Introduction
Prostate carcinoma is the most common type of
malignancy (28%) and the second leading cause of cancer-associate
mortality (10%) in men in the United States. In the United States,
~238,590 new cases of prostate cancer were diagnosed and ~29,720
deaths were attributable to the disease in 2013 (1,2).
This devastating disease has a significant impact on public health,
however, current prostate cancer therapies, including surgery,
chemotherapy and radiation therapy are of limited efficacy in
progressive, recurrent and metastatic prostate cancer, particularly
in hormone-refractory prostate cancer (HRPC) (3–6). In
investigations of alternative and preventive therapies for prostate
cancer, attention has focused on natural products. Plant-derived
compounds have been an important source of several clinically
useful anti-cancer agents (7).
Emodin (1,2,8-trihydroxy-6-methylanthraquinone), a natural compound
extracted from Rheum palmatum L. of the family Polygonaceae,
has received significant attention for its potent anti-cancer
activity. A number of studies have demonstrated that emodin can
cause a marked decrease in cell proliferation and an increase in
apoptosis in several types of cancer cells, including prostate
cancer (LNCaP), lung cancer (A549, H460, CH27 and WI-38), acute
myelogenous leukemia (HL-60), colorectal (WiDr) and pancreatic
cancer (PANC-1) cells (6,8–12).
The previous studies by Yu et al (6) and Cha el al (13) revealed that the emodin inhibits
prostate cancer cell growth and downregulates the androgen receptor
and p53-p21 pathways. Expression of the Notch receptor, which has
been widely demonstrated to be responsible for cell fate
determination during normal development, is implicated in human
T-cell leukemia and mouse mammary carcinoma, and can significantly
affect development of the prostate and cancer cell growth (14,15).
The present study aimed to elucidate the potential molecular
mechanisms of emodin-mediated inhibition of cell growth and
apoptotic induction in PC3 prostate cancer cells, and the
involvement of the Notch signaling pathway.
Materials and methods
Chemicals and reagents
Emodin was purchased from JRDUN biotechnology
(Shanghai, China). The 3-(4,5)-dimet
hylthiahiazo(-z-y1)-3,5-di-phenytetrazolium bromide (MTT), dimethyl
sulfoxide (DMSO), RNase and propidium iodide (PI) were obtained
from Sigma-Aldrich (St. Louis, MO, USA); M-MLV Reverse
Transcriptase was obtained from Toboyo Co., Ltd. (Osaka, Japan);
TaqplusDNA Polymerase, dNTP and TRIzol were obtained from
Invitrogen Life Technologies (Carlsbad, CA, USA); Goat anti-human
polyclonal antibodies against Notch1 (sc-6014), Jagged1 (sc-6011),
VEGF (sc-1836), bFGF (sc-1360), immunoglobulin (Ig)G-FITC and
peroxidase-conjugated goat anti-human IgG (H+L) (sc-2012) were
obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA); All
other chemicals were of reagent grade.
Cell culture
PC3 cells were obtained from the China Center for
Type Culture Collection (Wuhan, China) and were cultured in DMEM
supplemented with 10% FBS and 100 U/ml penicillin/streptomycin in a
humidified CO2 incubator at 37°C.
Cell viability assays
An MTT assay was used to assess the effect of emodin
on PC3 cell viability. In brief, the PC3 cells were seeded in
96-well culture plates at a cell density of 5×104
cells/ml and treated with emodin (10, 20, 40, 60 and 80
μg/ml) or remained untreated (control) for 12, 24, 36, 48,
72 and 96 h incubated at 37°C in a humidified atmosphere containing
5% CO2. Subsequently, the cell viability was evaluated
using an MTT assay (5 mg/ml MTT). The absorbance was measured at a
test wavelength of 490 nm using a Tecan Sunrise Elisa-Reader (Tecan
Group, Ltd., Mannedorf, Switzerland).
Assay to analyze cell cycle
distribution
The PC3 cells were cultured in triplicate in 96-well
plates at a density of 5×104 cells/ml and treated with
emodin (0, 20, 40, 60 and 80 μg/ml) or remained untreated
(control) for 24 h. The cells were trypsinized, and washed three
times with phosphate-buffered-saline (PBS), and then fixed in 75%
ethanol overnight at 4°C. The fixed cells were washed three times
with PBS, incubated with 10 μl RNase for 30 min at 37°C, and
stained with 10 μl PI, followed by incubation for 30 min at
4°C in the dark. Data acquisition and analysis were performed using
a FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
The percentage of apoptitic cells and cell cycle analysis were
performed using ModFit LT software for Windows (Version V3.2;
Verity Software House, Inc., Topsham, ME, USA).
