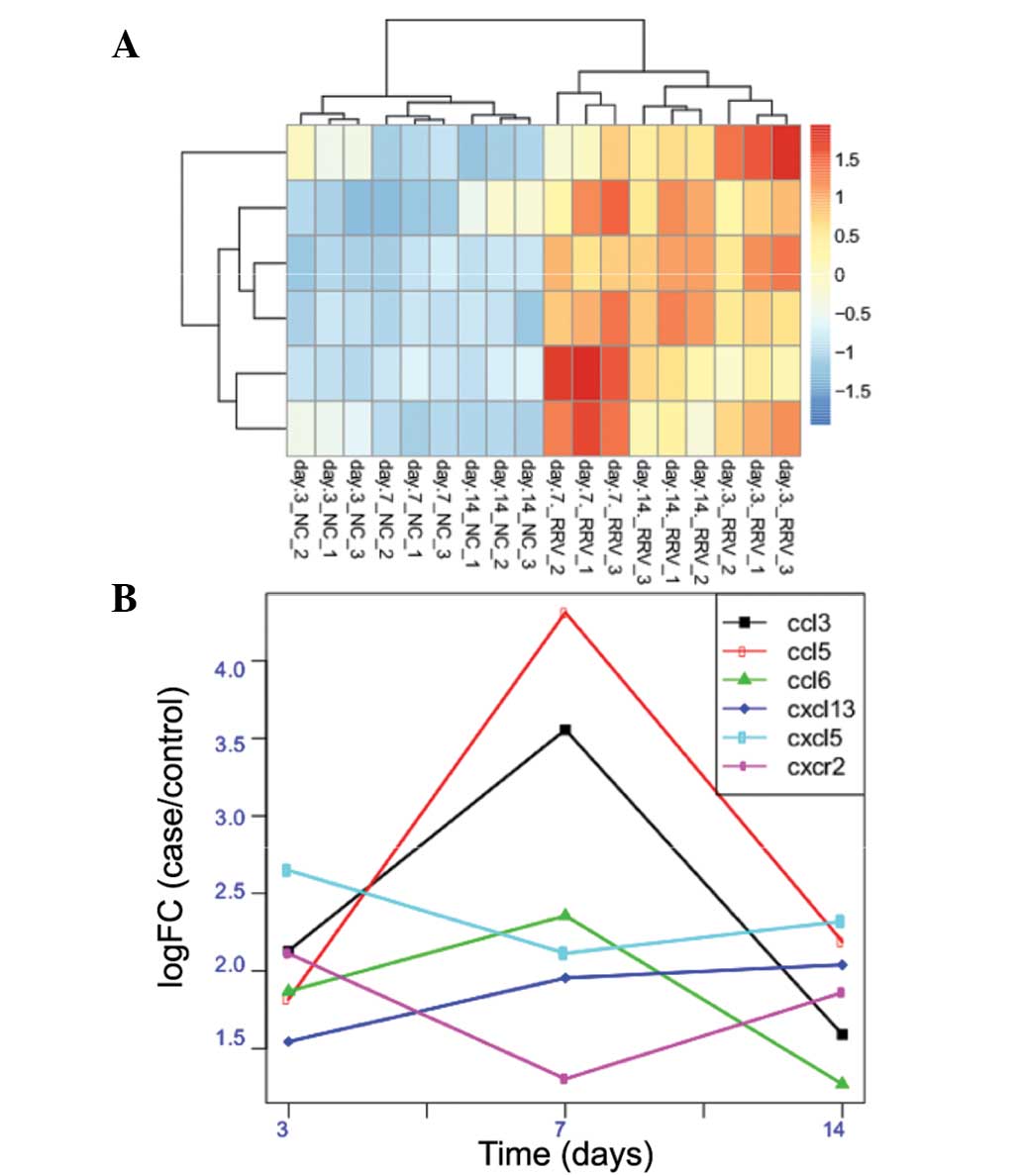
CXCR2, CCL5 and CCL6. The expression profile of these 6 DEGs in
each of the three samples were analyzed (Fig. 4A). It is observed in the figure
that all 6 of the DEGs in the three sample types were significantly
overexpressed, and CXCL13 was identified to have a consistent
overexpression from the 3rd day to the 14th day. CCL3, CCL5 and
CCL6; however, demonstrated the highest overexpression on the 7th
day, but were decreased on the 14th day. CXCL5 and CXCR2 presented
the highest overexpression on the 3rd day (Fig. 4B).
