Introduction
Osteonecrosis of the femoral head (ONFH) is a
debilitating bone disease, characterized by cellular death in the
bone tissue that can lead to collapse of the architectural bone
structure and loss of joint function (1). Although several conditions, including
steroids, alcoholism, coagulation defects, storage diseases,
marrow-infiltrating diseases and certain autoimmune diseases, can
increase the risk of ONFH, the pathogenesis remains to be fully
elucidated (1–5). Several hypotheses regarding its
pathogenesis, including intravascular coagulation (6,7),
apop-tosis of osteoblasts and osteocytes (8), and fatty necrosis of osteocytes
(9), have been suggested. Among
them, a vascular hypothesis is considered to be the most
persuasive, which implicates a decrease in blood flow to the local
femoral head in the pathogenesis of ONFH (10).
In support of this hypothesis, genetic mutations
associated with coagulation abnormalities, including the factor V
(F5) Leiden mutation (G1691A; Arg506Gln) and
5,10-methylene-tetrahydrofolate reductase gene polymorphism (C677T;
Ala222Val), have been used to assess the role of genetics in the
risk of ONFH (6,7). Of four studies investigating the role
of the F5 Leiden mutation, three demonstrated a positive
correlation with ONFH in Caucasian patients (6,11,12).
However, Lee et al (13)
reported no association between ONFH and increased thrombosis or
impaired fibrinolysis in Korean patients with ONFH. Additionally,
other studies have reported the absence of F5 Leiden and
20210A mutations in Koreans (14–16).
It is possible that there are geographic and ethnic differences in
the prevalence of these mutations.
Although the F5 Leiden mutation has not been
observed in the Korean population, the F5 gene is considered
to be important in ONFH. Therefore, the present study performed
extensive screening of the F5 gene by direct sequencing to
identify the polymorphisms and mutations, and examined the genetic
association with ONFH risk in a Korean population.
Patients and methods
Patients
Unrelated patients, diagnosed with ONFH (n=423; 342
male and 81 female; age, 47.0±14.0 years) and unrelated control
subjects (n=348; 298 male and 50 female; age, 51.9±10.7 years) were
recruited in the present study. All individuals were consecutively
enrolled at Kyungpook National University Hospital (Daegu, Korea)
between 2002 and 2006, and provided informed consent. The present
study was approved by the ethics committee of Kyungpook National
University Hospital (Daegu, Korea). The patients were diagnosed
using anteroposterior and lateral pelvic radiographs
(Innovision-SH; DK, Seoul, Korea), and magnetic resonance images
(Signa 1.5 Excite; GE Healthcare, Little Chalfont, UK). The
patients with available genotype data were sub-grouped, according
to etiological factors (17) into
alcohol-induced (206 cases), steroid-induced (77 cases) and
idiopathic (140 cases) ON groups. The controls were defined by a
lack of hip pain and by the absence of any lesion with a sclerotic
margin or subchondral collapse consistent with ONFH in the
anteroposterior and frog leg lateral pelvic radiographs. Any
relations of the patients were excluded from the control group.
Sequencing analysis of the human F5
gene
All 25 exons of the F5 gene and their
boundaries, including the promoter region (~1.5 kb), were sequenced
to identify single nucleotide polymorphisms (SNPs) in the 24 Korean
DNA samples using a MegaBace1000 DNA analyzer (GE Healthcare) with
a DYEnamic ET Dye terminator kit (GE Healthcare). The primer 3
online primer design tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi)
was used to design 35 primer sets of the F5 gene for PCR
amplification and sequencing analysis, based on GenBank (http://www.ncbi.nlm.nih.gov/genbank/)
sequences (Reference genome sequence for F5; NT_004487.18).
The sequence variants were confirmed using chromatograms.
Genotyping
Genomic DNA was isolated from the peripheral blood
of each individual using a FlexiGene DNA kit (Qiagen, Valencia, CA,
USA). The DNA concentration was determined using a Nanodrop 2000
spectrophotometer (Thermo Fisher Scientific, Waltham, MA USA). The
genotype was determined using a TaqMan™ fluorogenic 5′-nuclease
assay with pre-designed or custom TaqMan primer/probe sets (Applied
Biosystems, Foster City, CA, USA). The primer and probe sequences
are indicated in Table I. One
allelic probe was labeled with 6-carboxyfluorescein (FAM)™ dye and
the other was labeled with fluorescent VIC® dye.