Assessment of cell morphological
changes
The PC3 cells were plated at a density of
4×105 cells/well and treated with 60 μmol/l
emodin for 24 h at 37°C in a humidified atmosphere containing 5%
CO2, washed with PBS, collected in an Eppendorf tube and
fixed with 2.5% glutaraldehyde for 30 min. The PC3 cells were then
washed with PBS and resuspended, and the ultrastructure of the PC3
cells was observed under a transmission electron microscope
(JEM2000; JEOL, Ltd., Tokyo, Japan) at 100 kV.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The PC3 cells (5×106 cells/100-mm dish)
were treated with emodin (10, 20, 40, 60 and 80 μg/ml) or
remained untreated (control) for 24 h. The total RNA was isolated
from the PC3 cells using TRIzol reagent, according to the
manufacturer's instructions. Total RNA (2 μg) was converted
into cDNA in a series of standard RT reactions using M-MLV Reverse
Transcriptase. The resulting cDNA mixture (3 μl) was then
used for enzymatic amplification. The primer sequences of Notch1,
Jagged1, VEGF, bFGF and β-actin, and the thermal cycling conditions
are shown in Table I. The reaction
products were visualized by electrophoresis on a 1.2% agarose gel
(Promega Corporation, Madison, WI, USA), containing ethidium
bromide, followed by ultraviolet (UV) light illumination using a
UVP Gel Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
 | Table IPrimers and thermal cycling conditions
used for reverse transcription-quantitative polymerase chain
reaction analysis. |
Table I
Primers and thermal cycling conditions
used for reverse transcription-quantitative polymerase chain
reaction analysis.
| Target gene | Primer sequence | Length (bp) | Thermal cycling
conditions |
|---|
| Notch1 |
5′-GACATCACGGATCATATGGA-3′
5′-CTCGCA TTGACCA TTCAAAC-3′ | 666 | 95°C 1 min, 60°C 2
min, 72°C 1.5 min, 34 cycles |
| Jaggedl |
5′-AACTGGTACCGGTGCGAA-3′
5′-TGATGCAAGATCTCCCTGAAAC-3′ | 216 | 95°C 1 min, 54°C 1
min, 72°C 1 min, 34 cycles |
| VEGF |
5′-TCGGGCCTCCGAAACCATGA-3′
5′-CCTGGTGAGAGATCTGGTTC-3′ | 516 | 94°C 30 sec, 55°C 40
sec, 72°C 1 min, 34 cycles |
| bFGF |
5′-GGAGAAGAGCGACCCACA-3′
5′-CCAGTTCGTTTCAGTGCC-3′ | 234 | 94°C 30 sec, 43°C 30
sec, 72°C 30 min, 30 cycles |
| β-actin |
5′-TCTACAATGAGCTGCGTGTG-3′
5′-CAACTAAGTCATAGTCCGCC-3′ | 878 | 95°C 1 min, 60°C 2
min, 72°C 1.5 min, 34 cycles |
Western blot analysis
The expression levels of Notch1, Jagged1, VEGF and
bFGF apoptosis-associated proteins were detected using western blot
analysis to clarify the mechanism underlying the induction of PC3
cell apoptosis by emodin, according to the manufacturer's
instructions and the methods of a previous study (16). The PC3 cells (5×106
cells/100-mm dish) were treated with emodin (10, 20, 40, 60 and 80
μg/ml) for 24 h at 37°C in a humidified atmosphere
containing 5% CO2, and the protein extracts were
prepared, as described by Schreiber et al (17). In brief, the cells were washed
twice with cold PBS and lysed in radioimmunoprecipitation assay
buffer (1X TBS, 1% Nonidet-P40, 0.5% sodium deoxycholate, 0.1% SDS,
0.004% sodium azide, 1% MSF, 1% sodium orthovanadate and 1%
protease inhibitor cocktail; Beyotime Biotechnology, Shanghai,
China) following the manufacturer's instructions. The cell lysates
were agitated for 1 h at 4°C followed by a 15 min centrifugation at
10,000 x g. The protein concentrations were determined using a
Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc.). For western
blot analysis, the proteins were subjected to 12% SDS-PAGE at 60 V
for 1 h and 120 V for 2 h. The proteins were transferred onto
polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.) at
30 mA for 1.5 h. The membranes were then blocked with 5% skimmed
milk for 1 h and incubated with goat anti-human polyclonal
anti-Notch1 (1:1,000), anti-Jagged1 (1:1,000), anti-VEGF (1:1,000)
and anti-bFGF (1:1,000) antibodies. The membranes were washed three
times in Tris-buffered saline with 0.2% Tween 20 (TBST) for 10 min
and incubated with the appropriate peroxidase-conjugated secondary
antibodies in TBS. Following another three washes with TBST for 10
min, the membranes were visual-ized using 3,3′-diaminobenzidine
(Sigma-Aldrich), and were exposed to films (Kodak, Rochester, NY,
USA). The protein levels were quantified by scanning densitometry
using an Scanning Densitometry/Image Analysis System (Scion
Corporation, Frederick, MD, USA).