Discussion
BA is an infantile disorder in which there is
obliteration or discontinuity of the extrahepatic biliary tree. If
left untreated, it progresses to cirrhosis, liver failure and
eventually to mortality, and the current treatment strategies are
unsatisfactory (2,5). Previous studies using histological
sections of extrahepatic biliary tract tissue from patients, in
addition to experimental studies using the BA mice model, have
demonstrated that an inflammatory process appears to contribute to
the pathogenesis of BA (18,33).
Previous studies that further identify the relevant inflammatory
cascade revealed an upregulation in pro-inflammatory cytokines
(20). A total of 306 DEGs were
identified in the 3 day samples, 721 in the 7 day samples and 370
in the 14 day samples in mice with BA following injection with
rhesus rotavirus. In total, 74 common DEGs were selected from the
three sample types at different time points, which function
predominantly in the immune biological process, including the
defense response, leukocyte migration, cell chemotaxis and
leukocyte chemotaxis. The 'cytokine-cytokine receptor interaction'
and 'chemokine signaling pathway' were identified as the most
significantly enriched pathways. A total of six significantly
overexpressed genes (CCL3, CXCL5, CXCL13, CXCR2, CCL5 and CCL6)
associated with the immune response and cytokine pathways at the
three time points were screened, which were investigated in the
significant functions and pathways.
Chemokines are small proteins that are vital in
immune and inflammatory reactions. Induction of leukocyte migration
is the predominant function of the chemokines, which additionally
affect angiogenesis, collagen production and the proliferation of
hematopoietic precursors (34).
CCL3 (MIP-1α) is a chemokine produced by activated T-cells,
basophilic granulocytes and type 2 T helper cells (35). The chemokine CCL3 has several
functions, including serving a role in cell lysis mediated by
natural killer cells, participating in the inflammatory response
associated with immunological processes, and mediating the release
of the other cytokines (36). Seki
et al (37) reported that
MIP-1α had the ability to gather the hepatic stellate and
inflammatory cells migrating to the site of injury, so as to
function on the liver fibration. Zerfaoui et al (38) demonstrated that CCL3 was able to
activate T cells thus producing CCL5 (RANTES). CCL3 and CCL5 have
the ability to assemble and to activate eosinophilic granulocytes
to release histamine and leukotrienes so as to induce inflammatory
cell adherence to the vessel wall (38). Increased levels of CCL5 have been
observed in patients with primary biliary cirrhosis, and the
therapeutic benefits of fibrates in this biliary disease may be due
to the inhibition of this CCL5, with its resultant effects on
inflammatory cell migration (39).
From the results of the present study, it is suggested that the
expression of CCL3 and CCL5 are elevated in a time-dependent manner
until 7 days, indicating that these two molecules are critical for
the pathogenesis of BA in mice. Furthermore, previous studies have
demonstrated that the two cytokines are associated with the
migration and infiltration of monocytes, memory T lymphocytes and
natural killer cells (40–42), so it was speculated that they may
be involved in the early recruitment of different subsets of
leukocytes in the formation of BA.
