Introduction
Neuropathic pain is a common neurological disorder
resulting from peripheral nerve damage, which affects millions of
people worldwide (1). Wnt proteins
are a family of secreted lipid-modified proteins, which act as
signaling molecules in the regulation of cellular processes such as
cell proliferation, differentiation, migration, and polarity,
during the development of the cardiac and nervous systems (2,3). To
date, a total of 19 members of the human Wnt family have been
identified. Wnt ligands bind to the cysteine-rich domain of
frizzled receptors and co-receptors, in order to activate
intracellular signaling cascades (2). Wnt signaling includes both canonical
Wnt/β-catenin pathways, and non-canonical β-catenin-independent
pathways (2,4–8).
β-catenin is a multifunctional protein that interacts with
transcription factors in order to activate the transcription of a
target gene. Non-canonical β-catenin-independent pathways induce
the release of intracellular calcium, which in turn activates a
calcium-calmodulin-dependent kinase (CaMKII) (4,5).
Wnts are predominantly expressed in the brain, where they are
critical to various neurological developmental processes (2,3), as
well as the regulation of synaptic plasticity (3,5,9).
Dysregulation of Wnt signaling is responsible for poor axonal
regeneration, as well as the development of certain oncogenic
processes and mental disorders (10–13).
A recent study demonstrated the role of Wnt signaling in the
development and maintenance of neuropathic pain in various rodent
models (2). The present study
aimed to investigate the functional significance of spinal Wnt
signaling in a rodent model of neuropathic pain.
An increasing amount of evidence suggests that
epigenetic modifications of DNA and/or histones in the
transcriptional control region may significantly modulate the
transcription of specific loci within the genome, thus critically
influencing neuronal development, learning and memory, as well as
various neurological disorders including chronic pain (3). Previous studies have reported that
the suppression of histone acetylation via the enhanced activity of
histone deacetylase 2, was may significantly decrease the
expression levels of genes associated with synaptic plasticity, and
modify cognitive function (3). A
recent study also demonstrated the alteration of histone
acetylation and cytosine methylation during the induction and
maintenance of sustained pain-induced behavior, caused by
peripheral inflammation and nerve damage (3). These epigenetic modifications present
at specific genomic loci may significantly affect the expression
levels of certain proteins, thus contributing to the sensitization
of peripheral sensory neurons and central nociceptive neurons
(5). The present study aimed to
elucidate the epigenetic mechanisms underlying altered Wnt
signaling in the rat spinal dorsal horn.
Materials and methods
Animals and CCI surgery
Adult male Wistar rats weighing 180–220 g were
obtained from the Institutional Center of Experiment Animals
(Quingdao, China), and were housed in standard laboratory
conditions (22±2°C with a 12 h light/dark cycle). The rats were
provided ad libitum access to food and water. The 88 rats
were divided into four groups: control (n=35), CCI (n=35), CCI +
XAV939 (n=10) and control + XAV939 (n=9). All animal experiments
were approved by the Institutional Animal Care and Use Committee
(Qingdao University, Qingdao, China), and conformed to the
guidelines of the National Institution of Health (14).
For chronic constriction injury (CCI) surgery, all
rats were anesthetized with 1% isoflurane. The right sciatic nerve
was exposed and three Natural Chromic-Gut 4–0 absorbable sutures
(Stoelting Co., Dublin, Ireland) were placed around the nerve with
1 mm between each suture. The muscle and skin layers were closed
with silk sutures. In the rats of the sham (control) group, the
sciatic nerve was exposed but no sutures were placed around the
nerve.
The inhibitor of Wnt signaling XAV939, was purchased
from Stemgent Inc. (Cambridge, MA, USA), and unless otherwise
stated, all other chemicals and reagents were purchased from
Sigma-Aldrich (St. Louis, MO, USA).
Behavioral tests
Intrathecal cannulation was performed as previously
described (6). PE-10 polyethylene
intrathecal catheters were implanted one week prior to the surgical
procedures. The catheters were placed 8 cm into the caudal side of
the rats through an incision in the cisternal membrane, and secured
to the musculature at the incision site. Only the rats that showed
no evidence of neurological deficits following catheter insertion
were selected for further experimentation. A total of 5 µg
inhibitor of Wnt signaling XAV939 or vehicle (saline) in a 10
µl total volume were administered daily into the rats,
followed by a 10 µl flush with normal saline.
Mechanical allodynia was detected using a series of
von Frey filaments with logarithmic incremental stiffness
(Stoelting Co., Wood Dale, IL, USA), and the 50% probability of paw
withdrawal thresholds were calculated using the up-down method, as
previously described (7). The
filaments were applied to the plantar surface of the hind paws for
~6 sec in an ascending or descending order following a negative or
positive paw withdrawal response, respectively. Following the first
observed change in behavioral response, the subsequent six
consecutive responses were used to calculate the paw withdrawal
pressure threshold (in grams). The maximum pressure threshold was
set as 16 g.
The thermal nociceptive thresholds were measured
using radiant heat (8). Briefly,
the rats were placed into individual plastic cubicles mounted on a
glass surface maintained at 30°C, and allowed a 1 h habituation
period. A thermal stimulus from a radiant heat source was then
delivered to the plantar surface of the hind paws. The time taken
for the rat to remove the paw from the thermal stimulus was
electronically recorded and determined as the paw withdrawal
latency period. The stimulus ceased automatically when the hind paw
moved, or following 16 sec, in order to prevent tissue damage. The
intensity of the heat stimulus was maintained constant throughout
all experiments.