Immunofluorescence and confocal
microscopy analysis
The PC3 cells were maintained on glass coverslips in
24-well plates at a density of 1×105 cells/well for 24 h
at 37°C in a humidified atmosphere containing 5% CO2.
Following treatment with emodin (0, 10, 20, 40, 60 and 80
μg/ml), the cells were fixed with ice-cold dehydrated
ethanol/acetone. Subsequently, the cells were washed with cold PBS
for 10 min and blocked with 5% normal goat serum for 30 min at room
temperature, followed by incubation with primary anti-Notch1
(1:100) overnight at 4°C. The cells were washed again and then
incubated with FITC-conjugated goat anti-rabbit secondary antibody
(1:100) for 1 h in the dark. Following several additional washing
steps, the coverslips were mounted using Fluoromount-G™ mounting
media with DAPI, and fluorescence was visualized using Leica
confocal software (Leica Microsytems, Inc., Buffalo Grove, IL,
USA).
Statistical analysis
The data are presented as the mean ± standard
deviation (n=6). Differences between experimental groups were
assessed using one-way analysis of variance. P<0.05 was
considered to indicate a statistically significant difference.
Results
Effects of emodin on PC3 cell
proliferation
The proliferation of the cells was determined using
an MTT assay. As shown in Table
II, emodin treatment inhibited the proliferation of the PC3
cells at 40, 60, 80 μmol/l significantly between 12 and 96 h
(P<0.05). Treatment with emodin inhibited PC3 cell proliferation
in a time- and dose-dependent manner.
 | Table IIEffects of emodin on PC3 cell
proliferation. |
Table II
Effects of emodin on PC3 cell
proliferation.
| Dose
(μmol/l) | Time (h)
|
|---|
| 12 | 24 | 36 | 48 | 72 | 96 |
|---|
| 0 | 0.19±0.02 | 1.09±0.06 | 3.02±0.12 | 5.07±1.02 | 6.10±1.21 | 6.65±2.12 |
| 20 | 0.32±0.12 | 2.27±0.93 | 4.48±1.93 | 8.33±0.97 | 9.92±2.09 | 12.98±2.19 |
| 40 | 3.31±0.73a | 6.13±1.42a | 12.23±2.08a | 16.88±3.12a | 25.64±2.51a | 38.42±2.94a |
| 60 | 9.29±1.95a | 17.64±1.23a | 26.59±3.62a | 39.25±3.87a | 52.32±3.41a | 63.02±5.12a |
| 80 | 16.01±2.03a | 25.21±1.59a | 38.93±3.08a | 56.77±2.80a | 68.34±4.14a | 79.13±3.34a |
Effects of emodin on PC3 cell cycle and
apoptosis
To examine the mechanism responsible for cell
proliferation inhibition, apoptosis and cell cycle distribution
were evaluated using flow cytometry. As shown in Table III, a significant increase of
apoptosis was observed at 24 h following treatment with different
doses of emodin (0–60 μmol/l) in the PC3 cells. No
significant effect on the proportion of cells in the G0/G1 phase
was observed following treatment with different concentration of
emodin (0–80 μmol/l) for 24 h, however, the proportion of
the cells in the G2/M phase increased significantly in a
dose-dependent manner, and the proportion of cells in the S phase
decreased as the concentration of emodin increased. The cell cycle
analysis results revealed G2/M phase arrest in the emodin-treated
PC3 cells. Typical morphological features of apoptotic cell death,
including cell shrinkage, membrane blebbing and nuclear
fragmentation were also observed in the majority of the
emodin-treated cells on analysis using transmission electron
microscopy. A representative image containing these features is
shown in Fig. 1.
 | Table IIIEffects of emodin on PC3 cell cycle
and apoptosis. |
Table III
Effects of emodin on PC3 cell cycle
and apoptosis.