Genotyping was performed using TaqMan Universal master mix (Applied
Biosystems) with PCR primer concentrations of 900 nM and TaqMan MGB
probe concentrations of 200 nM and ABI 7500 real-time PCR system
(Applied Biosystems). The final reaction volume for PCR was 10 μl
containing 20 ng of genomic DNA. The thermal cycle conditions were
as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15
sec and at 60°C for 1 min as previously described (18). The fluorescence data files from
each plate were collected and analyzed using automated
allele-calling software (SDS 2.2; Applied Biosystems). Quality
control of the genotyping was performed on 10% of the samples by
examining duplicates (rate of concordance in duplicates
>99%).
 | Table ISequences of primer and Taqman probes
for genotyping of F5 single nucleotide polymorphisms |
Table I
Sequences of primer and Taqman probes
for genotyping of F5 single nucleotide polymorphisms
| TaqMan®
genotyping assays | Probes (ABI) | Context Sequence
(VIC/FAM) |
|---|
| rs6028 | C_8919464_20 |
TTTCAGGCTTACCTGAAATGGTAGA(C/T)T
GTGGTTTTTCTTTCTTAAAATATG |
| rs6029 | C_11975262_10 |
GAGCCACAGCGTCGTCCATCTTCTC(C/T)G
CAGGGAATGTGTGGTCAAGGTAAG |
| rs6022 | C_11975261_10 |
AGATAAGCAGGGGCCCAATCAGCCC(A/C)G
AGTTGAAATCCTCGATCAGATTTT |
| rs6032 | C_8919441_30 |
AGTAACAGATCACTAGGAGGGTCCT(C/T)C
CAGGGCCTCATTCTGGAAGGAGAA |
|
| Custom
Taqman® genotyping assays | Primer, Probe | Sequences |
|
| rs6672595 | Forward
Reverse
VIC
FAM |
GTGGTGTCACTACTGCAGTCA
TGAGAAGGGTTTGGCTGTGATTTT
TCCTACGTGTATACTACC
TCCTACATGTATACTACC |
| rs6672595 | Forward
Reverse
VIC
FAM |
GCCTTTGACCTCTTGCTTAAAAATGT
CCAATCCTCACTTCCAGTCCAAA
ACTTGGTGATGAAATC
CTTGGTGATAAAATC |
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) was used to
determine significant deviation of the genotypic frequency from
each SNP using a χ2 test. Statistical significance was
determined using the P-values obtained from the logistical
regression analysis, controlling for age and gender as covari-ates
using three alternative models (co-dominant, dominant and
recessive). To assess the risk of the phenotypes, odds ratios and
95% confidence intervals were also estimated using a logistic
regression procedure. The linkage disequilibrium (LD) between the
loci was measured using the absolute value of Lewontin's D′ (|D′|)
(19).
The haplotype structures and frequencies were
estimated from the genotyped data within the LD block using
Haploview 3.32 (http://www.broad.mit.edu/mpg/haploview/), which
estimates haplotypes using an accelerated expectation–maximization
(EM) algorithm (20). Fisher's
exact or χ2 test was used to compare the frequency of
discrete variables between the controls and the patients.
Continuous variables were compared using Student's t-test or
analysis of variance. Statistical analyses were performed using SAS
9.1 software (SAS Institute Inc., Cary, NC, USA) and P<0.05 was
considered to indicate a statistically significant difference.
Results
A total of 423 unrelated patients with ONFH and 348
control individuals were recruited for investigation in the present
study. The clinical characteristics of the controls and patients
are summarized in Table II. The
data revealed no significant differences between the patients and
the controls, with the exception of the mean age (P<0.05).