Protein extraction and
immunoblotting
Following behavioral testing at day 15, the rats
were anesthetized with 50 mg/kg pento-barbital sodium (P-010;
Sigma-Aldrich, St. Louis, MO, USA), and transcardially perfused
with 30 ml phosphate-buffered saline. The ipsilateral dorsal horn
tissue (L4-L6) was subsequently rapidly collected, and homogenized
(Thermo Fisher Scientific, Waltham, MA, USA) in a lysis buffer
supplemented with a protease inhibitor cocktail (1 mM
phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM NaVO3, 1
µg/ml leupeptin, 1 µg/ml pepstatin, and 1
µg/ml aprotinin). The lysates were centrifuged at 12,000 x g
for 10 min at 4°C, and the protein concentration was measured using
the Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The samples were treated with SDS sample buffer
at 95°C for 5 min, separated by 10% SDS-PAGE, and blotted to a
nitrocellulose membrane. The membranes were incubated overnight at
4°C with the following primary antibodies: Anti-active β-catenin
(1:1,000; cat. no. 07-1651, EMD Millipore, Billerica, MA, USA),
anti β-catenin (1:1,000; cat. no. 04-958, EMD Millipore),
anti-Wnt3a (1:1,000; cat. no. ABD124, EMD Millipore), and
anti-GAPDH (1:200; cat. no. sc-365062, Santa Cruz Biotechnology
Inc., Dallas, TX, USA). The membranes were subsequently incubated
with appropriate horseradish peroxidase-conjugated secondary
antibodies against mouse IgG (1:10,000; cat. no. 715-035-150;
Jackson ImmunoResearch, West Grove, PA, USA) or rabbit IgG
(1:10,000, cat. no. 711-035-152, Jackson ImmunoResearch) at room
temperature for 1 h and the blots were visualized using an enhanced
chemiluminescence system (GE Healthcare Life Sciences, Little
Chalfont, UK). The intensity of the bands were quantified using
ImageJ analysis software (v1.48; National Institute of Health,
Bethesda, MA, USA) and the ratios of the target proteins against
GAPDH band density were calculated accordingly.
Chromatin immunoprecipitation (ChIP) and
polymerase chain reaction (PCR)
A ChIP assay was performed as previously described
(9). Briefly, the rat ipsilateral
dorsal horn tissue samples (L4–L6) were collected, and fixed with
1% formaldehyde at 37°C for 5 min. The chromatin fraction was
extracted and fragmented to 400–500 bp by sonication, as previously
described (9). A total of 200
µl sonicated chromatin in lysis buffer supplemented with 1%
Triton X-100 was pre-cleared using 5 µg normal rabbit
immunoglobulin G (IgG) conjugated to 30 µl Protein A, and
subsequently incubated with either 5 µg polyclonal
acetylated histone H3 (EMD Millipore), or normal rabbit IgG
conjugated to Protein A at 4°C for 20 h. The immunoprecipitants
were consecutively washed with 1 ml of the following buffers: Low
Salt Wash Buffer, High Salt Wash Buffer, LiCl Wash Buffer, and TE
Buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA). Following reverse
crosslinking at 65°C for 4 h, the DNA was purified with ChIP DNA
Clean & Concentrator (Zymo Research, Irvine, CA, USA), and used
as a template for the PCR, which was carried out using the SYBR
Select PCR Master Mix (Applied Biosystems Life Technologies, Foster
City, CA, USA) and the 7500 Fast Real-Time PCR system (Applied
Biosystems Life Technologies). The PCR cycling conditions were as
follows: Heating at 50°C for 2 min followed by 95°C for 10 min, 40
cycles of 95°C for 15 sec, and 60°C for 1 min. The following
primers (SBS Genetech Co., Ltd., Beijing, China) were designed to
amplify ~200 bp fragments in the promoter region of the target
genes: Wnt3a forward, 5′-GAGCTCTGCCATTCACCCTA-3′, and reverse,
5′-GGCTGCAGAGCACTAAGAGG-3′; and GAPDH forward,
5′-AGACAGCCGCATCTTCTTGT-3′, and reverse 5′-CGTCCTCTACCATCCTCTGC-3′.
The ChIP signal was calculated according to the manufacturer's
instructions, using the following method (15): ΔCt[normalized ChIP] =
(Ct[ChIP] - (Ct[Input] -
Log2(input dilution factor))); and the ChIP/Input ratio
was calculated as 2(−ΔCt[normalized ChIP]).
Methylated DNA immunoprecipitation
(MeDIP) Assay
The MeDIP assay was performed as previously
described with minor modifications (10). Briefly, the rat ipsilateral dorsal
horn tissue samples (L4–L6) were collected, and the genomic DNA was
fragmented into 400–500 bp as described above. The sonicated
samples were centrifuged at 14,000 x g for 10 min, and the
supernatants were subsequently collected. The samples were then
incubated with a polyclonal antibody targeting 5-methylcytosine
(1:100, EMD Millipore) overnight at 4°C with gentle agitation. The
DNA-antibody complex was enriched with Protein A agarose beads, and
the DNA fragments in the input and pulled-down fractions were
purified. A PCR was performed to amplify the ~200 bp segments
corresponding to the CpG sites within the Wnt3a promoter region.
