Introduction
Breast cancer is the most frequently diagnosed
cancer and the leading cause of cancer mortality among females,
accounting for 23% of total cancer cases and 14% of cancer
mortalities (1). Breast cancer has
the best prognosis when detected at early stages. However, patients
diagnosed with advanced breast cancer or metastatic breast cancer
have a poor prognosis. Therefore, in order to improve the overall
survival rate of patients with breast cancer, it is of importance
to investigate factors that could promote the proliferation,
invasion and metastasis of human breast cancer cells.
A variety of genes are involved in the development
of breast cancer, including BRCA1, BRCA2, p53 (2) and c-erbB-2 (3). Previously, it has also been reported
that HOX genes are widely correlated with various types of cancer
(4–6).
The homeobox (HOX) subgroup of the HOX supergene
family encompasses 39 genes located in four contiguous clusters
(HOXA, HOXB, HOXC and HOXD) (7).
HOX genes containing a 183-bp DNA sequence coding for a
61-amino-acid domain defined as the homeodomain (HD) (8), which is responsible for the
recognition and binding of sequence-specific DNA motifs (9), are transcription factors that are
important in anterior-posterior body axis patterning during normal
embryonic development (7).
However, aberrant expression of HOX genes has been demonstrated in
different tumor types, including leukemia (10), ovarian carcinoma (11) and pancreatic cancer (12), suggesting that HOX genes are
important in various types of tumor.
HOXB7 is a newly identified member of the HOX gene
family. It is highly expressed in embryonic tissues and is limited
to specific tissues of the embryo during the late trimester of
pregnancy but is not detected in the majority of terminally
differentiated adult tissues under normal conditions. As a
transcription factor, HOXB7 is also important in embryogenesis and
tissue differentiation. However, deregulation of HOXB7 has been
observed in esophageal squamous cell carcinoma, oral cancer,
ovarian carcinoma and breast cancer (13–16).
In addition, it has been reported that increased expression of
HOXB7 was closely associated with tumor cell proliferation,
invasion and metastasis (7,14,17).
However, the role of HOXB7 in breast cancer has yet
to be characterized. In the current study, the mRNA and protein
expression levels of HOXB7 in breast cancer cells were investigated
and the effect of HOXB7-siRNA on the proliferation, apoptosis and
invasion capacity of breast cancer was determined using a CCK-8
assay, flow cytometry (FCM) and transwell chambers. It was
hypothesized that HOXB7 siRNA inhibits the proliferation and
invasion of breast cancer cells and that HOXB7 may have therapeutic
potential in breast cancer.
Materials and methods
Main reagents and instruments
Fetal bovine serum (FBS), RPMI-1640 medium,
penicillin, streptomycin, phosphate-buffered saline (PBS; used as a
solvent) and pancreatic enzymes were purchased from HyClone
Laboratories, Inc. (Logan, UT, USA). The primers for HOXB7 and
GAPDH, rabbit monoclonal anti-human HOXB7 antibody, Lipofectamine
2000 and Lipofectamine™ RNAi MAX transfection reagents were
obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). The
RevertAid™ First Strand cDNA Synthesis kit was purchased from
Fermentas (Vilnius, Lithuania). The mouse polyclonal anti-human
GAPDH antibody was obtained from Tianjin Sungene Biotech Co., Ltd.
(Tianjin, China), siRNA was obtained from Shanghai Genepharma Co.,
Ltd., (Shanghai, China), the Cell Counting kit-8 (CCK-8) was
purchased from Beyotime Institute of Biotechnology (Wuhan, China),
Transwell plates were obtained from Corning Inc. (Corning, NY,
USA), Matrigel was purchased from BD Biosciences (San Jose, CA,
USA) and the Annexin-V-EGFP/propidium iodide (PI) Apoptosis
Detection kit was obtained from EnoGene Biotech Co., Ltd. (Nanjing,
China).
Cell culture and treatments
MDA-MB-231 and MCF-7 human breast cancer cell lines
were purchased from the China Center for Type Culture Collection
(Wuhan, China). The cells were cultured in RPMI-1640 medium
containing 10% FBS, 1% penicillin/streptomycin (100 U/ml
penicillin-G and 100 μg/ml streptomycin) at 37°C in a
humidified incubator that was supplemented with 5% CO2.
The cells were then passaged at 80–90% confluence and digested with
0.25% trypsin.
RNA extraction and reverse transcription
quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from MDA-MB-231 and MCF-7
breast cancer cells using TRIzol reagent (Invitrogen Life
Technologies). Total RNA (1.0 μg) was transcribed into
complementary DNA (cDNA) using the RevertAid™ First Strand cDNA
Synthesis kit according to the manufacturer's instructions. The RT
reaction was performed at 25°C for 10 min, 42°C for 60 min and 70°C
for 10 min. The mRNA levels were analyzed by RT-qPCR using the SYBR
Premix Ex Taq System (Clontech Laboratories, Inc., Mountain View,
CA, USA). The following primers were used: GAPDH, forward
5′-AATCCCATCACCATCTTCCAG-3′ and reverse 5′-GAGCCCCAGCCTTCTCCAT-3′
(118 bp); HOXB7, forward 5′-TACCCCTGGATGCGAAGCTC-3′ and reverse
5′-AATCTTGATCTGTCTTTCCGTGA-3′ (171 bp). The reaction mixture,
including 10.0 μl of 2X All-in-One™ qPCR Master mix
(GeneCopoeia, Rockville, MD, USA; AOPR-1200), 0.4 μl of
forward primer, 0.4 μl of reverse primer, 5 μl of
cDNA (10X dilution) and 4.2 μl of ddH2O, was
incubated at 95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec
for 40 cycles. The mRNA level of each sample was measured by the
2−ΔΔCT method (18). In
addition, qPCR (Applied Biosystems StepOnePlus™ Real-Time PCR
System; Applied Biosystems, Foster City, CA, USA) was performed
three times independently.
Western blotting
Cells were extracted using radioimmunoprecipitation
assay protein lysis buffer and the level of protein expression was
measured using the bicinchoninic acid assay protein assay kit
(Thermo Fisher Scientific, Inc., Wuhan, China). Total protein (40
μg) was loaded and run on a 12.5% SDS-polyacrylamide
gradient gel and 4% SDS-polyacrylamide stacking gel. Subsequently,
protein was transferred onto nitrocellulose filter membranes for
1.5 h and then incubated with 5% non-fat milk at room temperature
for 2 h to block nonspecific binding. This was then washed three
times with Tris-buffered saline with Tween 20 (TBST; 5 min each
time) and then treated with rabbit monoclonal anti-human HOXB7
antibody (1:400; Invitrogen Life Technologies; cat. no. 40–2000)
and mouse polyclonal anti-human GAPDH antibody (1:10,000; Tianjin
Sungene Biotech Co., Ltd.; cat. no. KM9002) overnight at 4°C with
continuous agitation. Subsequently, membranes were combined with
corresponding secondary antibody, goat anti-rabbit immunoglobulin G
(IgG)/horseradish peroxidase (HRP; 1:10,000 dilution; MR Biotech,
Co., Ltd., Shanghai, China) and goat anti-mouse IgG/HRP; (1:10,000
dilution; MR Biotech, Co., Ltd.) at 37°C for 1 h after washing
three times in TBST (15 min each time). Finally, the protein levels
were analyzed using an enhanced chemiluminescence kit (Biossci
Biotechnology Co., Ltd., Wuhan, China) according to the
manufacturer's instructions. Gray scale images were determined by
Quantity One 4.62 software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) and analyzed using GraphPad Prism 5 software (GraphPad
Software Inc., La Jolla, CA, USA). The optical density for target
protein was shown as a proportion of GAPDH optical density. The
experiments were replicated three times.
