Introduction
Osteoarthritis (OA) is the most prevalent chronic
joint disease and results in a large economic burden due to the
associated costs of medical care and lost earnings (1). OA is characterized by the
disappearance of the cartilage, combined with sub-chondral bone
sclerosis, formation of osteophytes and a mild inflammation of the
synovial membrane (2).
The pathology of OA is complex and a large number of
studies have focused on the pathogenesis of OA to identify a
therapeutic method for patients with OA. The sub-chondral bone is
involved in the pathophysiology of OA through biochemical and
mechanical pathways (3). OA
microarray analysis has predominantly focused on the articular
cartilage, meniscus or synovium; however, only few studies on
sub-chondral bone are available (4,5).
Therefore, the present study assessed the sub-chondral bone in
order to enhance the understanding of the pathology of OA. Zhen
et al (6) reported that the
pathological changes of OA may be caused by high concentrations of
active transforming growth factor-β1 in the subchondral bone, and
that inhibition of this process may represent a potential
therapeutic method. According to a study by Valverde-Franco et
al (7), bone-specific
overexpression of EphB4 on subchondral bone and cartilage
has a protective effect on OA. With the number of studies
increasing, further genes associated with OA are being
identified.
Microarray analysis has been used to analyze the
gene expression of thousands of transcripts in OA samples. Several
novel candidate genes, including bone formation-associated genes
(CLEC3B, CDH11, GPNMB, CLEC3A,
CHST11, MSX1, MSX2) and genes encoding
collagens (COL13A1, COL14A1, COL15A1,
COL8A2), were identified by microarray analysis (8). Chou et al (4) only performed functional and pathway
enrichment analyses of the identified differentially expressed
genes (DEGs). In the present study, the microarray data (GSE51588)
(4) were downloaded from Gene
Expression Omnibus using additional bioinformatics analysis to gain
further insight into the molecular mechanisms of OA. Gene Ontology
(GO) functional terms and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis were performed for the DEGs. In
addition, a protein-protein interaction (PPI) network was
constructed and protein domain enrichment of the genes in the
modules of the PPI network were performed to screen the significant
genes, which were involved in the pathogenesis of OA.
Materials and methods
Microarray data and data
pre-processing
The microarray data (GSE51588) deposited by Chou
et al (4) were downloaded
from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The platform used
was the GPL13497 Agilent-026652 Whole Human Genome Microarray 4×44
K v2 (Agilent Technologies, Palo Alto, CA, USA). A total of 50
samples were available, including 20 OA knee lateral tibial plateau
samples, 20 OA knee medial plateau samples, 5 non-OA knee lateral
tibial plateau samples and 5 non-OA knee medial plateau
samples.
For data pre-processing, the expression profile chip
was pre-processed using the Affy package in Bioconductor
(http://www.bioconductor.org/packages/release/bioc/html/affy.html)
(9) and Affymetrix annotation
files from Brain Array Lab (Affymetrix, Santa Clara, CA, USA;
http://www.affymetrix.com/analysis/).
The background correction, quartile data normalization and probe
summarization were performed using the robust multiarray average
algorithm (http://www.bioconductor.org) (10) to obtain a gene expression
matrix.
Identification of DEGs
The expression values for the normalized data were
calculated using the Limma package in R (Affymetrix; http://www.affymetrix.com/analysis/) (11). DEGs were identified by Student's
t-test. The raw P-value was adjusted into false discovery rate
(FDR) using the Benjamini & Hochberg method (12). An FDR <0.01 and |log2fold
change|>1 were selected as cut-off criteria.
Enrichment analysis for DEGs
Functional enrichment of the DEGs in the biological
process, molecular function and cellular component categories was
performed using the GO database (http://geneontology.org/) (13). Based on the KEGG database
(http://www.genome.jp/kegg/pathway.html) (14), pathway enrichment of the DEGs was
performed. P<0.01 was selected as a cut-off criterion.
Construction of the PPI network and
module analysis
The identified DEGs were mapped to the Search Tool
for the Retrieval of Interacting Genes version 9.1 database
(http://www.string-db.org/) (15) to search for the interaction
associations between the proteins, and a confidence score >0.4
was selected as cut-off criterion. Cytoscape software (http://www.cytoscape.org/) (16) was subsequently used to visualize
the PPI network.
Module analysis of the PPI network was performed by
CFinder (http://www.cfnder.org/) (17). The search algorithm using the
Clique Percolation Method was used to identify the k-clique
percolation clusters of the network (18). Larger k-clique values represented
higher stringency during the identification of dense groups and
provided smaller groups with a higher density of links inside them.
A k-cliques value of 5 was selected as the cut-off criterion. The
protein domain enrichment analysis of genes in the module was
analyzed using the INTERPRO database (http://www.ebi.ac.uk/interpro/) (17,19)
and P<0.05 was selected as the cut-off criterion.
Results
DEG analysis
For microarray analysis, 1,377 DEGs were identified
between the OA group and the non-OA group. Of these genes, 869 DEGs
were upregulated and 508 DEGs were downregulated.