The following primers (SBS Genetech Co., Ltd.) were used to amplify
the fragments: Wnt3a forward, 5′-GAGCTCTGCCATTCACCCTA-3′, and
reverse, 5′-GGCTGCAGAGCACTAAGAGG-3′; and GAPDH forward,
5′-AGACAGCCGCATCTTCTTGT-3′, and reverse,
5′-CTGCGGGAGAAGAAAGTCAG-3′. All amplifications were carried out in
triplicate, and the PCR data were analyzed as described above.
Reverse transcription-quantitative PCR
(RT-qPCR)
The ipsilateral dorsal horn tissues (L4–L6) were
collected from the rats. Total RNA was extracted using
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA), and reverse transcribed using SuperScript III Reverse
Transcriptase (Invitrogen Life Technologies). The following primers
(SBS Genetech Co., Ltd.) were designed to amplify the ~200 bp
fragment within the cDNA sequence of the target genes: Wnt3a
forward, 5′-ATGGCTCCTCTCGGATACCT-3′, and reverse,
5′-GGGCATGATCTCCACGTAGT-3′; and GAPDH forward,
5′-AGACAGCCGCATCTTCTTGT-3′, and reverse 5′-CTTGCCGTGGGTAGAGTCAT-3′.
RT-qPCR was performed using SYBR Green qPCR SuperMix (Invitrogen
Life Technologies). The cycle threshold (Ct) values for each sample
were determined with logarithmic phase amplification plots. The
data were subsequently analyzed using the 2−ΔΔCt
method.
Bisulfite sequencing PCR
The ipsilateral dorsal horn tissue samples (L4–L6)
were collected from the rats. Total DNA was subsequently extracted,
fragmented to 400–500 bp, and purified. The bisulfite conversion of
the DNA was performed using the EZ DNA Methylation-Gold™ kit (Zymo
Research) as previously described (11). A ~250 bp fragment in the promoter
region of Wnt3a was amplified, independent of its methylation
status, using the following primers (SBS Genetech Co., Ltd.):
Forward, 5′-GGGTTTTAGTGTTATTTTAGGGTAAT-3′, and reverse,
5′-ACATAAACCTTCACTACCCTCTTTC-3′. The PCR product was then purified