| Dose
(μmol/l) | Apoptosis (%) | PC3 cell cycle
phase
|
|---|
| G0/G1 | S | G2/M |
|---|
| 0 | 1.17±0.47 | 45.12±1.24 | 53.91±0.93 | 0.37±0.02 |
| 10 | 2.03±0.77 | 43.96±1.91 | 49.53±1.21 | 6.51±0.31a |
| 20 | 5.32±1.97a | 44.18±1.25 | 41.72±2.12a | 14.1±1.01a |
| 40 | 10.21±1.31a | 41.01±1.93 | 36.42±1.83a | 22.57±1.54a |
| 60 | 21.15±1.54a | 43.37±1.42 | 18.24±1.39a | 38.39±1.91a |
| 80 | 15.61±1.17a | 42.71±1.76 | 11.63±1.69a | 45.66±2.07a |
Emodin increases the mRNA expression of
Notch1 and decreases the mRNA expression levels of Jagged1, VEGF
and bFGF in the PC3 cells
The mRNA expression levels of Notch1, Jagged1, VEGF
and bFGF were examined using RT-qPCR to access the effect of emodin
on the Notch pathway in PC3 cell apoptosis. As shown in Fig. 2, following treatment with different
concentrations of emodin (10–80 μmol/l) for 24 h, the mRNA
expression of Notch1 increased considerably, in a dose-dependent
manner, in the PC3 cells. However, following treatment with emodin
(10–80 μmol/l), the mRNA expression levels of Jagged1, VEGF
and bFGF decreased significantly, compared with the control (0
μmol/l) group, which also occured in a dose-dependent
manner. These results suggested that emodin may increase the mRNA
expression of Notch1 and inhibit the mRNA expression levels of
Jagged1, VEGF and bFGF at the transcriptional level in
emodin-induced PC3 cell apoptosis.
 | Figure 2Effects of emodin on the mRNA
expression levels of (A) Notch1, (B) Jagged1, (C) VEGF and (D) bFGF
in the PC3 cells. Following treatment with or without emodin (0,
10, 20, 40, 60 and 80 μmol/l), the mRNA expression levels of
Notch1, Jagged1, VEGF and bFGF in the PC3 cells were assessed using
western blot analysis. β-actin was used to normalize the quantity
of cDNA. VEGF, vascular endothelial growth factor; bFGF, basic
fibroblast growth factor. |
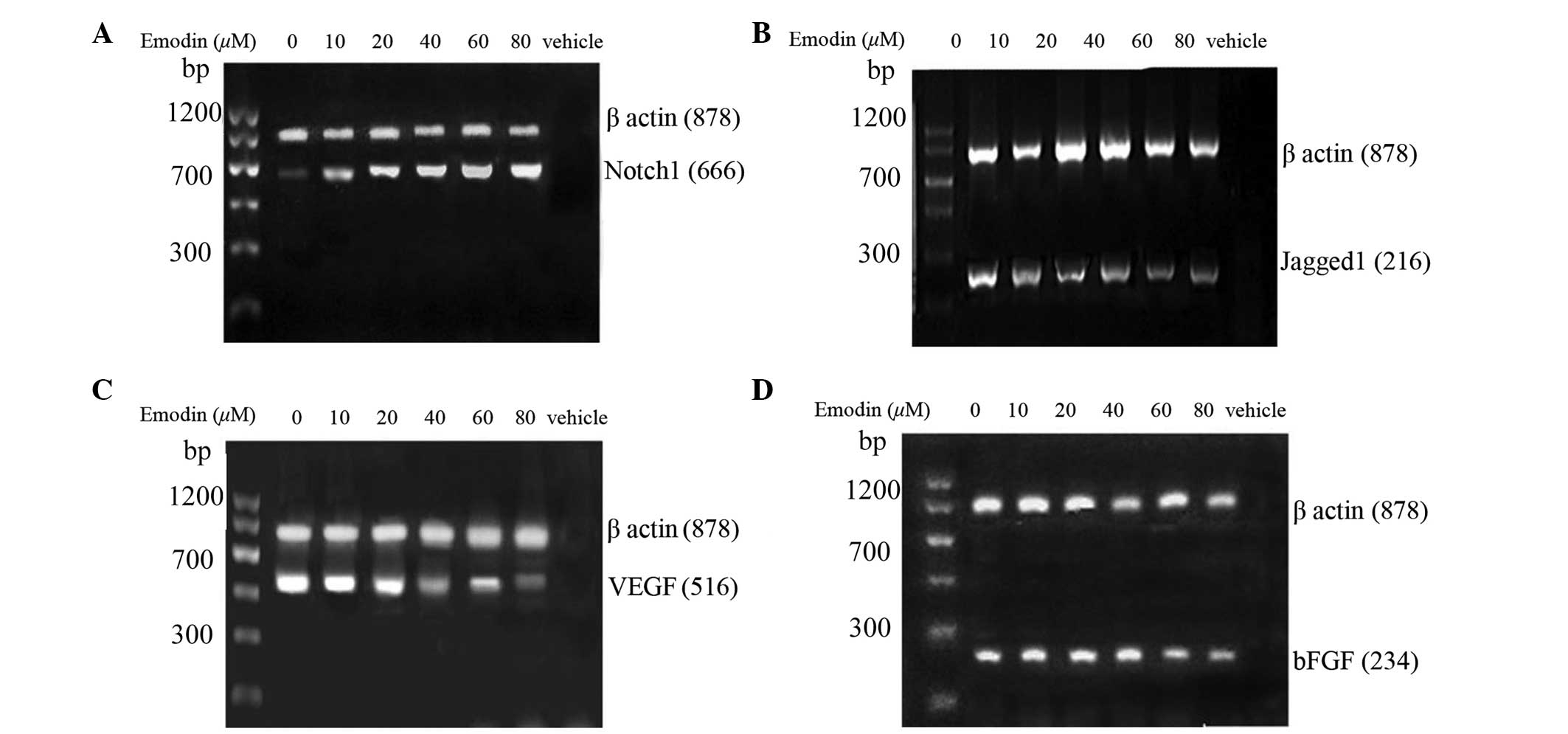
Emodin increases the protein expression
of Notch1 and decreases protein expression levels of Jagged1, VEGF
and bFGF in PC3 cells
The effect of emodin on the protein expression
levels of Notch1, Jagged1, VEGF and bFGF in the PC3 cells were