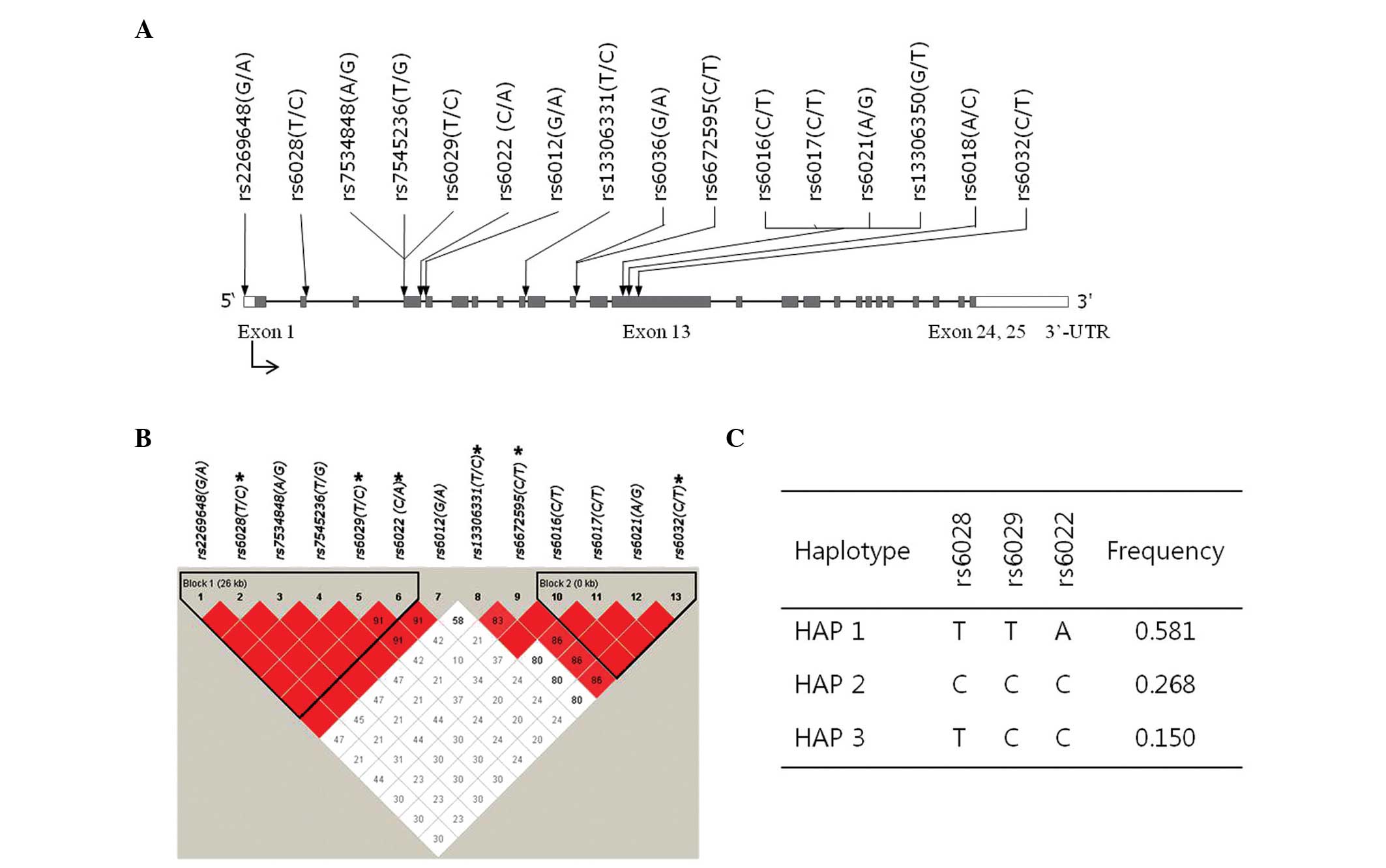
Direct DNA sequencing of 24 individuals revealed 16 SNPs within the
exons and flanking regions of F5, including a 1.5-kb
promoter region, 10 in the exons, five in the exonintron boundary
and one in the promoter region (Fig.