using a Gel Extraction kit (Qiagen, Inc., Valencia, CA, USA), and
sequenced using a reverse primer (Qiagen). The methylation
percentage of each CpG site within the amplified promoter region of
Wnt3a was determined by calculating the ratio between the guanine
(G) and adenine (A) peaks [G/(G+A)], and these electropherogram
ratios were subsequently analysed using Chromas software (v.2.1.1;
Technelysium Pty, Ltd., South Brisbane, Australia).
Statistical analysis
All data were presented as the mean ± standard error
of the mean, and were analyzed using a Student's t-test or two-way
analysis of variance, followed by Fisher's least significant
difference post hoc analysis. Statistical analysis was performed
using StatView 5.0 software (Cary, NC, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Increased Wnt signaling in the dorsal
horn of the rats with CCI
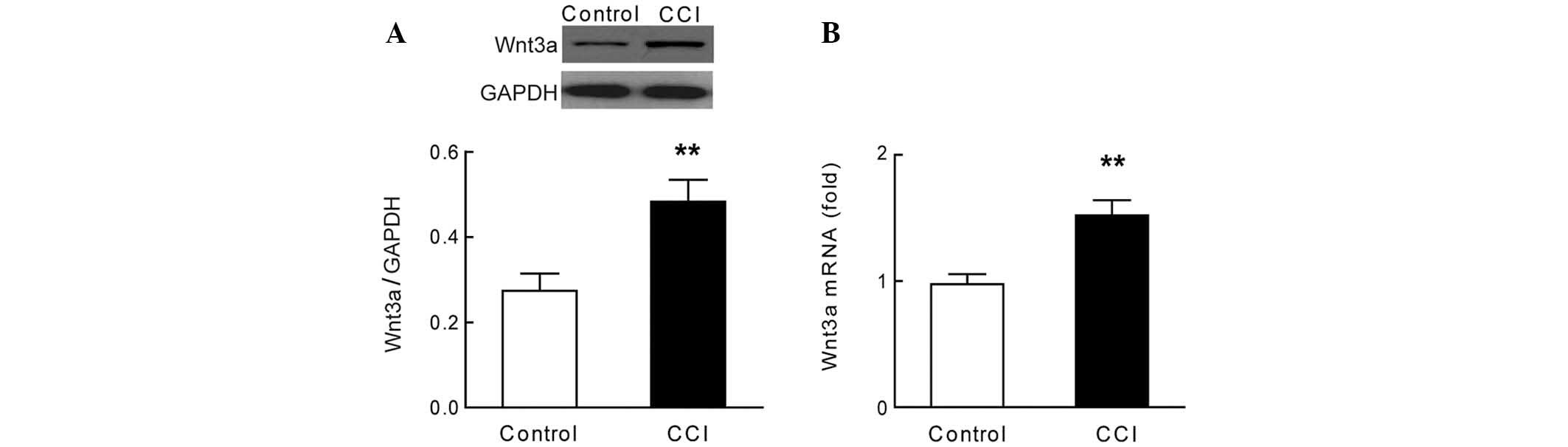
The expression levels of Wnt3a were significantly
increased in the dorsal horn (L4–L6) of the rats with CCI, as
compared with the rats in the sham group (Fig. 1A, P<0.01, n=7). These results
were concordant with the observed increase in mRNA expression
levels of Wnt3a in the dorsal horn of the rats with CCI (Fig. 1B, P<0.01, n=7). The results of
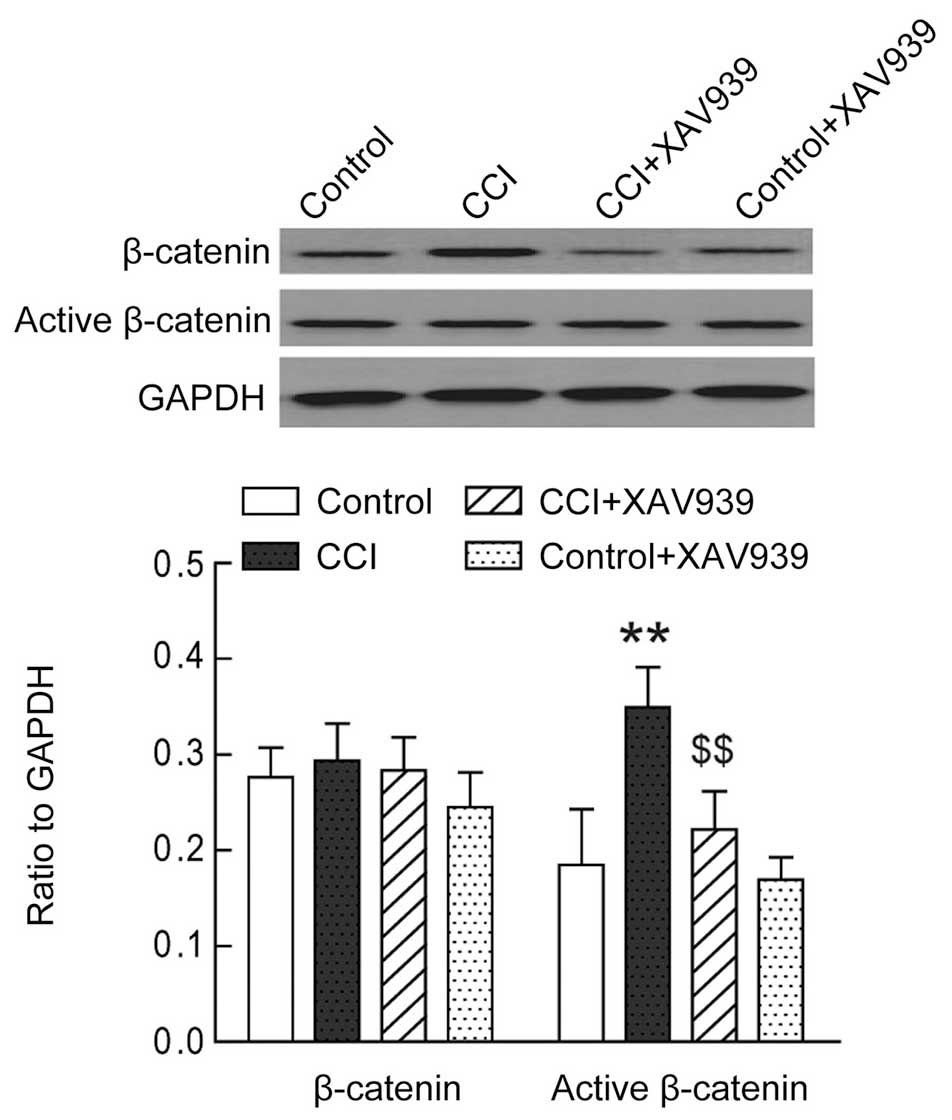
the present study also showed that the expression levels of active
β-catenin, the downstream effector of Wnt signaling, were
significantly increased in the dorsal horn (L4–L6) of the rats with
CCI (Fig. 2, P<0.01, n=7).
These results demonstrate the presence of increased Wnt signaling
in the dorsal horn of rats with CCI.
Epigenetic modifications of the Wnt3a
promoter region
Previous studies have demonstrated that the
epigenetic modification of histone acetylation and/or cytosine
methylation may significantly influence chromatin remodeling, thus
regulating the transcription of target genes (4,10).