Functional and pathway enrichment
analysis of the DEGs
The functional enrichment analysis of the
downregulated DEGs revealed that the top five enriched pathways
were associated with systemic lupus erythematosus
(P=1.17×10−13), the cell cycle (P=1.47×10−6),
complement and coagulation cascades (P=0.000685), nitrogen
metabolism (P=0.001086) and amoebiasis (P=0.001278) (Table I). BUB1, BUB1B,
CCNA2, CCNB1 and CCNE1 were the genes involved
in the cell cycle. The top five enriched GO terms included response
to stress (P = 4.91×10−14), involving CCNA2,
CCNB1, FGF19, KIF11 and KIF2C, immune
system-associated processes (P=1.87×10−12) involving
FGF19, KIF11 and KIF2C, response to wounding
(P=2.04×10−12), including CCNB1, KIF11 and
KIF2C, defense response (P=2.37×10−12), involving
FGF19, and mitotic cell cycle (P=2.60×10−12)
associated with BUB1, BUB1B, CCNA2,
CCNB1, CCNE1, KIF20, KIF11 and
KIF2C (Table II).
 | Table ITop five enriched KEGG pathways for
downregulated differentially expressed genes. |
Table I
Top five enriched KEGG pathways for
downregulated differentially expressed genes.
| KEGG pathway | Gene count | P-value | Genes |
|---|
| Systemic lupus
erythematosus | 27 |
1.17×10−13 | C9,
CTSG, ELANE, H2AFX, HIST1H2AC |
| Cell cycle | 17 |
1.47×10−6 | BUB1,
BUB1B, CCNA2, CCNB1, CCNE1 |
| Complement and
coagulation cascades | 9 | 0.000685 | C5AR1,
C9, CR1, F12, F3 |
| Nitrogen
metabolism | 5 | 0.001086 | CA1,
CA2, CA8, GLUL, HAL |
| Amoebiasis | 11 | 0.001278 | ARG1,
ARG2, C9, COL4A6, CTSG |
 | Table IITop five enriched GO terms for the
downregulated differentially expressed genes. |
Table II
Top five enriched GO terms for the
downregulated differentially expressed genes.
| GO ID | Term | Gene count | P-value | Genes |
|---|
| 0006950 | Response to
stress | 155 |
4.91×10−14 | KIF11,
KIF2C, FGF19, ADORA3, AEN |
| 0002376 | Immune system
process | 111 |
1.87×10−12 | KIF11,
KIF2C, FGF19, AHCY, APOBEC3B |
| 0009611 | Response to
wounding | 75 |
2.04×10−12 | KIF11,
KIF2C, ADM, ADORA3, AHCY |
| 0006952 | Defense
response | 85 |
2.37×10−12 | FGF19,
AHCY, ALOX5, ALOX5AP, APOBEC3B |
| 0000278 | Mitotic cell
cycle | 61 |
2.60×10−12 | KIF11,
KIF2C, KIF20A, AURKA, AURKB |
The functional enrichment analysis of the
upregulated DEGs demonstrated that the top five enriched pathways
were associated with extracellular matrix (ECM)-receptor
interaction (P=3.01×10−5), axon guidance (P=0.000472),
amoebiasis (P=0.003458), focal adhesion (P=0.003709) and
cancer-associated pathways (P=0.00407) (Table III). Certain genes, including
CD36, COL11A2, COL1A1, COL2A1 and
COL3A1, were involved in the ECM-receptor interaction
pathway. In addition, the GO functional enrichment analysis
demonstrated that ACAN, ADAMTS10 and BGN were
involved in proteinaceous ECM (P=1.11×10−15) and ECM
(P=2.22×10−15) terms, A2M, ABCC6 and
ACAN were involved in single-multicellular organism process
(P=4.66×10−15), and multicellular organism process
(P=8.27×10−14) terms, while ADAMTS10,
COL10A1 and COL11A2 were involved in the ECM part
(P=5.17×10−14) (Table
IV).
 | Table IIITop five enriched KEGG pathways for
the upregulated differentially expressed genes. |
Table III
Top five enriched KEGG pathways for
the upregulated differentially expressed genes.
| KEGG pathway | Gene count | P-value | Genes |
|---|
| ECM-receptor
interaction | 12 |
3.01×10−5 | CD36,
COL11A2, COL1A1, COL2A1, COL3A1 |
| Axon guidance | 13 | 0.000472 | EPHB3,
LRRC4C, NTN1, NTN4, PLXNA3 |
| Amoebiasis | 10 | 0.003458 | COL11A2,
COL1A1, COL2A1, COL3A1, COL4A5 |
| Focal adhesion | 15 | 0.003709 | AKT2,
COL11A2, COL1A1, COL2A1, COL3A1 |
| Pathways in
cancer | 21 | 0.00407 | AKT2,
ARNT2, COL4A5, CSF1R, DAPK2 |
 | Table IVTop five enriched GO terms for the
upregulated differentially expressed genes. |
Table IV
Top five enriched GO terms for the
upregulated differentially expressed genes.