Synthesis of HOXB7-siRNA
Three sequences of siRNA oligonucleotides targeting
HOXB7 synthesized by Shanghai Genepharma Co., Ltd. were identified
and matched with the following HOXB7 cDNA sequences obtained from
GenBank through a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi): Sequence 1
(S1), sense (HOXB7-homo-462) 5′-GAGAGUAACUUCCGGAUCUTT-3′; sequence
2 (S2), sense (HOXB7-homo-633): 5′-CGGAAA GACAGAUCAAGAUTT-3′;
sequence of siRNA3, (S3), s en s e (HOX B7-homo -10 83): 5′-G C UAU
UGUA AGGUCUUUGUTT-3′. The siRNA sequence that exhibited the highest
interfering efficiency was selected to continue the study. By
contrast, one sequence of negative control siRNA (Sn) was
synthesized as: 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense).
siRNA transfection
The MCF-7 breast cancer cells cultured as described
were divided into the following five groups: Con-B group, blank
control group; Sn group, negative control siRNA; S1 group, S1
transfection group; S2 group, S2 transfection group and S3 group,
S3 transfection group. Cells were seeded on 6-well plates at a
density of 1×106 cells/well 24 h prior to transfection.
When 80–90% confluence was reached, HOXB7-siRNA or negative control
siRNA were transfected into MCF-7 cells at three different
concentrations (0.1, 0.2 and 0.4 μM, respectively) using
Lipofectamine 2000 (Invitrogen Life Technologies) in RPMI-1640
medium. Fluorescence, which was detected under a fluorescence
microscope (VHB100F; Olympus Corporation, Shanghai, China) after
transfection for 4–6 h, was used to optimize the condition of
transfection.
Detection of interference efficiency by
RT-qPCR and western blotting
Following transfection with HOXB7-siRNA for 48 and
72 h, five groups of MCF-7 cells were harvested to detect the mRNA
and protein expression levels of HOXB7 by RT-qPCR and western
blotting as described above, respectively. Interference
efficiencies were calculated as follows: Interference efficiency =
(mRNA expression of HOXB7 in the control group - mRNA expression of
HOXB7 in the treatment group) / mRNA expression of HOXB7 in the
control group × 100%. Each experiment was repeated three times.
Cells transfected with siRNA3 had the lowest expression levels of
HOXB7 mRNA and protein and, therefore, S3 was used in the following
experiments.
CCK-8 cell proliferation assay
According to the manufacturer's instructions for the
CCK-8 assay, the three groups of MCF-7 cells, including the Con-B
group (blank control group), Sn group (negative control siRNA) and
S3 group (siRNA3 transfection group) were collected at the
exponential phase and inoculated onto a 96-well plate at a density
of 5×103 cells/well. Following inoculation for 24, 48,
72 or 96 h, each well was replaced with 100 μl RPMI-1640
supplemented with 10% FBS, 1% penicillin/streptomycin and 10
μl CCK. Subsequently, the cells were incubated at 37°C away
from light for 4 h. The absorbance was then measured at 450 nm for
each well using a microplate reader (EL×808™; BioTek Instruments,
Inc., Winooski, VT, USA). Cell viability was calculated using the
following formula: Cell viability (%) = (Absorbance in treated
sample / Absorbance in control) × 100%. Three independent
experiments were performed with triplicate wells.
Flow cytometric analysis
Following the transfection of MCF-7 cells with
HOXB7-S3 and Sn for 48 h, 1×106 cells were collected and
then resuspended in 500 μl binding buffer containing 5
μl Annexin V-EGFP conjugated antibody and 5 μl PI for
exactly 5 min in the dark at room temperature. Cells were
transferred into flow test tubes within 1 h and then detected on a
BD FACSCalibur flow cytometer (Becton-Dickinson, San Jose, CA,
USA). The data were analyzed using FlowJo software version 6.0
(Tree Star, Inc., Ashland, OR, USA).
Cell invasion assay
Transwell membranes were purchased from Corning Inc.
Matrigel (100 μl), which was diluted in serum-free RPMI-1640
medium following thawing at 4°C overnight, was placed into the
upper chamber of a 24-well transwell and the transwell was
incubated at 37°C for at least 4–5 h for gelling. Three groups of
MCF-7 cells grouped as described above with a given concentration
of 5×103 cells/ml were prepared, 100 μl of which
was added to the upper chamber at 37°C. At 24 and 48 h
post-incubation, the cells were fixed with formaldehyde, stained
and counted under a microscope (37XB; Zhonghen Instrument, Co.,
Ltd., Shanghai, China). The invasion activity of the carcinoma
cells was characterized with the average transmembrane cell
numbers. Each experiment was repeated three times.
Statistical analysis
Each experiment was repeated three times. All data
are expressed as the mean ± standard deviation and undertaken using
the statistical software SPSS 18.0 (SPSS, Inc., Chicago, IL, USA).
Comparisons were made using an independent samples t-test.
P<0.05 was considered to indicate a statistically significant
difference (*P<0.05, **P<0.01 and
***P<0.001).
Results
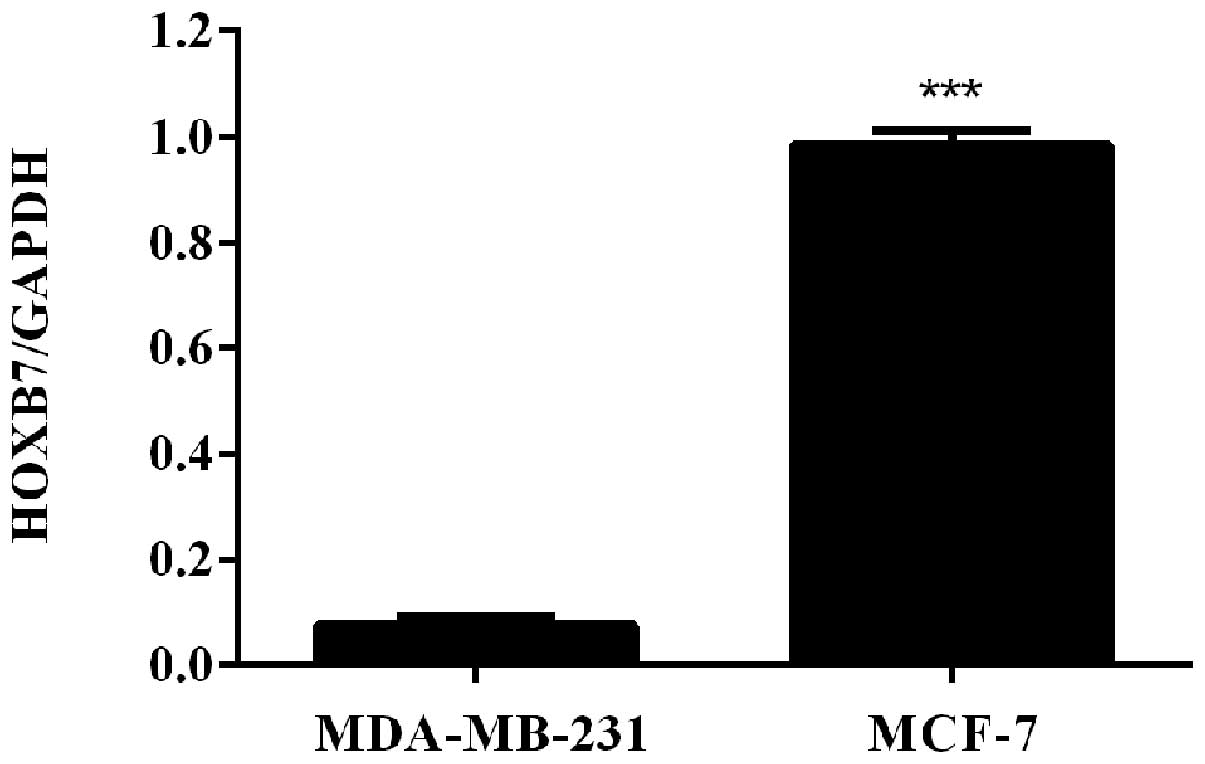
HOXB7 mRNA is overexpressed in two breast
cancer cells
To detect the mRNA expression level of HOXB7 in
breast cancer cells, RT-qPCR was used in two human breast cancer
cell lines, including MDA-MB-231 and MCF-7 cells. The results of
the RT-qPCR revealed that HOXB7 mRNA was overexpressed in
MDA-MB-231 and MCF-7 breast cancer cell lines and the expression
level in MCF-7 breast cancer cells was significantly higher
compared with that in MDA-231 breast cancer cells
(***P<0.001; Fig.