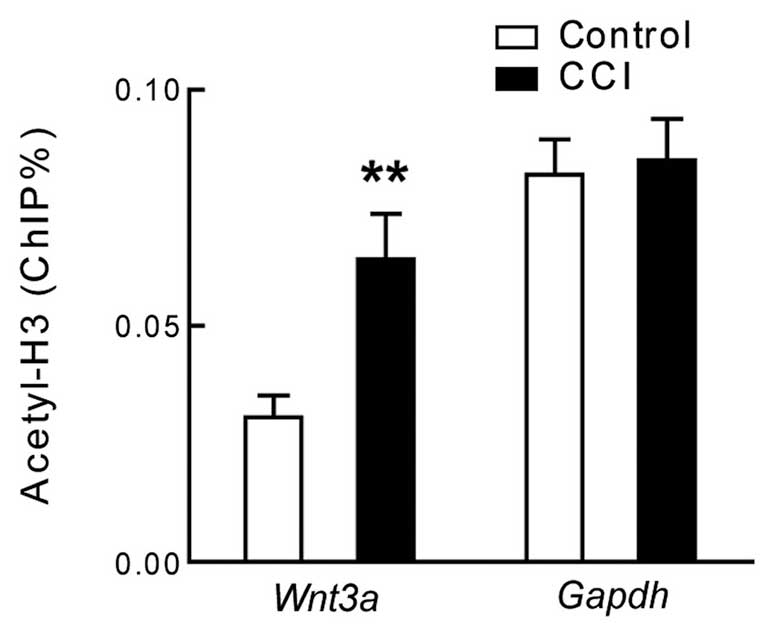
The present study examined the extent of histone H3 acetylation and
cytosine methylation in the promoter region of Wnt3a, in the lumbar
dorsal horn of rats with CCI. The ChIP assay used polyclonal
antibodies targeting acetylated histone H3 and demonstrated that
the acetylation of histone H3 was significantly increased in the
promoter region of Wnt3a, but not GAPDH, in the lumbar dorsal horn
of the rats with CCI (Fig. 3;
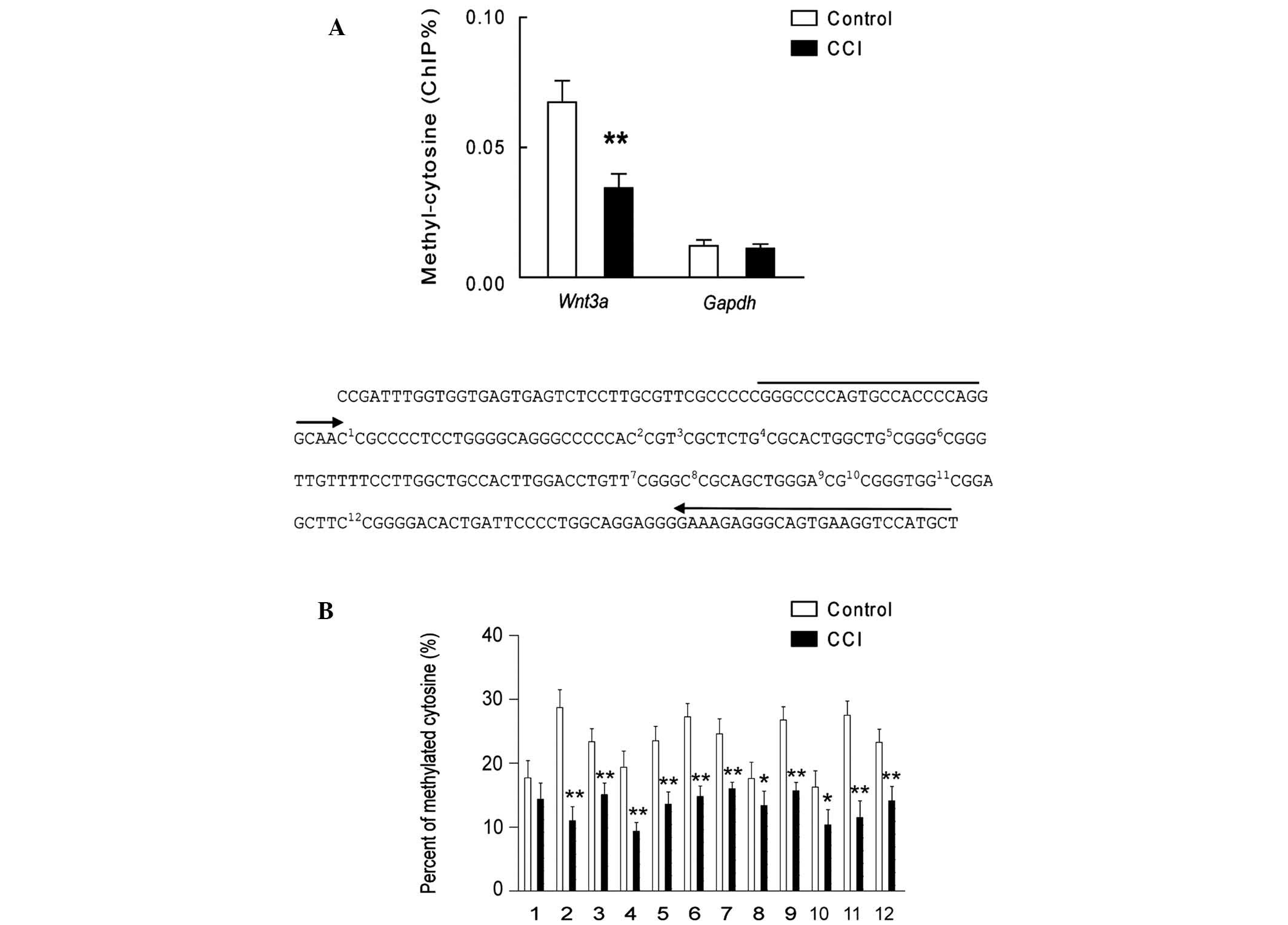
P<0.01, n=7). Conversely, the results showed that when using the
polyclonal antibodies targeting 5′-cytosine, cytosine methylation
was significantly decreased in the promoter region of Wnt3a, but
not GAPDH, in the lumbar dorsal horn of the rats with CCI (Fig. 4A; P<0.01; n=7). Furthermore, the
results of the bisulfite sequencing PCR demonstrated the alteration
of methylation at specific CpG sites in the Wnt3a promoter region
in the lumbar dorsal horn of the rats with CCI (Fig. 4B, n=6). These results demonstrate
the presence of substantial epigenetic modification within the
Wnt3a promoter region, which may contribute to the upregulation of
Wnt3a signaling in the lumbar dorsal horn of rats with CCI.