1A). The frequencies of the identified SNPs in the Korean
population are shown in Table
III. Of the SNPs, rs6036, rs13306350 and rs6018 were excluded
from further analysis, as they did not fulfill the criteria of a
call rate >95.0, minor allele frequency >0.05 and
HWE>0.01. Pair-wise comparisons among the 13 SNPs revealed two
haplotype blocks, with each block exhibiting a marked LD spine
(Fig. 1B). From these
polymorphisms, six were selected for larger-scale genotyping by
considering their location, allele frequencies and LD coefficients
among polymorphisms (Table III),
and were genotyped for association analysis in the 423 patients and
348 controls. Logistic regression analysis in all analysis models
(co-dominant, dominant and recessive) revealed no significant
association between the individual F5 SNPs and the risk of
ONFH (Table IV).
 | Table IIClinical profiles of patients with
osteonecrosis of the femoral head and healthy controls. |
Table II
Clinical profiles of patients with
osteonecrosis of the femoral head and healthy controls.
| Characteristic | Control
(n=348) | Patients
|
|---|
Total
(n=423) |
Idiopathic
(n=140) |
Alcohol-induced
(n=206) |
Steroid-induced
(n=77) |
|---|
| Age (mean ±
SD)a | 51.9±10.7 | 47.0±14.0 | 43.9±15.2 | 50.6±12.5 | 42.9±13.4 |
| Gender
(male/female) | 298/50 | 342/81 | 91/49 | 198/8 | 53/24 |
| Involvement
(uni/bilateral) | 0 | 182/241 | 80/60 | 74/132 | 28/49 |
| BMI
(kg/m2; mean ± SE) | 23.8±0.17 | 24.1±0.63 | 23.4±0.38 | 24.5±0.89 | 22.9/0.53 |
 | Table IIIFrequency of factor V gene
polymorphisms in a Korean population by sequencing and/or
genotyping. |
Table III
Frequency of factor V gene
polymorphisms in a Korean population by sequencing and/or
genotyping.
| Locus | Amino acid
change | Position | rs number | Genotype
| MAF | HWE |
|---|
| CC | CR | RR | N |
|---|
| −426 G>A | | 5′-UTR | rs2269648 | 8 | 11 | 5 | 24 | 0.438 | 0.915 |
| +3943 T>C | Gln79Gln | Exon 2 | rs6028 | 399 | 301 | 46 | 746 | 0.263 | 0.890 |
| +25532 A>G | | Intron 3 | rs7534848 | 8 | 11 | 5 | 24 | 0.438 | 0.915 |
| +25555 T>G | | Intron 3 | rs7545236 | 8 | 11 | 5 | 24 | 0.438 | 0.915 |
| +25652 T>C | Ala135Ala | Exon 4 | rs6029 | 265 | 346 | 142 | 753 | 0.418 | 0.023 |
| +25799 C>A | Ser184Ser | Exon 4 | rs6022 | 269 | 354 | 143 | 766 | 0.418 | 0.035 |
| +27045 G>A | | Intron 4 | rs6012 | 9 | 10 | 5 | 24 | 0.417 | 0.692 |
| +35860 T>C | | Intron 9 |
rs13306331 | 311 | 381 | 74 | 766 | 0.345 | 0.129 |
| +39899 G>A | Glu572Glu | Exon 11 | rs6036 | 23 | 1 | 0 | 24 | 0.021 | 0.000 |
| +40089 C>T | | Intron 11 |
rs6672595 | 472 | 266 | 25 | 763 | 0.207 | 0.579 |
| +43505 C>T | Ile736Ile | Exon 13 | rs6016 | 11 | 9 | 2 | 22 | 0.295 | 0.728 |
| +43532 T>C | Asn745Asn | Exon 13 | rs6017 | 12 | 10 | 2 | 24 | 0.292 | 0.710 |
| +43598 A>G | Ser767Ser | Exon 13 | rs6021 | 10 | 9 | 2 | 21 | 0.310 | 0.682 |
| +43640 T>G | Ser781Arg | Exon 13 | rs13306350 | 22 | 2 | 0 | 24 | 0.042 | 0.023 |
| +43747 A>C | Asn817Thr | Exon 13 | rs6018 | 23 | 1 | 0 | 24 | 0.021 | 0.000 |
| +44070 C>T | Glu925Lys | Exon 13 | rs6032 | 442 | 280 | 20 | 742 | 0.216 | 0.180 |
 | Table IVAssociation analyses of factor V gene
polymorphisms between patients with osteonecrosis of the femoral
head and healthy controls. |
Table IV
Association analyses of factor V gene
polymorphisms between patients with osteonecrosis of the femoral
head and healthy controls.