1).
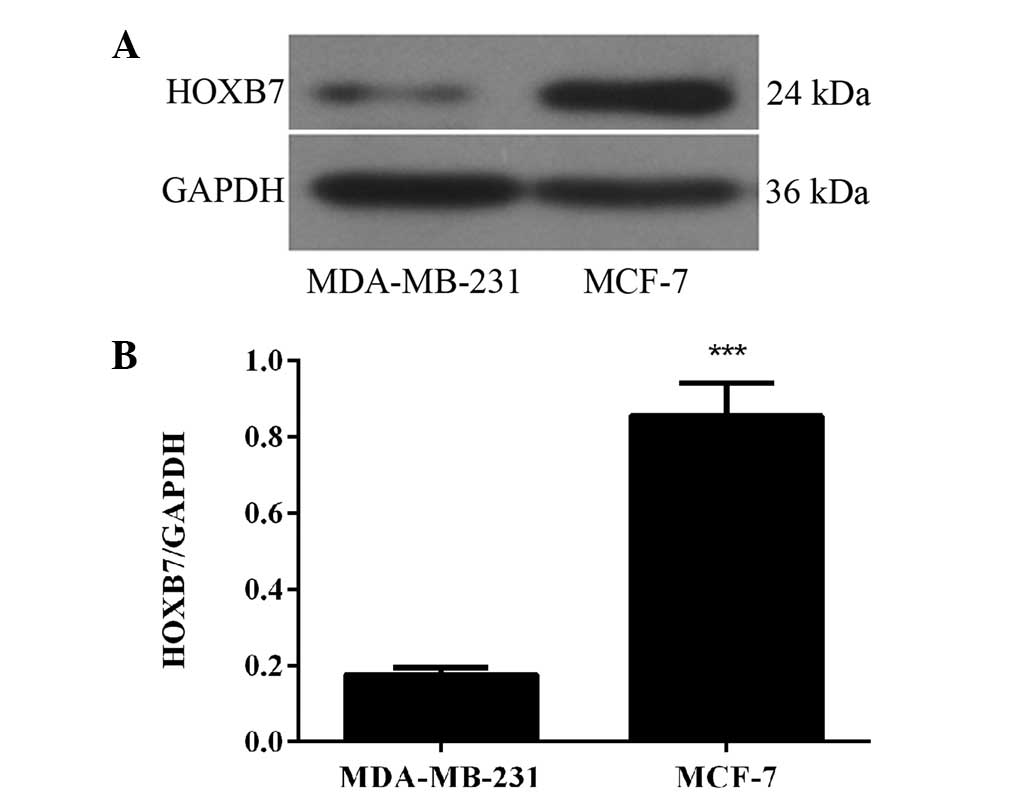
HOXB7 protein is overexpressed in two
breast cancer cells
To detect whether the protein expression level of
HOXB7 was in accordance with the results of the mRNA expression
level in breast cancer cells, western blotting was used in the same
two human breast cancer cell lines, namely MDA-MB-231 and MCF-7
cells. The results of the western blotting revealed that HOXB7
protein was also overexpressed in MDA-MB-231 and MCF-7 breast
cancer cell lines and the expression level in MCF-7 breast cancer
cells was significantly higher compared with that in MDA-231 breast
cancer cells (***P<0.001; Fig. 2). Therefore, combined with the
result in Fig. 1, the MCF-7 human
breast cancer cell line was selected for further experiments.
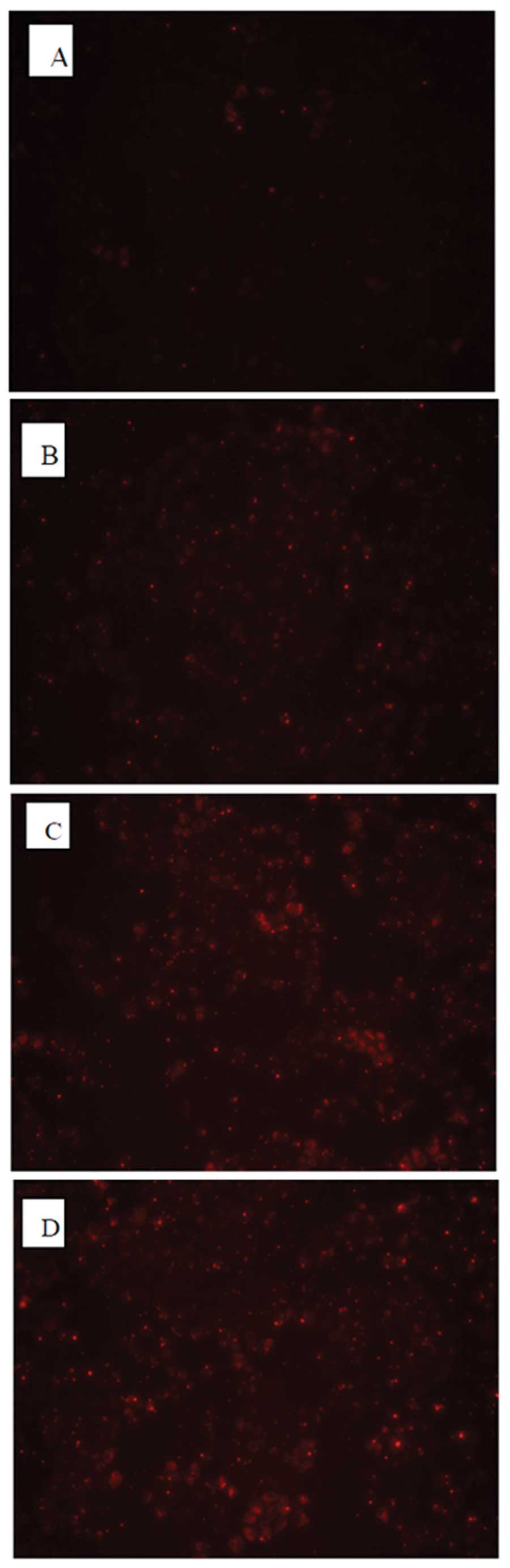
Detection of the optimal conditions of
transfection into MCF-7 cells with HOXB7-siRNA
To optimize conditions of transfection, three
different concentrations of HOXB7-siRNA-NC-CY3 (fluorescent
negative control) were transfected into MCF-7 breast cancer cells.