Inhibition of Wnt3a signaling attenuates
the pain-induced behaviors of rats with CCI
In order to investigate the functional significance
of increased Wnt3a expression in the pain-induced behavior of the
rats with CCI, 5 µg of either XAV939 (Wnt3a inhibitor), or
vehicle were administered to the rats with CCI via an intrathecal
catheter. Treatment with XAV939 significantly attenuated the
expression levels of active β-catenin in the lumbar dorsal horn of
the rats with CCI (Fig. 2;
P<0.01; n=7), whereas no change in the expression levels of
active β-catenin was observed in the sham group rats. The
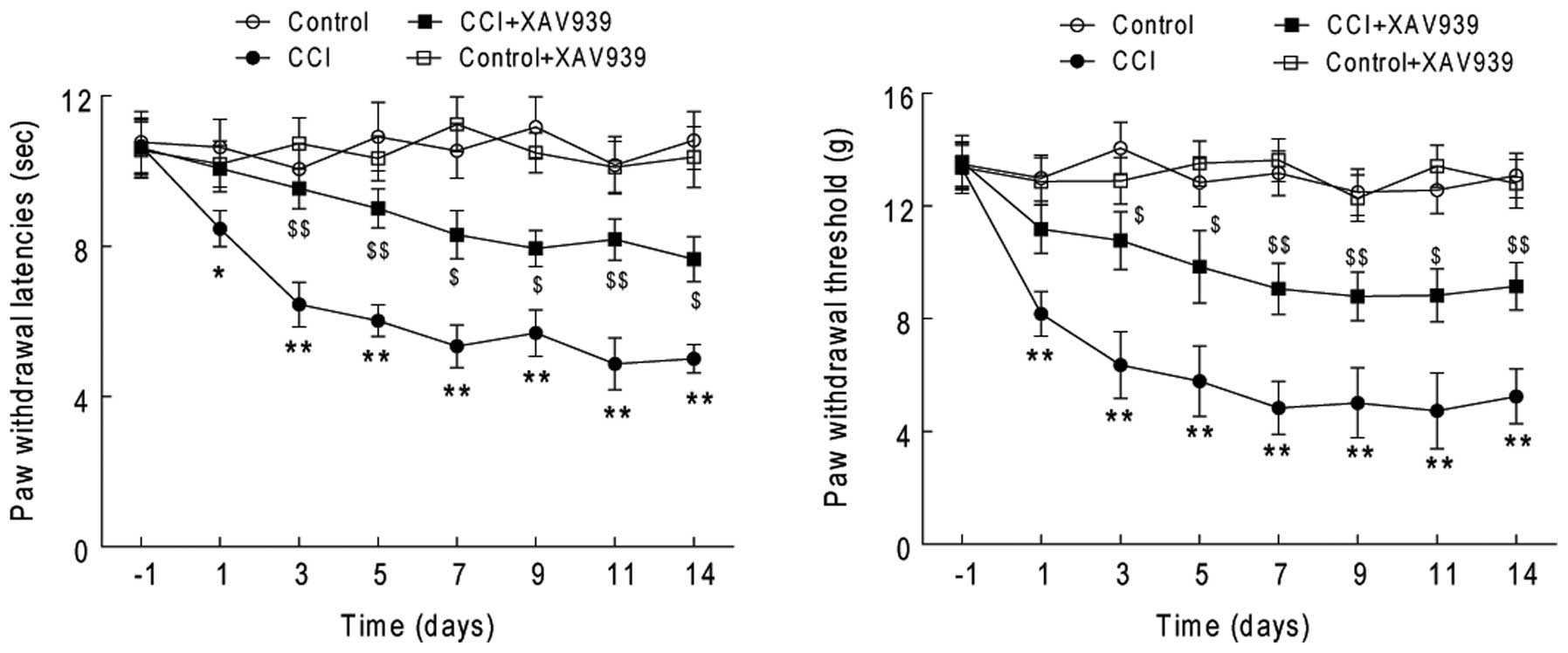
behavioral studies also showed that treatment with XAV939
significantly attenuated the behavioral responses to mechanical and
radiant thermal stimuli of the rats with CCI (Fig. 5, P<0.01, n=10), whereas it did
not significantly change the behavioral responses of the rats in
the sham (control) group. These results indicate that the
upregulation of spinal Wnt3a signaling may significantly contribute
to the pain-induced behavior of the rats with CCI.