| rs number | Position | Genotype | Frequency
| Co-dominant
| Dominant
| Recessive
| Allele
|
|---|
| Control (%) | Patient (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| rs6028 | Exon 2 | TT | 180 (53.25) | 219 (53.68) | | | | | | | | |
| TC | 134 (39.65) | 167 (40.93) | 1.04
(0.83–1.32) | 0.7189 | 0.98
(0.74–1.31) | 0.9084 | 0.75
(0.41–1.36) | 0.3356 | 0.95
(0.75–1.19) | 0.6848 |
| CC | 24 (7.10) | 22 (5.39) | | | | | | | | |
| rs6029 | Exon 4 | TT | 122 (35.47) | 143 (34.96) | | | | | | | | |
| TC | 148 (43.02) | 198 (48.41) | 1.00
(0.82–1.21) | 0.9838 | 1.02
(0.76–1.38) | 0.8858 | 0.73
(0.50–1.05) | 0.0885 | 0.91
(0.74–1.12) | 0.4201 |
| CC | 74 (21.51) | 68 (16.63) | | | | | | | | |
| rs6022 | Exon 4 | AA | 124 (35.63) | 145 (34.69) | | | | | | | | |
| AC | 151 (43.39) | 203 (48.57) | 0.96
(0.79–1.17) | 0.6878 | 1.04
(0.77–1.40) | 0.7854 | 0.76
(0.53–1.09) | 0.1353 | 0.93
(0.76–1.15) | 0.5502 |
| CC | 73 (20.98) | 70 (16.75) | | | | | | | | |
| rs13306331 | Intron 9 | CC | 141 (40.87) | 170 (40.38) | | | | | | | | |
| CT | 169 (48.99) | 212 (50.36) | 1.01
(0.81–1.27) | 0.897 | 1.02
(0.76–1.36) | 0.8908 | 0.90
(0.56–1.46) | 0.6813 | 0.99
(0.80–1.23) | 0.9791 |
| TT | 35 (10.15) | 39 (9.26) | | | | | | | | |
| rs6672595 | Intron 11 | CC | 214 (61.49) | 258 (62.17) | | | | | | | | |
| CT | 120 (34.48) | 146 (35.18) | 0.97
(0.75–1.25) | 0.7851 | 0.97
(0.72–1.30) | 0.8485 | 0.65
(0.29–1.45) | 0.2922 | 0.94
(0.73–1.20) | 0.6687 |
| TT | 14 (4.02) | 11 (2.65) | | | | | | | | |
| rs6032 | Exon 13 | TT | 205 (59.94) | 237 (59.25) | | | | | | | | |
| TC | 125 (36.55) | 155 (38.75) | 1.09
(0.84–1.42) | 0.5084 | 1.03
(0.77–1.38) | 0.8483 | 0.56
(0.23–1.39) | 0.2117 | 0.98
(0.76–1.25) | 0.8986 |
| CC | 12 (3.51) | 8 (0.02) | | | | | | | | |
Haplotype frequencies were estimated from the
genotyped SNP data within LD block 1 using the EM algorithm. This
revealed three common haplotypes (frequency>0.05) accounting for
>99% of the observed haplotypes (Fig. 1C), which were used for subsequent
association analysis. No significant association was observed
between the haplotypes and the risk of ONFH (Table V).
 | Table VAssociation analysis of factor V gene
haplotypes in patients with osteonecrosis of the femoral head and
healthy controls. |
Table V
Association analysis of factor V gene
haplotypes in patients with osteonecrosis of the femoral head and
healthy controls.