As shown in Fig. 3, all MCF-7
cells transfected with HOXB7-siRNA for 4–6 h exhibited red
fluorescence under a fluorescence microscope. It was observed that
the untransfected MCF-7 exhibited no red fluorescence (Fig. 3A) and the MCF-7 cells transfected
with 0.1 μM siRNA demonstrated a little red fluorescence
(Fig. 3B). By contrast, ~80%
infected cells exhibited red fluorescence at an siRNA concentration
of 0.2 μM as shown in Fig.
3C, while 90% exhibited red fluorescence at an siRNA
concentration of 0.4 μM as shown in Fig. 3D, suggesting successful
transfection. Thus, the concentration of 0.4 μM siRNA that
exhibited the highest transfection efficiency was selected for the
following experiments.
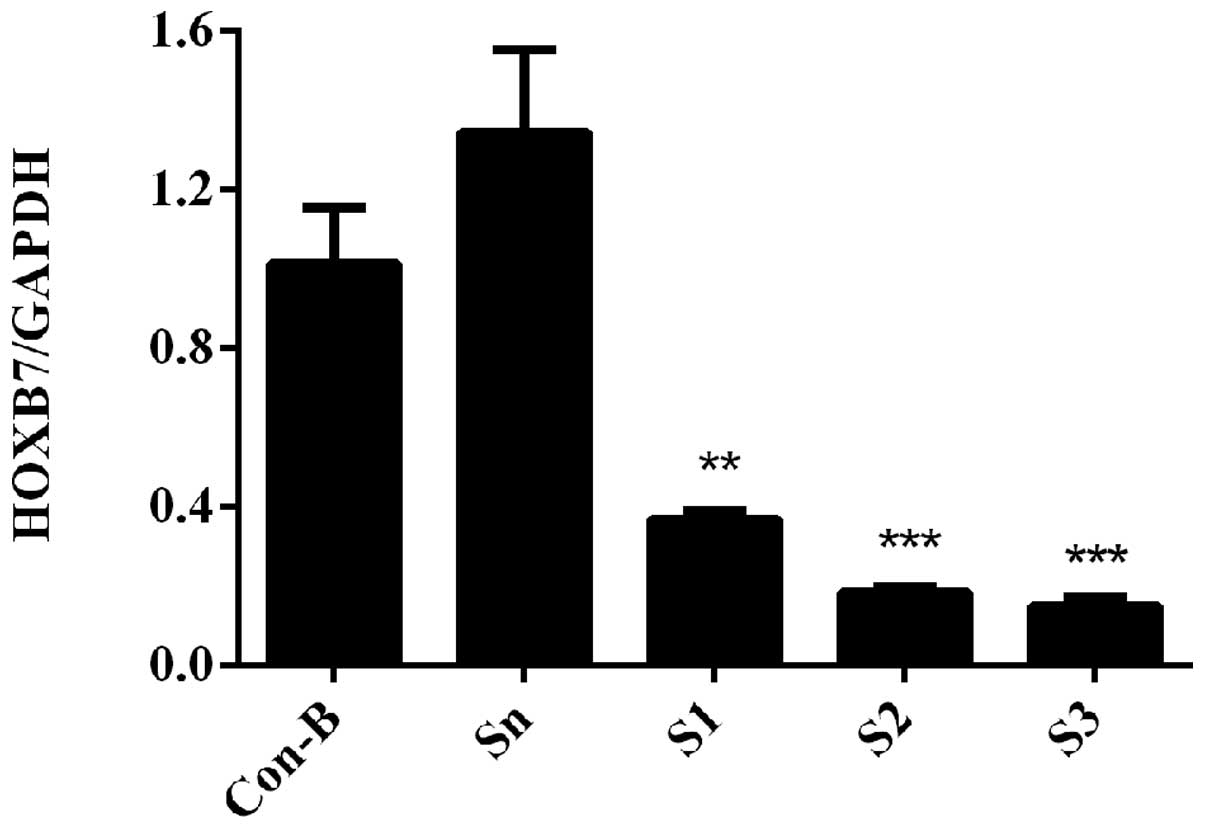
HOXB7 mRNA expression level is
downregulated following siRNA transfection
To determine the interference efficiency of
HOXB7-siRNA on MCF-7 cells, RT-qPCR was used following transfection
for 48 h. The results demonstrated that HOXB7 mRNA was
overexpressed in Con-B and Sn groups following transfection and no
clear difference was observed in the two groups (P>0.05).
However, the mRNA expression levels of HOXB7 in the S1, S2 and S3
groups were significantly downregulated compared with the Con-B and
Sn groups with interference efficiencies of 63.38±0.02, 81.53±0.01
and 84.87±0.02%, respectively. The differences were statistically
significant (**P<0.01 and ***P<0.001;
Fig. 4). The mRNA expression level
of HOXB7 was the lowest in the S3 group and differed markedly from
that in the Con-B and Sn groups (***P<0.001; Fig. 4).