examined using western blot analysis. As shown in Fig. 3, treatment with emodin (10–80
μmol/l) for 24, led to a significant increase in the protein
expression of Notch1 and a significant decrease in the protein
expression levels of Jagged1, VEGF and bFGF. The increasing mRNA
and protein expression levels of Notch1 and the decreasing mRNA and
protein expression levels of Jagged1, VEGF and bFGF indicated that
the Notch signaling pathway is involved in emodin-suppressing
prostate tumor mechanisms.
 | Figure 3Effects of emodin on the protein
expression levels of (A) Notch1, (B) Jagged1, (C) VEGF and (D) bFGF
in the PC3 cells. Following treatment with or without (control)
emodin (0, 10, 20, 40, 60 and 80 μmol/l), the protein
expression levels of Notch1, Jagged1, VEGF and bFGF in PC3 cells
were assessed using western blot analysis. The data are expressed
as the mean ± standard deviation. ##P<0.01 and
#P<0.05, vs. control group. VEGF, vascular
endothelial growth factor; bFGF, basic fibroblast growth
factor. |
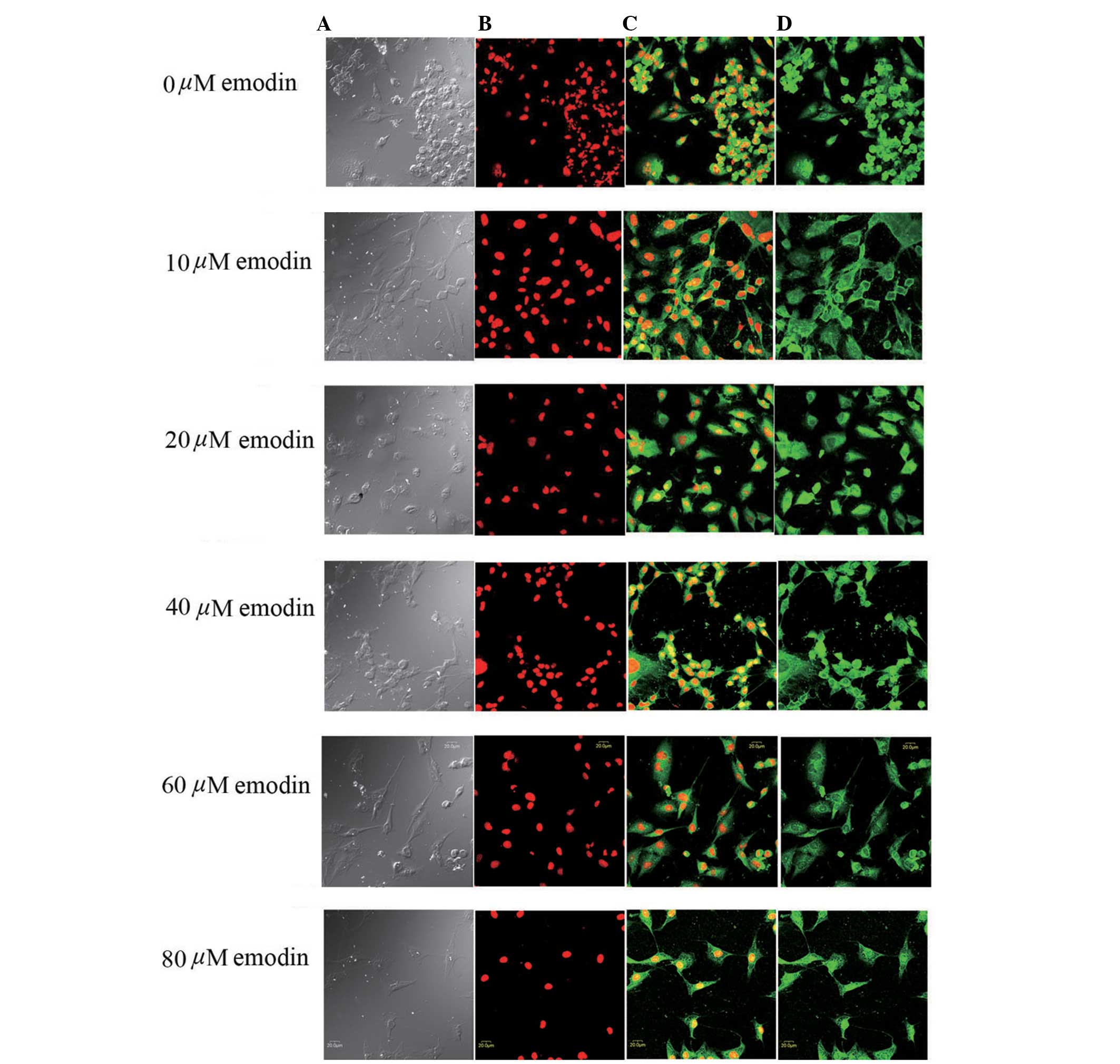
Immunofluorescence and confocal
microscopic analysis of Notch1 subcellular localization
The effects of emodin on the expression and
subcellular localization of Notch1 receptor were examined using
immunofluorescence and confocal microscopy. The confocal microscopy
revealed pseudopodal shrinkage, cells turned round with poor
adherence, clustering and apoptosis in the PC3 cells following
treatment with emodin (0–80 μmol/l) for 24 h (Fig. 4). The protein expression of Notch1
was detected in all the groups of PC3 cells, however, the
fluorescent intensity increased gradually following treatment with
emodin for 24 h, which occurred in a dose-dependent manner. The
fluorescence intensities of the PC3 cells treated with emodin at 0,
10, 20, 40, 60 and 80 μmol/l, were 8.73±2.54, 15.31±3.47,
22.64±4.34, 66.32±12.92, 112.64±16.26 and 163.21±19.18,
respectively. Treatment with emodin (10-80 μmol/l) increased
the fluorescence intensity significantly, compared with the control
group (P<0.05). The intracellular location assay revealed