The CXCR2 gene was first cloned by Murphy and
Tiffany (43) and the chemokine
CXCR2, which is produced on the surface of neutrophils and natural
killer cells, is also a member of the G-protein superfamily. A
previous study demonstrated that CXCR2 may combine with the
chemokine CXCL5 in order to activate the downstream effector
molecules, and may subsequently mediate signal transduction and
cell migration via the allosteric G-protein (44). CXCL5 is a small-molecule type
secretory protein, which has a direct chemotaxis for neutrophils
and is able to mediate the infiltration of neutrophils, thus
promoting the metastasis of liver cancer cells (45). Li et al (46) reported that CXCL5 contributed to
the formation of tumor vessels by activating certain signaling
pathways, such as the protein kinase B, signal transduction and
activating transcription factor pathways (47). In the present study, the variation
tendency of the CXCL5 expression level was similar with that of
CXCR2 and their expression peaked at 3 days. This phenomenon may
indicate that the chemokine receptor CXCR2 and CXCL5 may combine,
in order to affect inflammation in mice with BA by collecting
leukocytes.
CXCL13 is a chemokine produced by B-cells and is
critical for targets seeking B-cell and B-cell zone formation in
lymphatic tissue (48). A previous
study indicated that modulation of the inflammatory signaling
cascade, particularly cytokine secretion, may attenuate the
severity of BA in the murine model. Shivakumar et al
demonstrated that RRV-infected, interferon γ-deficient Bulb/c mice
had a lower mortality rate than wild type animals (48). Anesl et al (49) demonstrated that the natural
antibody production in mice deficient in CXCL13 was markedly
reduced, and no response was triggered to a bacterial antigen by
intraperitoneal injection in this case. In addition, the roles of
CXCL13 have been observed in other immune system-associated
diseases. Hjelmstrom et al (50) observed that CXCL13 was
overexpressed in transgenic mice suffering from chronic
inflammation induced by lymphotoxin-α (50). Based on the data from the current
study, the expression levels of CXCL13 were increased until 14 days
in mice with BA. Elevated levels of CXCL13 is not unique to active
BA, but it may serve as a marker of disease activity or response to
treatment in combination with other diagnostic markers.
Notably, low expression levels of CCL6 were observed
in BA-positive mice. It was hypothesized that the chemokine CCL6
may serve an important function in the progression of BA, however
further study is required in order to further elucidate the
mechanisms.
In conclusion, CCL3, CCL5, CXCL5, CXCR2, CXCL13 and
CCL6, are suggested to be involved in the pathogenesis of BA, but
serve different roles. The current study provided a basis for
future experiments with the potential use of RRV-injected mice,
which may enhance understanding of the pathogenesis of BA and lead
to the development of novel therapeutic strategies.
References
|
1
|
Mack CL, Feldman AG and Sokol RJ: Clues to
the etiology of bile duct injury in biliary atresia. Semin Liver
Dis. 32:307–316. 2012. View Article : Google Scholar
|
|
2
|
Petersen C: Pathogenesis and treatment
opportunities for biliary atresia. Clin Liver Dis. 10:73–88. 2006.
View Article : Google Scholar
|
|
3
|
Jimenez-Rivera C, Jolin-Dahel KS,
Fortinsky KJ, Gozdyra P and Benchimol EI: International incidence
and outcomes of biliary atresia. J Pediatr Gastroenterol Nutr.
56:344–354. 2013. View Article : Google Scholar
|
|
4
|
Davenport M, Ong E, Sharif K, et al:
Biliary atresia in England and Wales: results of centralization and
new benchmark. J Pediatr Surg. 46:1689–1694. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mendoza MM, Chiang JH, Lee SY, et al:
Reappraise the effect of redo-Kasai for recurrent jaundice
following Kasai operation for biliary atresia in the era of liver
transplantation. Pediatr Surg Int. 28:861–864. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bijl EJ, Bharwani KD, Houwen RH and de Man
RA: The long-term outcome of the Kasai operation in patients with
biliary atresia: a systematic review. Neth J Med. 71:170–173.
2013.PubMed/NCBI
|
|
7
|
Cronin DC, Squires J, Squires R,
Mazariegos G and Lantos JD: Parental refusal of a liver transplant
for a child with biliary atresia. Pediatrics. 131:141–146. 2013.