Discussion
Wnt signaling was originally thought to be
exclusively involved in the regulation of cellular processes such
as proliferation, differentiation, and migration during neuronal
development; however, recent studies have demonstrated the
important role of Wnt signaling in the pathogenesis of neuropathic
and bone cancer-induced pain (2,12). A
previous study reported that treatment with Wnt3a significantly
enhanced Aδ fiber response to mechanical stimuli, and induced
murine hypersensitivity to mechanical and thermal stimuli (13). Nerve injury and bone cancer cause
rapid and long-lasting upregulation of Wnts, such as Wnt3a, as well
as activation of the Wnt-Frizzled-β-catenin signaling pathway in
primary sensory neurons and spinal dorsal horn nociceptive neurons,
which contribute to the development and maintenance of thermal
hyperalgesia and mechanical allodynia. These responses may be
substantially attenuated by the spinal injection of inhibitors of
Wnt signaling (2,16). A recent study also showed that
partial sciatic nerve ligation induced the upregulation of spinal
Wnt3a expression, and the inhibition of Wnt/β-catenin signaling,
which attenuated the induction of neuropathic pain (17). Activation of Wnt signaling may
contribute to synaptic plasticity in dorsal horn neurons, and may
be involved in the pathogenesis of human immunodeficiency
virus-associated chronic pain (18). The present study detected increased
expression levels of Wnt3a in the ipsilateral dorsal horn of a
rodent model of CCI, and demonstrated that inhibition of Wnt
signaling significantly attenuated the rat behavioral responses to
mechanical and radiant thermal stimuli. These results further
confirmed the involvement of Wnt3a signaling in the pathogenesis of
CCI-induced neuropathic pain.
A previous study showed that epigenetic mechanisms
were critically involved in the development and maintenance of
persisted pain-induced behavior in peripheral inflammation and
nerve injury (3). Peripheral
inflammation also reportedly induced significant hypermethylation
of CpG islands in the miR-219 promoter, which in turn led to the
upregulation of spinal CaMKIIγ, and the increase in pain-induced
behavior of rats injected with complete Freud's adjuvant (19). Persistent peripheral inflammation
has been shown to upregulate the expression of transcriptional
regulator MeCP2 in the central nucleus of the mouse amygdala, which
consequently resulted in repression of the expression of histone
dimethyltransferase G9a, which resulted in increased central
expression of brain-derived neurotrophic factor, thus inducing
mouse hypersensitivity to painful stimuli (20). A previous study also reported that
the epigenetic suppression of GAD65 via histone hypoacetylation in
the brain stem neurons had a critical role in the induction and
maintenance of pain-induced behavior caused by peripheral
inflammation and nerve injury (21). The present study demonstrated the
presence of increased expression levels of spinal Wnt3a due to
significant epigenetic modifications in the promoter region of
Wnt3a, which contributed to the pain-induced behavior of rats with
CCI. These results, together with the findings of previous studies,
strongly suggest that the epigenetic modification of specific genes
may substantially contribute to the sensitization of peripheral and
central nociceptive circuits, and to the induction and maintenance
of pain-induced behavior in a rodent pain model.
The addition of a methyl group to the 5 position of
cytosine in the CpG sites of a promoter region, catalyzed by DNA
methyltransferases, may lead to transcriptional silencing via the
recruitment of transcriptional repressors, and to chromatin
condensation, thus restricting the access of transcriptional
proteins, and masking the binding sites of transcriptional
activators (22). Conversely,
histone lysine acetylation has been associated with active
transcription of target genes, whereas deacetylated histones were
mostly detected within transcriptionally silenced domains (23). The alteration of histone
acetylation and/or cytosine methylation in the transcriptional
control region of specific genes has been extensively investigated
in numerous neurological disorders (24), including neuropathic pain (5). Increased cytosine methylation of the
transcriptional control region of the gene encoding endo-thelin-1 B
receptor in sensory neurons has previously been shown to be
associated with pain-induced behavior in a murine model of oral
squamous cell carcinoma (25). In
addition, the expression levels of lysine 9-acetylated histone H3
in the promoter regions of the chemokine CCL2 and CCL3 genes have
been shown to be increased in injured sciatic nerves, which was
associated with pain-induced behavior (26). The present study demonstrated an
increase in the expression levels of spinal Wnt3a caused by
increased histone H3 acetylation, and decreased cytosine
methylation in the Wnt3a promoter region of rats with CCI, which
may contribute to the development and maintenance of pain-induced
behaviors in a rodent model of neuropathic pain.
In conclusion, the present study demonstrated an
increase in Wnt3a-mediated signaling, resulting from increased
histone acetylation, and decreased cytosine methylation in the
promoter region of Wnt3a, in the dorsal horn of rats with CCI. In
addition, the results of the present study showed that the
inhibition of Wnt3a signaling was able to significantly attenuate
the pain-induced behavior of rats with CCI. Therefore, these
results provide a novel insight into the development of therapeutic
strategies for the treatment of neuropathic pain.