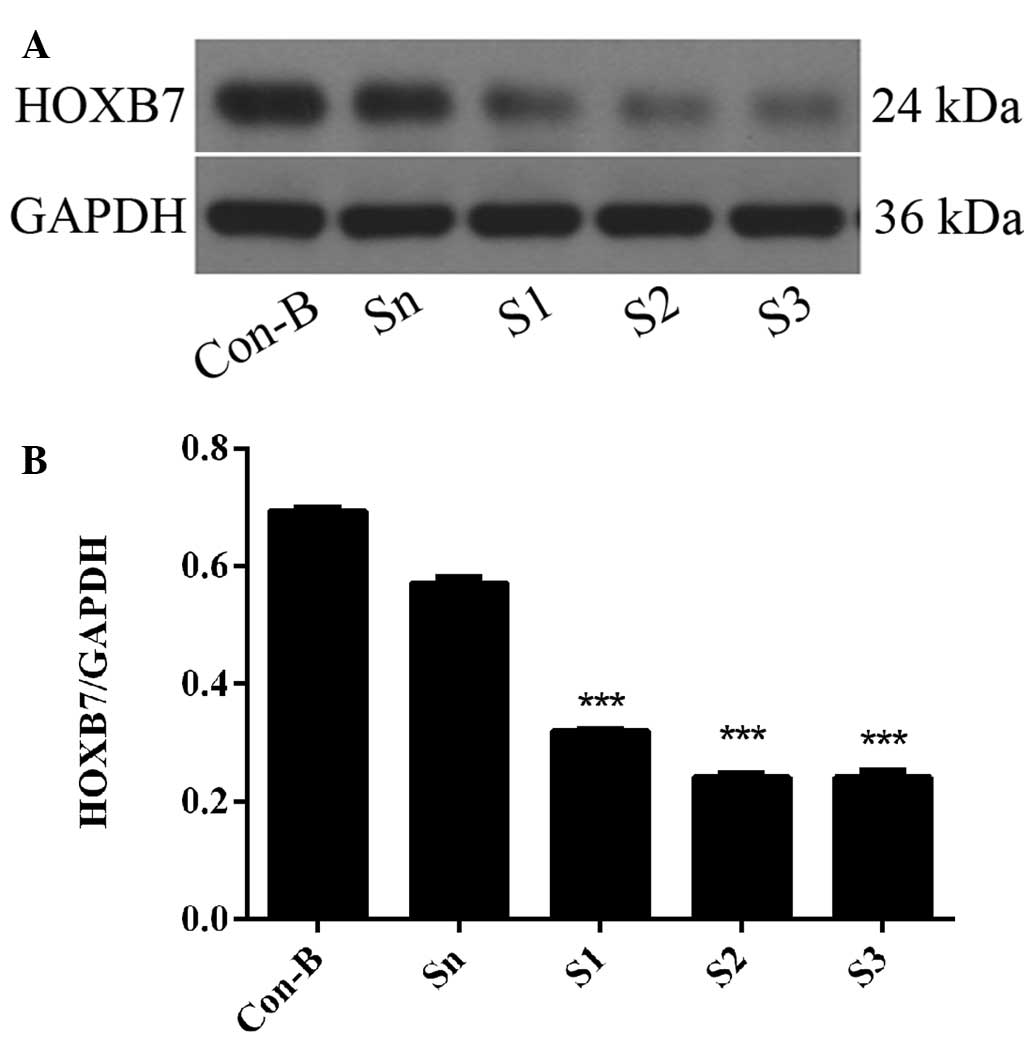
HOXB7 protein expression level is
downregulated following siRNA transfection
To further determine the interference efficiency of
HOXB7-siRNA on MCF-7 cells, western blotting was used following
transfection for 72 h. The results demonstrated that HOXB7 protein
was overexpressed in the Con-B and Sn groups following transfection
and no differences were observed between the two groups
(P>0.05). However, the expression of HOXB7 protein in the S1, S2
and S3 groups was significantly downregulated compared with the
Con-B and Sn groups with interference efficiencies of 53.88±0.0001,
65.22±0.004 and 65.25±0.001%, respectively. The differences were
all statistically significant (***P<0.001; Fig. 5). The HOXB7 protein expression
level was the lowest in the S3 group and differed markedly from
that in the Con-B and Sn groups (***P<0.001; Fig. 5). Combined with the result in
Fig. 4, S3 had the maximum
inhibition efficiency of HOXB7 and therefore S3 was used in the
following experiments.
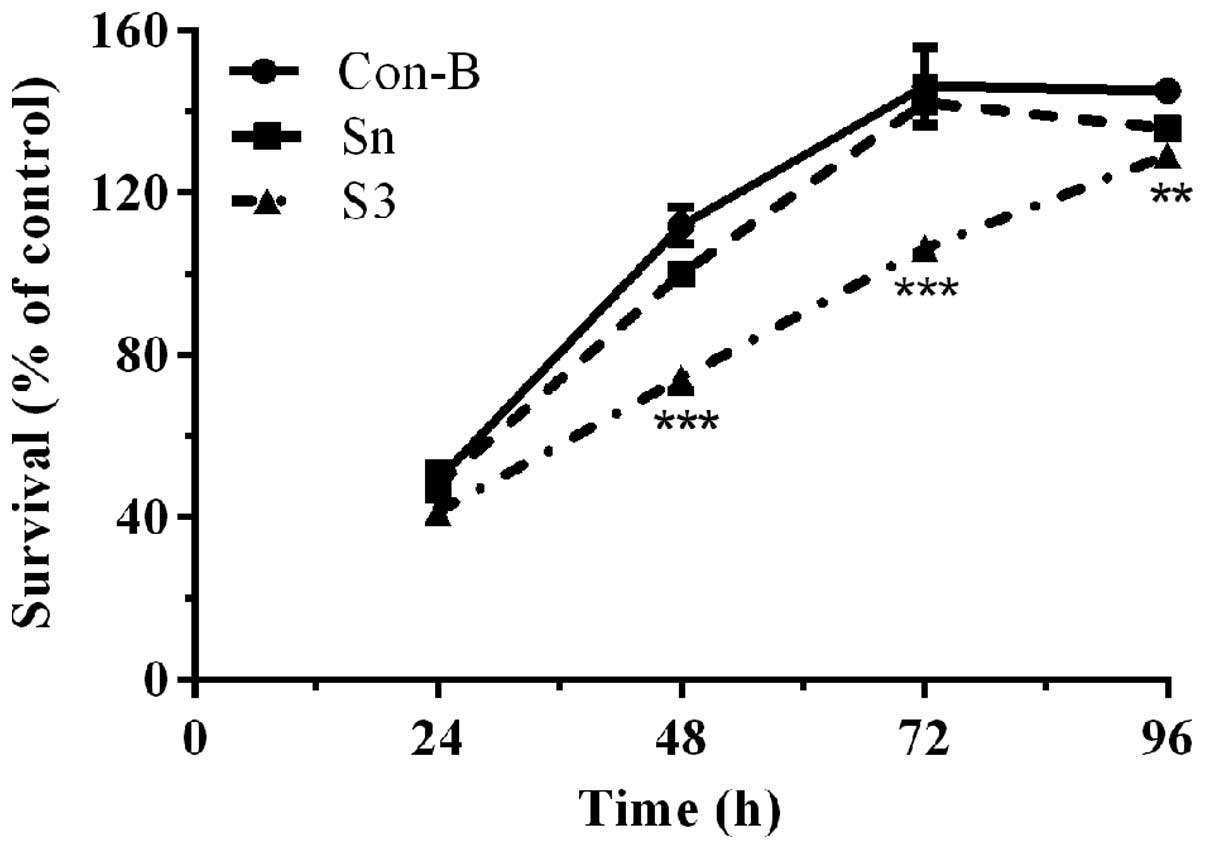
Downregulation of HOXB7 gene expression
effectively inhibits MCF-7 cell proliferation
To determine the effect of HOXB7-S3 on cell
proliferation in MCF-7 breast cancer cells, a CCK-8 assay was used.
As shown in Fig. 6, the cell
viability of the three groups demonstrated no significant
difference following transfection with HOXB7-S3 or HOXB7-Sn for 24
h. Between 48 and 72 h, the cell viability of the S3 group was
significantly decreased compared with that of the Con-B groups. In
particular, the cell viability of the S3 group (74.43±2.55%) at 48
h post-transfection was significantly decreased compared with that
of the Con-B group (111.97±3.66%) and Sn group (100±1.58%). The
difference was statistically significant
(***P<0.001). The cell viability of the S3 group at
96 h post-transfection demonstrated a partially decreased cell
survival, however, this decease was not so pronounced. This result
demonstrated that HOXB7-S3 could effectively and significantly
inhibit the proliferation of human breast cancer MCF-7 cells.