View Article : Google Scholar
|
|
8
|
Shimadera S, Iwai N, Deguchi E, Kimura O,
Fumino S and Yokoyama T: The inv mouse as an experimental model of
biliary atresia. J Pediatr Surg. 42:1555–1560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mack CL, Falta MT, Sullivan AK, et al:
Oligoclonal expansions of CD4+ and CD8+
T-cells in the target organ of patients with biliary atresia.
Gastroenterology. 133:278–287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Carvalho E, Ivantes CA and Bezerra JA:
Extrahepatic biliary atresia: current concepts and future
directions. J Pediatr (Rio J). 83:105–120. 2007. View Article : Google Scholar
|
|
11
|
Soomro GB, Abbas Z, Hassan M, Luck N,
Memon Y and Khan AW: Is there any association of extra hepatic
biliary atresia with cytomegalovirus or other infections. J Pak Med
Assoc. 61:281–283. 2011.PubMed/NCBI
|
|
12
|
Mahjoub F, Shahsiah R, Ardalan FA, et al:
Detection of Epstein Barr virus by chromogenic in situ
hybridization in cases of extra-hepatic biliary atresia. Diagn
Pathol. 3:192008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schaffer K, Hassan J, Staines A, et al:
Surveillance of Epstein-Barr virus loads in adult liver
transplantation: associations with age, sex, posttransplant times,
and transplant indications. Liver Transpl. 17:1420–1426. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Whitington PF, Malladi P, Melin-Aldana H,
Azzam R, Mack CL and Sahai A: Expression of osteopontin correlates
with portal biliary proliferation and fibrosis in biliary atresia.
Pediatr Res. 57:837–844. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mack CL: The pathogenesis of biliary
atresia: evidence for a virus-induced autoimmune disease. Semin
Liver Dis. 27:233–242. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leonhardt J, Stanulla M, Von Wasielewski
R, et al: Gene expression profile of the infective murine model for
biliary atresia. Pediatr Surg Int. 22:84–89. 2006. View Article : Google Scholar
|
|
17
|
Schreiber RA and Kleinman RE: Biliary
atresia. J Pediatr Gastroenterol Nutr. 35(Suppl 1): S11–S16. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mack CL, Tucker RM, Sokol RJ, et al:
Biliary atresia is associated with CD4+ Th1
cell-mediated portal tract inflammation. Pediatr Res. 56:79–87.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lages CS, Simmons J, Chougnet CA and
Miethke AG: Regulatory T cells control the CD8 adaptive immune
response at the time of ductal obstruction in experimental biliary
atresia. Hepatology. 56:219–227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bezerra JA, Tiao G, Ryckman FC, et al:
Genetic induction of proinflammatory immunity in children with
biliary atresia. Lancet. 360:1653–1659. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakamura K and Tanoue A: Etiology of
biliary atresia as a developmental anomaly: recent advances. J
Hepatobiliary Pancreat Sci. 20:459–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirschfield GM, Liu X, Xu C, et al:
Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2
variants. N Engl J Med. 360:2544–2555. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrett T, Troup DB, Wilhite SE, et al:
NCBI GEO: mining tens of millions of expression profiles - database
and tools update. Nucleic Acids Res. 35:D760–D765. 2007. View Article : Google Scholar
|
|
24
|
Fujita A, Sato JR, Rodrigues Lde O,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Anders S, McCarthy DJ, Chen Y, et al:
Count-based differential expression analysis of RNA sequencing data
using R and Bioconductor. Nat Protoc. 8:1765–1786. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
27
|
Langfelder P and Horvath S: Fast R
functions for robust correlations and hierarchical clustering. J
Stat Softw. 46:112012.
|
|
28
|
Mukherjee S, Chen Z and Gangopadhyay A: A
privacy-preserving technique for Euclidean distance-based mining
algorithms using Fourier-related transforms. VLDB J. 15:293–315.