mottled expression of Notch1 in the nucleus in addition to the
cytoplasm and cell membrane. Notch1 localization was gradually
increased in the nuclear membrane and nucleus at emodin
concentrations >40 μmol/l (Fig. 4). It has been suggested that
nuclear translocation or activation of Notch1 is occurring when
nuclear fluorescence intensity is increased (18).
Discussion
Emodin is a natural anthraquinone derivative,
isolated from Rheum palmatum L (19). Pharmacological studies have
demonstrated that emodin has antitumor, antibacterial, diuretic and
vasorelaxant effects, however, its antitumor activity has received
more attention (20–23). The curative effects and mechanisms
of emodin in prostate cancer cells have been previously
investigated to a certain extent, and include the inhibition of
proliferation and induction of apoptosis (24), regulation of the cell cycle
(25), enhanced cytotoxicity of
chemotherapeutic drugs in cells (26), suppression of angiogenesis and
metastasis (27) and
downregulation of androgen receptors (13). However, the molecular mechanisms
underlying the induction of apoptosis by emodin in prostate cancer
cells remain to be fully elucidated.
Notch-mediated cell-cell interaction and signaling
are important for stem cell maintenance, cell fate determination,
cell proliferation, differentiation and apoptosis in a variety of
tissues (28). Cross talk between
Notch and nuclear factor-κB (NF-κB) has been extensively
investigated, and has been found to have important roles in tissue
development and disease progression (29,30).
It has been revealed that the gene expression of Notch1 and its
effector, Hey-1, is significantly downregulated in human prostate
adenocarcinoma tissue, compared with normal prostate tissue, and
constitutively overexpressed active Notch1 can inhibit the
proliferation of various prostate cancer cell lines (31,32).
The results of the present study suggested that emodin upregulated
the expression of Notch1, and downregulated the expression levels
of Jagged1, VEGF and bFGF, which may have contributed to the
induction of proliferation inhibition and apoptosis in the PC3
cells.