| Locus | Genotype | Frequency
| Co-dominant
| Dominant
| Recessive
|
|---|
| Control (%) | Patient (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| HAP 1 | −/− | 74
(21.20) | 70
(16.59) | | | | | | |
| T-T-A | ht1/− | 151 (43.27) | 207 (49.05) | 1.07
(0.88–1.31) | 0.505 | 1.35
(0.94–1.95) | 0.102 | 0.95
(0.71–1.28) | 0.734 |
| ht1/ht1 | 124 (35.53) | 145 (34.36) | | | | | | |
| HAP 2 | −/− | 184 (52.72) | 225 (53.32) | | | | | | |
| C-C-C | ht2/− | 139 (39.83) | 172 (40.76) | 0.95
(0.75–1.19) | 0.635 | 0.98
(0.73–1.30) | 0.869 | 0.78
(0.44–1.38) | 0.397 |
| ht2/ht2 | 26 (7.45) | 25 (5.92) | | | | | | |
| HAP 3 | −/− | 248 (71.06) | 307 (72.75) | | | | | | |
| T-C-C | ht3/− | 94 (26.93) | 107 (25.36) | 0.93
(0.70–1.24) | 0.618 | 0.92
(0.67–1.26) | 0.603 | 0.94
(0.34–2.63) | 0.912 |
| ht3/ht3 | 7 (2.01) | 8 (1.90) | | | | | | |
Discussion
Among the pathogenic mechanisms of ONFH that have
been suggested to date, a vascular hypothesis is considered to be
the most compelling. It suggests that, if thrombosis occurs, it is
followed by a sequential process of blood flow obstruction,
increased venous pressure, impaired arterial flow, osseous hypoxia
and bone death (11,21), which appear to be important in the
development of ONFH. Previous studies have claimed that
thrombophilia due to inherited defects, including protein C
deficiency, protein S deficiency or activated protein C resistance,
as a result of the F5 Leiden mutation, may result in venous
occlusion of the femoral head, leading to ONFH in adults or
Legg-Calve-Perthes disease in children (22,23).
F5 Leiden generates coagulation F5, which is degraded
less effectively by activated protein C, resulting in a
hypercoagulable state.
Although an increased tendency for intravascular
coagulation has been suggested as a pathogenic mechanism of ONFH,
the association between genetic predisposition and thrombotic
tendency may differ between ethnic groups. A number of previous
studies have reported that the F5 Leiden mutation (G1691A;
Arg506Gln) increases the risk of primary ON (6,11,12),
however other studies have failed to observe these associations
(24,25). Additionally, neither F5
Leiden, nor the prothrombin G20210A mutation have been identified
in the Korean population (14,16).
Although the F5 Leiden mutation is not
observed in Koreans, it is likely that the F5 gene is
important with respect to ONFH. Therefore, the present study aimed
to identify novel common SNPs in the Korean population and to
perform an association analysis. In 24 samples, >90% of common
SNPs with a frequency of >0.05 were expected (26). Among the 16 SNPs identified, 6 SNPs
were genotyped. Comparison between the ONFH patients and control
subjects revealed no significant differences in the distribution of
the F5 genotypes and haplotypes, suggesting that F5
polymorphisms are not involved in susceptibility to ONFH in the
Korean population.
SNP markers provide the primary data for population
investigations. At present, >10,000,000 SNPs have been
identified in the human genome. A large portion of these SNPs have
a frequency <5% and are, therefore, private or common in only a
single population. The fixation index (FST) is a measure
of population differentiation due to genetic structure. Previous
data suggested the population exhibiting the highest level of
differentiation worldwide as the Amerindians (FST;
0.295), whereas no difference was observed among populations from
East Asian (FST; 0.01), including Koreans, Japanese and
Chinese populations (27,28). The F5 Leiden (G1691A) allele
frequency among European populations and American population of
European descent is high (2–8%) (29,30),
whereas those among native populations of the Far East or Africa
are markedly lower or even absent (30,31).
F5 Leiden and prothrombin G20210A mutations, which have been
associated with ONFH in Caucasian or other population, were not
observed in previous investigations of Korean populations (14–16),
nor were they observed in the population in present study.
As described above, previous studies have suggested
that thrombotic and fibrinolytic disorders may be etiological
causes of ONFH (5,32). However, in one small case-control
investigation of a Korean population, no significant differences
were observed in the levels of several thrombotic factors,
including protein C and S activity, antithrombin and
anticardiolipin antibody, and fibrinolytic factors, including
tissue plasminogen activator and plasminogen activator inhibitor-1,
and the data failed to confirm an etiological role for thrombotic
and fibrino-lytic disorders in East Asian patients with ONFH
(13). A possible explanation for
the conflicting results between populations may lie in geographic
and ethnic differences in the prevalence of disease and/or
associated SNPs.