2006. View Article : Google Scholar
|
|
29
|
Pirooznia M, Nagarajan V and Deng Y:
GeneVenn - A web application for comparing gene lists using Venn
diagrams. Bioinformation. 1:420–422. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: two different approaches for Gene Ontology
analysis. BMC Proc. 3(Suppl 4): S102009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
32
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
33
|
Mack CL, Tucker RM, Sokol RJ and Kotzin
BL: Armed CD4+ Th1 effector cells and activated
macrophages participate in bile duct injury in murine biliary
atresia. Clin Immunol. 115:200–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Carmeliet P: Angiogenesis in health and
disease. Nat Med. 9:653–660. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ramos CDL, Canetti C, Souto JT, et al:
MIP-alpha[CCL3] acting on the CCR1 receptor mediates neutrophil
migration in immune inflammation via sequential release of
TNF-alpha and LTB4. J Leukoc Biol. 78:167–177. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ekman AK, Fransson M, Rydberg C, Adner M
and Cardell LO: Nasal challenge with LPS stimulates the release of
macrophage inflammatory protein 1alpha. Int Arch Allergy Immunol.
149:154–160. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Seki E, De Minicis S, Gwak GY, et al: CCR1
and CCR5 promote hepatic fibrosis in mice. J Clin Invest.
119:1858–1870. 2009.PubMed/NCBI
|
|
38
|
Zerfaoui M, Naura AS, Errami Y, et al:
Effects of PARP-1 deficiency on airway inflammatory cell
recruitment in response to LPS or TNF: differential effects on
CXCR2 ligands and Duffy antigen receptor for chemokines. J Leukoc
Biol. 86:1385–1392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hirano Y, Hirano F, Fujii H and Makino I:
Fibrates suppress chenodeoxycholic acid-induced RANTES expression
through inhibition of NF-kappaB activation. Eur J Pharmacol.
448:19–26. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamamoto S, Shimizu S, Kiyonaka S, et al:
TRPM2-mediated Ca2+ influx induces chemokine production
in monocytes that aggravates inflammatory neutrophil infiltration.
Nat Med. 14:738–747. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schlecker E, Stojanovic A, Eisen C, et al:
Tumor-infiltrating monocytic myeloid-derived suppressor cells
mediate CCR5-dependent recruitment of regulatory T cells favoring
tumor growth. J Immunol. 189:5602–5611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Robertson MJ: Role of chemokines in the
biology of natural killer cells. J Leukoc Biol. 71:173–183.
2002.PubMed/NCBI
|
|
43
|
Murphy PM and Tiffany HL: Cloning of
complementary DNA encoding a functional human interleukin-8
receptor. Science. 253:1280–1283. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stillie R, Farooq SM, Gordon JR and
Stadnyk AW: The functional significance behind expressing two IL-8
receptor types on PMN. J Leukoc Biol. 86:529–543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Okabe H, Beppu T, Ueda M, et al:
Identification of CXCL5/ENA-78 as a factor involved in the
interaction between cholangiocarcinoma cells and cancer-associated
fibroblasts. Int J Cancer. 131:2234–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li A, King J, Moro A, et al:
Overexpression of CXCL5 is associated with poor survival in
patients with pancreatic cancer. Am J Pathol. 178:1340–1349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
van de Pavert SA, Olivier BJ, Goverse G,
et al: Chemokine CXCL13 is essential for lymph node initiation and
is induced by retinoic acid and neuronal stimulation. Nat Immunol.
10:1193–1199. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shivakumar P, Campbell KM, Sabla GE, et
al: Obstruction of extrahepatic bile ducts by lymphocytes is
regulated by IFN-γ in experimental biliary atresia. J Clin Invest.
114:322–329. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ansel KM, Harris RB and Cyster JG: CXCL13
is required for B1 cell homing, natural antibody production, and
body cavity immunity. Immunity. 16:67–76. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hjelmström P, Fjell J, Nakagawa T, Sacca
R, Cuff CA and Ruddle NH: Lymphoid tissue homing chemokines are
expressed in chronic inflammation. Am J Pathol. 156:1133–1138.
2000. View Article : Google Scholar : PubMed/NCBI
|