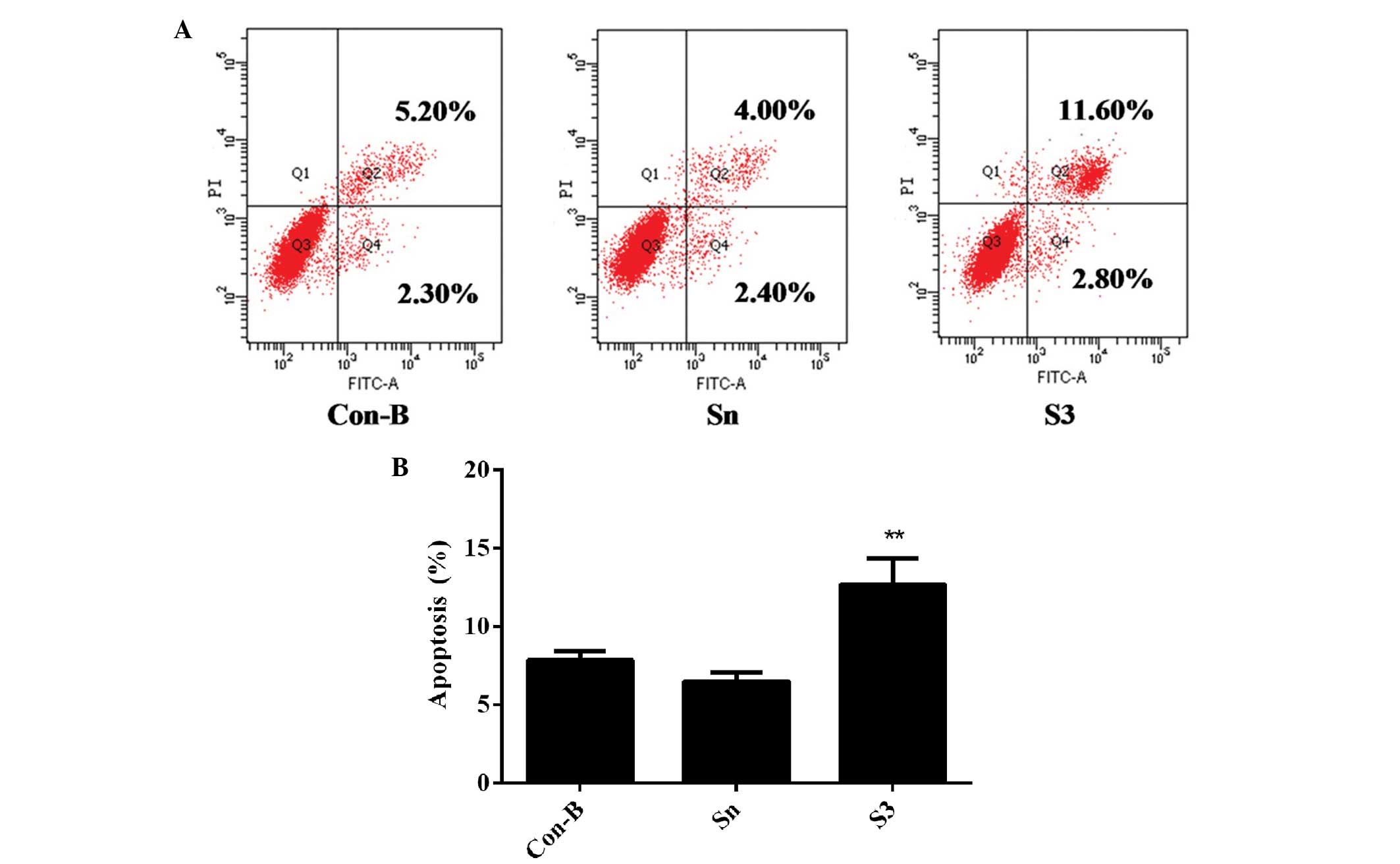
Downregulation of HOXB7 gene expression
effectively enhances MCF-7 cell apoptosis
To detect whether the effect of HOXB7-S3 on cell
proliferation in MCF-7 breast cancer cells occurred by apoptosis,
the percentage of early-stage apoptotic cells and late-stage
apoptotic cells was measured in three groups by FCM. At 48 h
post-transfection of siRNA, the total number of apoptotic cells
(12.70±1.75%) was markedly increased in the S3 group, compared with
7.83±0.47% in the Con-B group and 6.46±0.49% in the Sn group,
respectively (Fig. 7). The
differences between the three groups were statistically significant
(**P<0.01). These results indicated that HOXB7-S3
enhanced MCF-7 cell apoptosis.
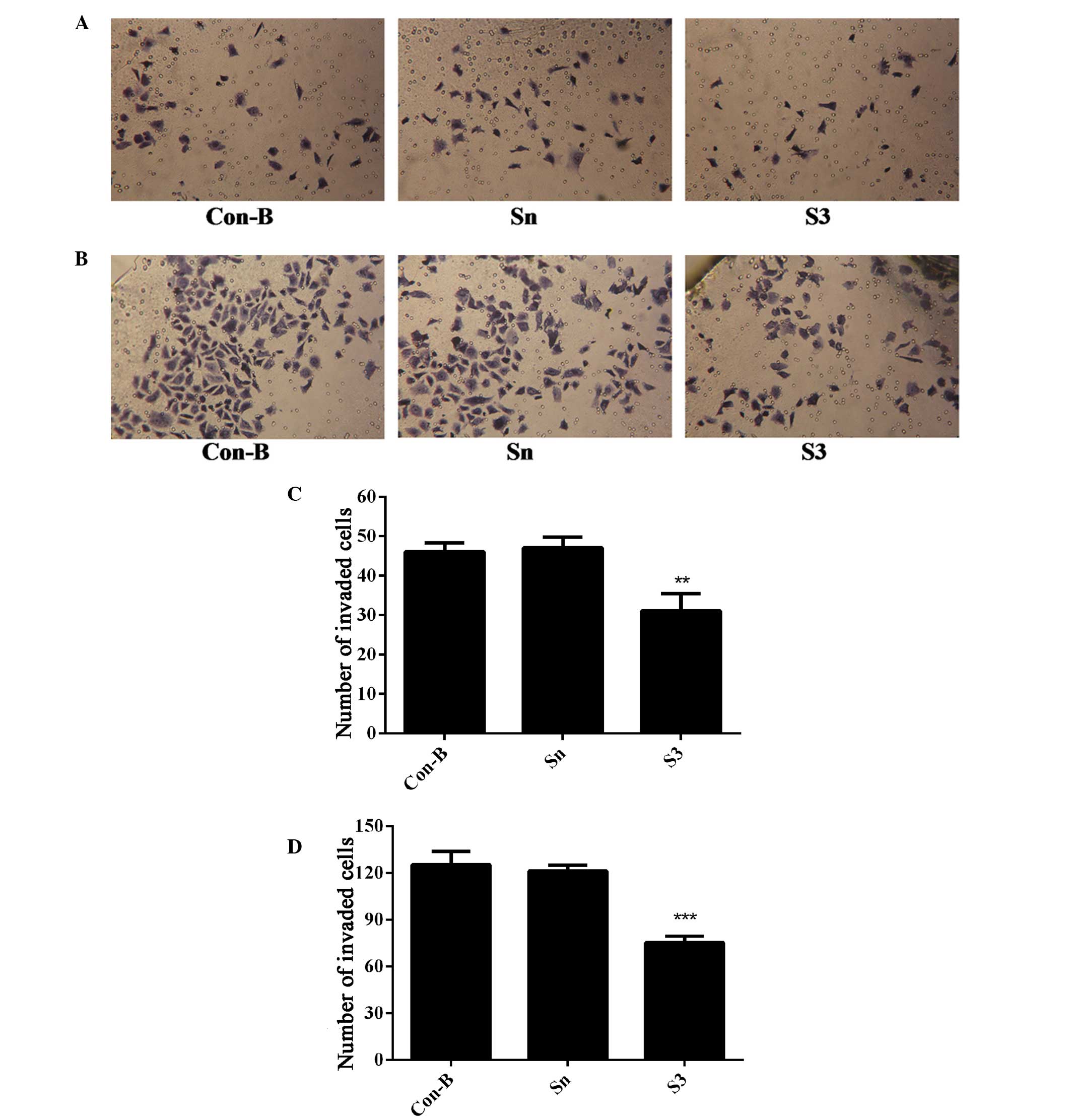
Downregulation of HOXB7 gene expression
inhibits MCF-7 cell invasion
To determine the effect of HOXB7-S3 on cell
proliferation in MCF-7 breast cancer cells, the number of cells
that had migrated through the Matrigel membrane was measured at 24
and 48 h post-transfection of MCF-7 cells with HOXB7-S3 by
Transwell assay. As shown in Fig. 8A
and B, the number of migrated cells in the S3 group was
markedly reduced compared with in the Sn and Con-B groups. The
number of migrated cells in the S3, Con-B and Sn groups 24 h
post-transfection was 31±3.63, 46±1.87 and 47±2.27, respectively.