References
|
1
|
Gilron I, Baron R and Jensen T:
Neuropathic pain: Principles of diagnosis and treatment. Mayo Clin
Proc. 90:532–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang YK, Huang ZJ, Liu S, Liu YP, Song AA
and Song XJ: WNT signaling underlies the pathogenesis of
neuropathic pain in rodents. J Clin Invest. 123:2268–2286. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Denk F and McMahon SB: Chronic pain:
Emerging evidence for the involvement of epigenetics. Neuron.
73:435–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guan JS, Haggarty SJ, Giacometti E,
Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X,
Mazitschek R, et al: HDAC2 negatively regulates memory formation
and synaptic plasticity. Nature. 459:55–60. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Denk F, McMahon SB and Tracey I: Pain
vulnerability: A neurobiological perspective. Nat Neurosci.
17:192–200. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou HY, Chen SR, Chen H and Pan HL: The
glutamatergic nature of TRPV1-expressing neurons in the spinal
dorsal horn. J Neurochem. 108:305–318. 2009. View Article : Google Scholar :
|
|
7
|
Zhang J, Wu D, Xie C, Wang H, Wang W,
Zhang H, Liu R, Xu LX and Mei XP: Tramadol and propentofylline
coadministration exerted synergistic effects on rat spinal nerve
ligation-induced neuropathic pain. PLoS One. 8:e729432013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hargreaves K, Dubner R, Brown F, Flores C
and Joris J: A new and sensitive method for measuring thermal
nociception in cutaneous hyperalgesia. Pain. 32:77–88. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fujita T, Ryser S, Tortola S, Piuz I and
Schlegel W: Gene-specific recruitment of positive and negative
elongation factors during stimulated transcription of the MKP-1
gene in neuroendocrine cells. Nucleic Acids Res. 35:1007–1017.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Provençal N, Suderman MJ, Caramaschi D,
Wang D, Hallett M, Vitaro F, Tremblay RE and Szyf M: Differential
DNA methylation regions in cytokine and transcription factor
genomic loci associate with childhood physical aggression. PLoS
One. 8:e716912013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayatsu H, Tsuji K and Negishi K: Does
urea promote the bisulfite-mediated deamination of cytosine in DNA?
Investigation aiming at speeding-up the procedure for DNA
methylation analysis. Nucleic Acids Symp Ser (Oxf). 50:69–70. 2006.
View Article : Google Scholar
|
|
12
|
Ji RR, Xu ZZ and Gao YJ: Emerging targets
in neuroinflammation-driven chronic pain. Nat Rev Drug Discov.
13:533–548. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Simonetti M, Agarwal N, Stösser S, Bali
KK, Karaulanov E, Kamble R, Pospisilova B, Kurejova M, Birchmeier
W, Niehrs C, et al: Wnt-Fzd signaling sensitizes peripheral sensory
neurons via distinct noncanonical pathways. Neuron. 83:104–121.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guide for the care and use of laboratory
animals. 8th Edition. (Committee for the update of the guide for
the care and use of laboratory animals). National Research Council
of the National Academies. National Academies Press; 2013,
Washongton, D.C:
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Tang SJ: Synaptic activity-regulated Wnt
signaling in synaptic plasticity, glial function and chronic pain.
CNS Neurol Disord Drug Targets. 13:737–744. 2014. View Article : Google Scholar
|
|
17
|
Itokazu T, Hayano Y, Takahashi R and
Yamashita T: Involvement of Wnt/β-catenin signaling in the
development of neuropathic pain. Neurosci Res. 79:34–40. 2014.
View Article : Google Scholar
|
|
18
|
Shi Y, Shu J, Gelman BB, Lisinicchia JG
and Tang SJ: Wnt signaling in the pathogenesis of human
HIV-associated pain syndromes. J Neuroimmune Pharmacol. 8:956–964.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan Z, Zhu LJ, Li YQ, Hao LY, Yin C, Yang
JX, Guo Y, Zhang S, Hua L, Xue ZY, et al: Epigenetic modification
of spinal miR-219 expression regulates chronic inflammation pain by
targeting CaMKIIγ. J Neurosci. 34:9476–9483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Z, Tao W, Hou YY, Wang W, Kenny PJ
and Pan ZZ: MeCP2 repression of G9a in regulation of pain and
morphine reward. J Neurosci. 34:9076–9087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Z, Cai YQ, Zou F, Bie B and Pan ZZ:
Epigenetic suppression of GAD65 expression mediates persistent
pain. Nat Med. 17:1448–1455. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng X: Structural and functional
coordination of DNA and histone methylation. Cold Spring Harb
Perspect Biol. 6:a0187472014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sultan FA and Day JJ: Epigenetic
mechanisms in memory and synaptic function. Epigenomics. 3:157–181.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klein CJ and Benarroch EE: Epigenetic
regulation: basic concepts and relevance to neurologic disease.
Neurology. 82:1833–1840. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Viet CT, Ye Y, Dang D, Lam DK, Achdjian S,
Zhang J and Schmidt BL: Re-expression of the methylated EDNRB gene
in oral squamous cell carcinoma attenuates cancer-induced pain.
Pain. 152:2323–2332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kiguchi N, Kobayashi Y, Saika F and
Kishioka S: Epigenetic upregulation of CCL2 and CCL3 via histone
modifications in infiltrating macrophages after peripheral nerve
injury. Cytokine. 64:666–672. 2013. View Article : Google Scholar : PubMed/NCBI
|