The basic reason for the malignant proliferation of
tumor cells is cell cycle imbalance. Notch1 inhibits the expression
of antiapoptotic proteins and cell proliferation-associated
proteins, including the downregulation of cyclin D1 and Bcl-2, and
the upregulation of p21Cip1 and p27Kip1, which induce cell cycle
arrest and apoptosis (33). In the
present study, the results demonstrated that upregulated expression
of Notch1 suppressed proliferation of the PC3 cells by inducing
G2/M arrest and inducing apoptosis in a time- and dose-dependent
manner. VEGF and bFGF are the most important factors in the
promotion of endothelial angiogenesis and differentiation, and can
be found in several types of solid tumor, including mammary cancer,
hepatocarcinoma, gastric carcinoma, rectal cancer and prostate
cancer (34). Previous evidence
indicates that Notch signaling is important in determining the way
in which an endothelial cell responds to VEGF (35). The results of the present study
suggested that the Notch pathway is involved in downregulating the
expression levels of VEGF and bFGF, which may contribute to the
inhibition of prostate cancer angiogenesis and metastasis by
emodin.
The present study demonstrated that the Notch
pathway was involved in the inhibition proliferation by emodin on
PC3 cells. Regarding the localization of Notch1, as the emodin
concentration increased, the distribution in the nuclear membrane
and nucleus was gradually strengthened, which indicated the nuclear
translocation or activation of Notch1.
In conclusion, emodin significantly inhibited PC3
human prostate cancer cell growth and induced apoptosis, in which
the Notch1 and Jagged1-mediated classical pathway was important.
Emodin may be a prospective candidate drug for HRPC and merits
further investigation.
References
|
1
|
McDonnell TJ, Troncoso P, Brisbay SM,
Logothetis C, Chung LW, Hsieh JT, Tu SM and Campbell ML: Expression
of the protooncogene bcl-2 in the prostate and its association with
emergence of androgen-independent prostate cancer. Cancer Res.
52:6940–6944. 1992.PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Petrylak DP: Chemotherapy for advanced
hormone refractory prostate cancer. Urology. 54(Suppl): 30–35.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pisters LL: The challenge of locally
advanced prostate cancer. Semin Oncol. 26:202–216. 1999.PubMed/NCBI
|
|
5
|
Richie JP: Anti-androgens and other
hormonal therapies for prostate cancer. Urology. 54(Suppl): 15–18.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu CX, Zhang XQ, Kang LD, Zhang PJ, Chen
WW, Liu WW, Liu QW and Zhang JY: Emodin induces apoptosis in human
prostate cancer cell LNCaP. Asian J Androl. 10:625–634. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cragg GM and Newman DJ: Plants as a source
of anti-cancer agents. J Ethnopharmacol. 100:72–79. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin SZ, Wei WT, Chen H, Chen KJ, Tong HF,
Wang ZH, Ni ZL, Liu HB, Guo HC and Liu DL: Antitumor activity of
emodin against pancreatic cancer depends on its dual role:
Promotion of apoptosis and suppression of angiogenesis. PLoS One.
7:e421462012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su YT, Chang HL, Shyue SK and Hsu SL:
Emodin induces apoptosis in human lung adenocarcinoma cells through
a reactive oxygen species-dependent mitochondrial signaling
pathway. Biochem Pharmacol. 70:229–241. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen YC, Shen SC, Lee WR, Hsu FL, Lin HY,
Ko CH and Tseng SW: Emodin induces apoptosis in human
promyeloleukemic HL-60 cells accompanied by activation of caspase 3
cascade but independent of reactive oxygen species production.
Biochem Pharmacol. 64:1713–1724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Damodharan U, Ganesan R and Radhakrishnan
UC: Expression of MMP2 and MMP9 (gelatinases A and B) in human
colon cancer cells. Appl Biochem Biotechnol. 165:1245–1252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo HC, Bu HQ, Luo J, Wei WT, Liu DL, Chen
H, Tong HF, Wang ZH, Wu HY, Li HH, et al: Emodin potentiates the
antitumor effects of gemcitabine in PANC-1 pancreatic cancer
xenograft model in vivo via inhibition of inhibitors of apoptosis.
Int J Oncol. 40:1849–1857. 2012.PubMed/NCBI
|
|
13
|
Cha TL, Qiu L, Chen CT, Wen Y and Hung MC:
Emodin down-regulates androgen receptor and inhibits prostate
cancer cell growth. Cancer Res. 65:2287–2295. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shou J, Ross S, Koeppen H, de Sauvage FJ
and Gao WQ: Dynamics of notch expression during murine prostate
development and tumorigenesis. Cancer Res. 61:7291–7297.
2001.PubMed/NCBI
|
|
15
|
Carvalho FL, Simons BW, Eberhart CG and
Berman DM: Notch signaling in prostate cancer: A moving target.