The difference was statistically significant
(**P<0.01; Fig. 8C).
At 48 h post-transfection, the number of migrated cells in the S3
group was 75±3.49, which was apparently reduced compared with
125±7.18 in the Con-B group and 121±3.25 in the Sn group (Fig. 8B). The difference was statistically
significant (***P<0.001; Fig. 8D). This suggested that HOXB7-S3
markedly inhibited the invasiveness of MCF-7 cells.
Discussion
Breast cancer is characterized by a multiphasic
process in which a series of changes occur in sequence, leading to
the loss of control of cell proliferation, differentiation,
apoptosis and DNA repair (19). To
date, there have been numerous studies demonstrating a possible
association between the abnormal expression of HOX genes and breast
cancer. For example, Ramen et al and Chu et al found
that HOXA5 and HOXA10 are underexpressed in breast cancer (20,21).
Conversely, Jansen et al suggested that HOXB13 is
overexpressed in breast cancer (22,23).
Initially identified in Drosophila (24), the HOX genes encode a family of
highly conserved transcription factors that normally regulate
temporospatial development of the extremities and organs (25). Aberrant expression of these genes
in different tissues has been demonstrated to be associated with
tumorigenesis (26,27), particularly HOXB7, a member of the
HOX gene family, which is reported to be overexpressed in
numerous cancer cells, including melanoma cells, ovarian epithelial
cells and SkBr3 breast carcinoma cells (15,28,29)
has a key role in tumorigenesis. To the best of our knowledge, the
current study is the first to demonstrate that the mRNA and protein
expression of HOXB7 was overexpressed in MDA-MB-231 and MCF-7
breast cancer cell lines.
Additionally, it was reported that as a
transcription factor, HOXB7 has two opposite functions in different
cellular contexts. The majority of studies supported that HOXB7 may
be important in promoting the multistep process of tumor formation
and progression, including transformation, proliferation, survival,
angiogenesis, invasion and metastasis (12,14,17,30,31).
By contrast, another study observed a promoting role of HOXB7 in
differentiation in hematopoietic stem cells and multipotent
mesenchymal cells (32). In order
to investigate the role HOXB7 in breast cancer cells, three pairs
of HOXB7-siRNA were transfected into MCF-7 breast cancer cells and
the mRNA and protein expression levels of HOXB7 were effectively
downregulated. In particular, HOXB7-S3 significantly and
specifically inhibited HOXB7 expression at the mRNA and protein
levels with interference efficiencies of 84.87±0.02 and
65.25±0.001%, respectively. Thus, it was concluded that HOXB7-S3
could effectively induce gene RNA interference (RNAi) and HOXB7-S3
was selected to downregulate HOXB7 gene expression in the following
experiments. The results of the CCK-8 assay and transwell chambers
demonstrated that downregulation of HOXB7 gene expression
effectively inhibited MCF-7 cell proliferation and invasion in
MCF-7 cells, which contributed to malignant transformation and
tumorigenesis. The present data predominantly support the
pro-tumorigenic function of HOXB7. In addition, understanding the
molecular abnormalities of HOXB7 involved in the pathogenesis of
breast cancer cells may reveal new targets for therapy and
HOXB7-siRNA and antagonists could be used to inhibit the
proliferation and invasion capacity of breast cancer cells.
Although HOXB7 has been associated with the
regulation of proliferation and invasion of cancer cells, the
molecular mechanisms remain poorly identified. Certain studies
reported that bFGF, one of the direct targets of HOXB7, contributed
to HOXB7-induced cellular proliferation and transformation
(14,15). In addition to bFGF, Carè et
al found that HOXB7 can also induce the expression of other
genes, particularly those associated with angiogenesis and tumor
invasion, including vascular endothelial growth factor,
interleukin-8, angiopoietin-2 and metalloproteases 2 and 9
(31). Wu et al
demonstrated that HOXB7 could activate the Ras-RAF-MAPK pathway in
breast cancer cell lines, thereby promoting cell proliferation
(16). In the current study,
whether the effect of HOXB7-S3 on cell proliferation in MCF-7
breast cancer cells occurred by apoptosis was detected using FCM
and the results demonstrated that at 48 h post-transfection of
siRNA, the total number of apoptotic cells (12.70±1.75%) was
markedly increased in the S3 group, compared with 7.83±0.47% in the
Con-B groups and 6.46±0.49% in the Sn groups, respectively
(Fig. 7), which was in accordance
with the result of the CCK-8 assay demonstrating that the cell
viability of the S3 groups (74.43±2.55%) at 48 h post-transfection
was significantly decreased compared with that of the Con-B groups
(111.97±3.66%) and Sn groups (100±1.58%; Fig. 6), which indicated that HOXB7-S3 may
inhibit the proliferation of MCF-7 breast cancer cells through
enhancing the apoptotic rate.
In conclusion, the results of the present study
demonstrated that HOXB7-S3 decreased the mRNA and protein
expression levels of HOXB7 and inhibited the cell proliferation and
invasion of MCF-7 breast cancer cells, indicating that HOXB7 RNAi
is a potential treatment for breast cancer. HOXB7 may be a
potential and valuable therapeutic target in human breast
cancer.
Acknowledgments
This study was supported by grants from the Science
and Technology of Hubei Province Funds (no. 2011 CHB 020[SP1]). The
authors would like to thank all members of our laboratories for
their technical assistance and advice during the experiments.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Walsh T, Casadei S, Coats KH, Swisher E,
Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, et
al: Spectrum of mutations in BRCA1, BRCA2, CHEK2 and TP53 in
families at high risk of breast cancer. JAMA. 295:1379–1388. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mantzourani M, Gogas H, Katsandris A and
Meletis J: Severe thrombocytopenia related to trastuzumab infusion.