In conclusion, the present study performed direct
sequencing to detect polymorphisms, and case-control association
analyses in patients with ONFH and normal control individuals using
six selected SNPs of the F5 gene. No evidence was obtained
to support an association of F5 genetic polymorphisms with
ONFH haplotypes. Although it is important in Caucasian individuals,
the results of the present study suggested that coagulation
F5 is not a genetic risk factor for ONFH in the Korean
population. Further investigations using larger sample sizes and
detailed coagulation profiling are necessary to fully assess the
signifi-cance of this gene in ONFH.
Acknowledgments
This study was supported by a grant from the Korea
Health Technology R&D Project through the Korea Health Industry
Development Institute, funded by the Ministry of Health &
Welfare, Republic of Korea (grant no. HI15C0001).
References
|
1
|
Assouline-Dayan Y, Chang C, Greenspan A,
Shoenfeld Y and Gershwin ME: Pathogenesis and natural history of
osteonecrosis. Semin Arthritis Rheum. 32:94–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fisher DE: The role of fat embolism in the
etiology of corticosteroid-induced avascular necrosis: clinical and
experimental results. Clin Orthop Relat Res. 130:68–80.
1978.PubMed/NCBI
|
|
3
|
Wang Y, Li Y, Mao K, Li J, Cui Q and Wang
GJ: Alcohol-induced adipogenesis in bone and marrow: a possible
mechanism for osteonecrosis. Clin Orthop Relat Res. 410:213–224.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abu-Shakra M, Buskila D and Shoenfeld Y:
Osteonecrosis in patients with SLE. Clin Rev Allergy Immunol.
25:13–24. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jones LC, Mont MA, Le TB, et al:
Procoagulants and osteonecrosis. J Rheumatol. 30:783–791.
2003.PubMed/NCBI
|
|
6
|
Bjorkman A, Burtscher IM, Svensson PJ,
Hillarp A, Besjakov J and Benoni G: Factor V Leiden and the
prothrombin 20210A gene mutation and osteonecrosis of the knee.
Arch Orthop Trauma Surg. 125:51–55. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zalavras CG, Vartholomatos G, Dokou E and
Malizos KN: Factor V Leiden and prothrombin gene mutations in
femoral head osteonecrosis. Thromb Haemost. 87:1079–1080.
2002.PubMed/NCBI
|
|
8
|
Gohel A, McCarthy MB and Gronowicz G:
Estrogen prevents glucocorticoid-induced apoptosis in osteoblasts
in vivo and in vitro. Endocrinology. 140:5339–5347. 1999.PubMed/NCBI
|
|
9
|
Kawai K, Tamaki A and Hirohata K:
Steroid-induced accumulation of lipid in the osteocytes of the
rabbit femoral head. A histochemical and electron microscopic
study. J Bone Joint Surg Am. 67:755–763. 1985.PubMed/NCBI
|
|
10
|
Kerachian MA, Harvey EJ, Cournoyer D, Chow
TY and Séguin C: Avascular necrosis of the femoral head: vascular
hypotheses. Endothelium. 13:237–244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zalavras CG, Vartholomatos G, Dokou E and
Malizos KN: Genetic background of osteonecrosis: associated with
thrombophilic mutations? Clin Orthop Relat Res. 422:251–255. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bjorkman A, Svensson PJ, Hillarp A,
Burtscher IM, Runow A and Benoni G: Factor V leiden and prothrombin
gene mutation: risk factors for osteonecrosis of the femoral head
in adults. Clin Orthop Relat Res. 425:168–172. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee JS, Koo KH, Ha YC, et al: Role of
thrombotic and fibrinolytic disorders in osteonecrosis of the
femoral head. Clin Orthop Relat Res. 417:270–276. 2003.PubMed/NCBI
|
|
14
|
Chang JD, Hur M, Lee SS, Yoo JH and Lee
KM: Genetic background of nontraumatic osteonecrosis of the femoral
head in the Korean population. Clin Orthop Relat Res.