Prostate. 74:933–945. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Y, Yang LF, Ye M, Gu HH and Cao Y:
Induction of apoptosis by epigallocatechin-3-gallate via
mitochondrial signal transduction pathway. Prev Med. 39:1172–1179.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schreiber E, Harshman K, Kemler I,
Malipiero U, Schaffner W and Fontana A: Astrocytes and glioblastoma
cells express novel octamer-DNA binding proteins distinct from the
ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res.
18:5495–5503. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weijzen S, Rizzo P, Braid M, Vaishnav R,
Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC,
et al: Activation of Notch-1 signaling maintains the neoplastic
phenotype in human Ras-transformed cells. Nat Med. 8:979–986. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsai T and Chen C: Ultraviolet spectrum
identification of emodin in rabbit plasma by HPLC and its
pharmacokinetics application. Asia Pac J Pharmacol. 1:53–56.
1992.
|
|
20
|
Koyama M, Kelly TR and Watanabe KA: Novel
type of potential anticancer agents derived from chrysophanol and
emodin. Some structure-activity relationship studies. J Med Chem.
31:283–284. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang HC, Chu SH and Chao PD:
Vasorelaxants from Chinese herbs, emodin and scoparone, possess
immunosuppressive properties. Eur J Pharmacol. 198:211–213. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou XM and Chen QH: Biochemical study of
Chinese rhubarb. XXII. Inhibitory effect of anthraquinone
derivatives on Na+-K+-ATPase of the rabbit renal medulla and their
diuretic action. Yao Xue Xue Bao. 23:17–20. 1988.In Chinese.
PubMed/NCBI
|
|
23
|
Lee HZ: Effects and mechanisms of emodin
on cell death in human lung squamous cell carcinoma. Br J
Pharmacol. 134:11–20. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jing X, Ueki N, Cheng J, Imanishi H and
Hada T: Induction of apoptosis in hepatocellular carcinoma cell
lines by emodin. Jpn J Cancer Res. 93:874–882. 2002. View Article : Google Scholar
|
|
25
|
Shieh DE, Chen YY, Yen MH, Chiang LC and
Lin CC: Emodin-induced apoptosis through p53-dependent pathway in
human hepatoma cells. Life Sci. 74:2279–2290. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang XZ, Wang J, Huang C, Chen YY, Shi
GY, Hu QS and Yi J: Emodin enhances cytotoxicity of
chemotherapeutic drugs in prostate cancer cells: The mechanisms
involve ROS-mediated suppression of multidrug resistance and
hypoxia inducible factor-1. Cancer Biol Ther. 7:468–475. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kwak HJ, Park MJ, Park CM, Moon SI, Yoo
DH, Lee HC, Lee SH, Kim MS, Lee HW, Shin WS, et al: Emodin inhibits
vascular endothelial growth factor-A-induced angiogenesis by
blocking receptor-2 (KDR/Flk-1) phosphorylation. Int J Cancer.
118:2711–2720. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou ZD, Kumari U, Xiao ZC and Tan EK:
Notch as a molecular switch in neural stem cells. IUBMB Life.
62:618–623. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lai EC: Notch signaling: Control of cell
communication and cell fate. Development. 131:965–973. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ang HL and Tergaonkar V: Notch and
NFkappaB signaling pathways: Do they collaborate in normal
vertebrate brain development and function? BioEssays. 29:1039–1047.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chau MD, Tuft R, Fogarty K and Bao ZZ:
Notch signaling plays a key role in cardiac cell differentiation.
Mech Dev. 123:626–640. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang XD, Leow CC, Zha J, Tang Z, Modrusan
Z, Radtke F, Aguet M, de Sauvage FJ and Gao WQ: Notch signaling is
required for normal prostatic epithelial cell proliferation and
differentiation. Dev Biol. 290:66–80. 2006. View Article : Google Scholar
|
|
33
|
Yao J, Duan L, Fan M, Yuan J and Wu X:
Notch1 induces cell cycle arrest and apoptosis in human cervical
cancer cells: Involvement of nuclear factor kappa B inhibition. Int
J Gynecol Cancer. 17:502–510. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seghezzi G, Patel S, Ren CJ, Gualandris A,
Pintucci G, Robbins ES, Shapiro RL, Galloway AC, Rifkin DB and
Mignatti P: Fibroblast growth factor-2 (FGF-2) induces vascular
endothelial growth factor (VEGF) expression in the endothelial
cells of forming capillaries: An autocrine mechanism contributing
to angiogenesis. J Cell Biol. 141:1659–1673. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Siekmann AF, Covassin L and Lawson ND:
Modulation of VEGF signalling output by the Notch pathway.
BioEssays. 30:303–313. 2008. View Article : Google Scholar : PubMed/NCBI
|