Med Sci Monit. 17:CS85–CS87. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nunes FD, de Almeida FC, Tucci R and de
Sousa SC: Homeobox genes: A molecular link between development and
cancer. Pesqui Odontol Bras. 17:94–98. 2003. View Article : Google Scholar
|
|
5
|
Grier DG, Thompson A, Kwasniewska A,
McGonigle GJ, Halliday HL and Lappin TR: The pathophysiology of HOX
genes and their role in cancer. J Pathol. 205:154–171. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhatlekar S, Fields JZ and Boman BM: HOX
genes and their role in the development of human cancers. J Mol Med
(Berl). 92:811–823. 2014. View Article : Google Scholar
|
|
7
|
Nguyen Kovochich A, Arensman M, Lay AR,
Rao NP, Donahue T, Li X, French SW and Dawson DW: HOXB7 promotes
invasion and predicts survival in pancreatic adenocarcinoma.
Cancer. 119:529–539. 2013. View Article : Google Scholar
|
|
8
|
Cantile M, Pettinato G, Procino A,
Feliciello I, Cindolo L and Cillo C: In vivo expression of the
whole HOX gene network in human breast cancer. Eur J Cancer.
39:257–264. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGinnis W and Krumlauf R: Homeobox genes
and axial patterning. Cell. 68:283–302. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pineault N, Abramovich C, Ohta H and
Humphries RK: Differential and common leukemogenic potentials of
multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell
Biol. 24:1907–1917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong JH, Lee JK, Park JJ, Lee NW, Lee KW
and Na JY: Expression pattern of the class I homeobox genes in
ovarian carcinoma. J Gynecol Oncol. 21:29–37. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chile T, Fortes MA, Corrêa-Giannella ML,
Brentani HP, Maria DA, Puga RD, de Paula Vde J, Kubrusly MS, Novak
EM, Bacchella T, et al: HOXB7 mRNA is overexpressed in pancreatic
ductal adenocarcinomas and its knockdown induces cell cycle arrest
and apoptosis. BMC Cancer. 13:4512013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen KN, Gu ZD, Ke Y, Li JY, Shi XT and Xu
GW: Expression of 11 HOX genes is deregulated in esophageal
squamous cell carcinoma. Clin Cancer Res. 11:1044–1049.
2005.PubMed/NCBI
|
|
14
|
De Souza Setubal Destro MF, Bitu CC,
Zecchin KG, Graner E, Lopes MA, Kowalski LP and Coletta RD:
Overexpression of HOXB7 homeobox gene in oral cancer induces
cellular proliferation and is associated with poor prognosis. Int J
Oncol. 36:141–149. 2010.
|
|
15
|
Naora H, Yang YQ, Montz FJ, Seidman JD,
Kurman RJ and Roden RB: A serologically identified tumor antigen
encoded by a homeobox gene promotes growth of ovarian epithelial
cells. Proc Natl Acad Sci USA. 98:4060–4065. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu X, Chen H, Parker B, Rubin E, Zhu T,
Lee JS, Argani P and Sukumar S: HOXB7, a homeodomain protein, is
overexpressed in breast cancer and confers epithelial-mesenchymal
transition. Cancer Res. 66:9527–9534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen H, Lee JS, Liang X, Zhang H, Zhu T,
Zhang Z, Taylor ME, Zahnow C, Feigenbaum L, Rein A, et al: Hoxb7
inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but
promotes progression and lung metastasis. Cancer Res. 68:3637–3644.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
19
|
Vogelstein B and Kinzler KW: The multistep
nature of cancer. Trends Genet. 9:138–141. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raman V, Martensen SA, Reisman D, Evron E,
Odenwald WF, Jaffee E, Marks J and Sukumar S: Compromised HOXA5
function can limit p53 expression in human breast tumours. Nature.
405:974–978. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chu MC, Selam FB and Taylor HS: HOXA10
regulates p53 expression and matrigel invasion in human breast
cancer cells. Cancer Biol Ther. 3:568–572. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jansen MP, Sieuwerts AM, Look MP, Ritstier
K, Meijer-van Gelder ME, van Staveren IL, Klijn JG, Foekens JA and
Berns EM: HOXB13-to-IL17BR expression ratio is related with tumor
aggressiveness and response to tamoxifen of recurrent breast
cancer: A retrospective study. J Clin Oncol. 25:662–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shah N, Jin K, Cruz LA, Park S, Sadik H,
Cho S, Goswami CP, Nakshatri H, Gupta R, Chang HY, et al: HOXB13
mediates tamoxifen resistance and invasiveness in human breast
cancer by suppressing ERα and inducing IL-6 expression. Cancer Res.
73:5449–5458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krumlauf R: Hox genes in vertebrate
development. Cell. 78:191–201. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garaulet DL, Castellanos MC, Bejarano F,
Sanfilippo P, Tyler DM, Allan DW, Sánchez-Herrero E and Lai EC:
Homeotic function of Drosophila Bithorax-complex miRNAs mediates
fertility by restricting multiple Hox genes and TALE cofactors in
the CNS. Dev Cell. 29:635–648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhatlekar S, Addya S, Salunek M, Orr CR,
Surrey S, McKenzie S, Fields JZ and Boman BM: Identification of a
developmental gene expression signature, including HOX genes, for
the normal human colonic crypt stem cell niche: Overexpression of
the signature parallels stem cell overpopulation during colon
tumorigenesis. Stem Cells Dev. 23:167–179. 2014. View Article : Google Scholar :
|
|
27
|
Cantile M, Scognamiglio G, La Sala L, La
Mantia E, Scaramuzza V, Valentino E, Tatangelo F, Losito S,
Pezzullo L, Chiofalo MG, et al: Aberrant expression of posterior
HOX genes in well differentiated histotypes of thyroid cancers. Int
J Mol Sci. 14:21727–21740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caré A, Silvani A, Meccia E, Mattia G,
Stoppacciaro A, Parmiani G, Peschle C and Colombo MP: HOXB7
constitutively activates basic fibroblast growth factor in
melanomas. Mol Cell Biol. 16:4842–4851. 1996.PubMed/NCBI
|
|
29
|
Caré A, Silvani A, Meccia E, Mattia G,
Peschle C and Colombo MP: Transduction of the SkBr3 breast
carcinoma cell line with the HOXB7 gene induces bFGF expression,
increases cell proliferation and reduces growth factor dependence.
Oncogene. 16:3285–3289. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao WT, Jiang D, Yuan J, Cui YM, Shi XW,
Chen CM, Bian XW, Deng YJ and Ding YQ: HOXB7 as a prognostic factor
and mediator of colorectal cancer progression. Clin Cancer Res.
17:3569–3578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carè A, Felicetti F, Meccia E, Bottero L,
Parenza M, Stoppacciaro A, Peschle C and Colombo MP: HOXB7: A key
factor for tumor-associated angiogenic switch. Cancer Res.
61:6532–6539. 2001.PubMed/NCBI
|
|
32
|
Boström K, Tintut Y, Kao SC, Stanford WP
and Demer LL: HOXB7 overexpression promotes differentiation of
C3H10T1/2 cells to smooth muscle cells. J Cell Biochem. 78:210–221.
2000. View Article : Google Scholar : PubMed/NCBI
|