466:1041–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hessner MJ, Luhm RA, Pearson SL, Endean
DJ, Friedman KD and Montgomery RR: Prevalence of prothrombin
G20210A, F5 G1691A (Leiden) and methylenetetrahydrofolate reductase
(MTHFR) C677T in seven different populations determined by
multiplex allele-specific PCR. Thromb Haemost. 81:733–738.
1999.PubMed/NCBI
|
|
16
|
Kim SY, Suh JS, Park EK, et al: Factor V
Leiden gene mutation in femoral head osteonecrosis. J Korean Ortho
Res Soc. 6:259–264. 2003.
|
|
17
|
Hong JM, Kim TH, Chae SC, et al:
Association study of hypoxia inducible factor 1α (HIF1α) with
osteonecrosis of femoral head in a Korean population.
Osteoarthritis Cartilage. 15:688–694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee HJ, Choi SJ, Hong JM, et al:
Association of a polymorphism in the intron 7 of the SREBF1 gene
with osteonecrosis of the femoral head in Koreans. Ann Hum Genet.
73:34–41. 2009. View Article : Google Scholar
|
|
19
|
Hedrick PW: Gametic disequilibrium
measures: proceed with caution. Genetics. 117:331–341.
1987.PubMed/NCBI
|
|
20
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar
|
|
21
|
Glueck CJ, Freiberg R, Tracy T, Stroop D
and Wang P: Thrombophilia and hypofibrinolysis: pathophysiologies
of osteonecrosis. Clin Orthop Relat Res. 334:43–56. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Glueck CJ, Brandt G, Gruppo R, et al:
Resistance to activated protein C and Legg-Perthes disease. Clin
Orthop Relat Res. 338:139–152. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gruppo R, Glueck CJ, Wall E, Roy D and
Wang P: Legg-Perthes disease in three siblings, two heterozygous
and one homozygous for the F5 Leiden mutation. J Pediatr.
132:885–888. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Glueck CJ, Fontaine RN, Gruppo R, et al:
The plasminogen activator inhibitor-1 gene, hypofibrinolysis and
osteonecrosis. Clin Orthop Relat Res. 366:133–146. 1999. View Article : Google Scholar
|
|
25
|
Celik A, Tekis D, Saglam F, et al:
Association of corticosteroids and F5, prothrombin and MTHFR gene
mutations with avascular osteonecrosis in renal allograft
recipients. Transplant Proc. 38:512–516. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eberle MA and Kruglyak L: An analysis of
strategies for discovery of single-nucleotide polymorphisms. Genet
Epidemiol. 19(Suppl 1): 29–35. 2000. View Article : Google Scholar
|
|
27
|
Adachi N, Shinoda K, Umetsu K and
Matsumura H: Mitochondrial DNA analysis of Jomon skeletons from the
Funadomari site, Hokkaido and its implication for the origins of
Native American. Am J Phys Anthropol. 138:255–265. 2009. View Article : Google Scholar
|
|
28
|
Jung J, Kang H, Cho YS, et al: Gene flow
between the Korean peninsula and its neighboring countries. PloS
one. 5:e118552010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bowen DJ, Bowley S, John M and Collins PW:
Factor V Leiden (G1691A), the prothrombin 3′-untranslated region
variant (G20210A) and thermolabile methylenetetrahydrofolate
reductase (C677T): a single genetic test genotypes all three
loci-determination of frequencies in the S. Wales population of the
UK. Thromb Haemost. 79:949–954. 1998.PubMed/NCBI
|
|
30
|
Rees DC, Cox M and Clegg JB: World
distribution of F5 Leiden. Lancet. 346:1133–1134. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pepe G, Rickards O, Vanegas OC, et al:
Prevalence of F5 Leiden mutation in non-European populations.
Thromb Haemost. 77:329–331. 1997.PubMed/NCBI
|
|
32
|
Glueck CJ, Freiberg RA and Wang P:
Heritable thrombo-philia-hypofibrinolysis and osteonecrosis of the
femoral head. Clin Orthop Relat Res. 466:1034–1040. 2008.
View Article : Google Scholar : PubMed/NCBI
|