| GO ID | Term | Gene count | P-value | Genes |
|---|
| 0005578 | Proteinaceous
extracellular matrix | 51 |
1.11×10−15 | ACAN, ADAMTS10,
BGN, C1QTNF8, C1QT- |
| NF9 | | | | |
| 0031012 | Extracellular
matrix | 55 |
2.22×10−15 | ACAN,
ADAMTS10, BGN, C1QTNF8, C1QTNF9 |
| 0044707 |
Single-multicellular organism process | 305 |
4.66×10−15 | A2M,
ABCC6, ACAN, ADAM20, ADAP2 |
| 0044420 | Extracellular
matrix part | 33 |
5.17×10−14 | ADAMTS10,
C1QTNF8, C1QTNF9, COL10A1/2 |
| 0032501 | Multicellular
organismal process | 309 |
8.27×10−14 | A2M,
ABCC6, ACAN, ADAM20, ADAP2 |
Analysis of PPI network and modules
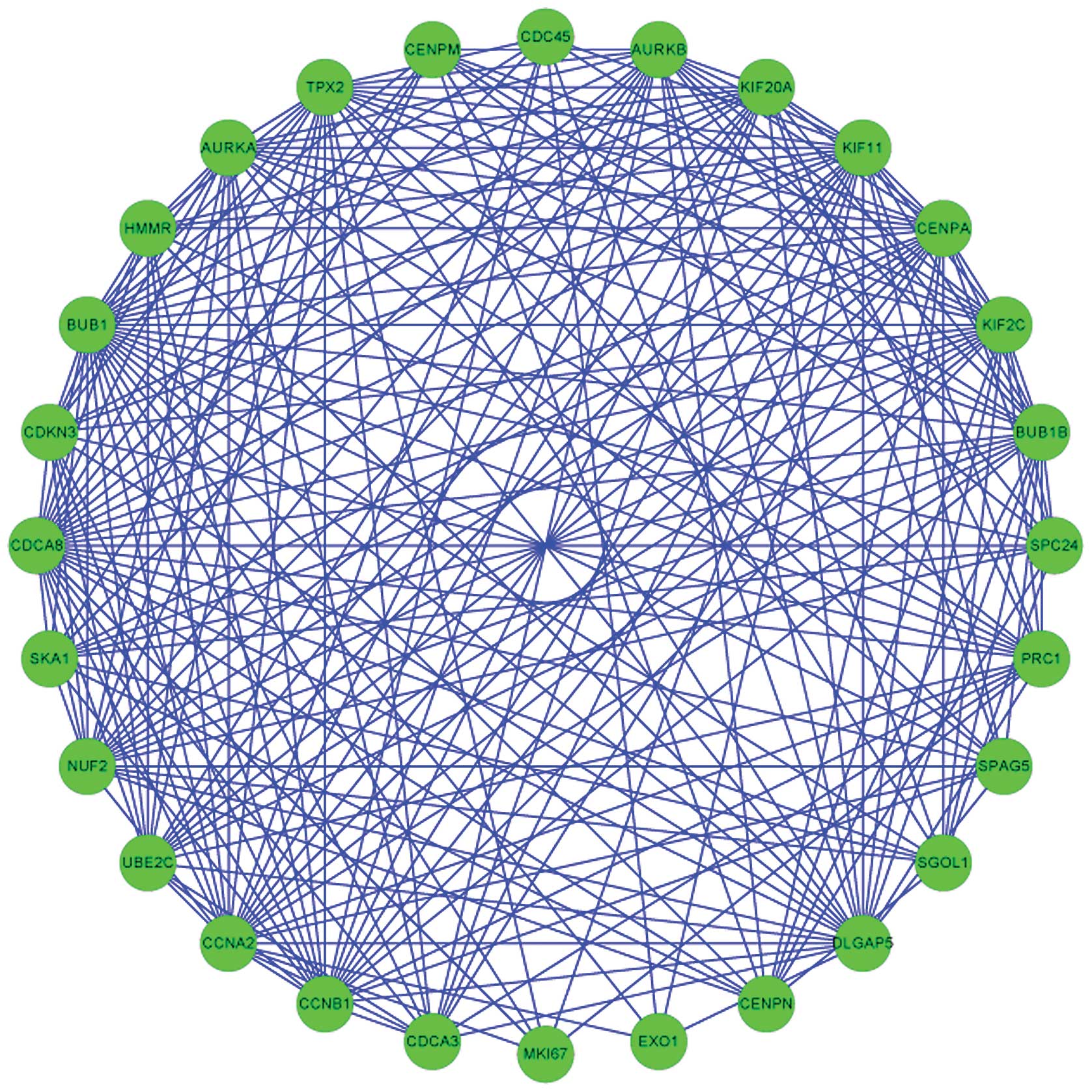
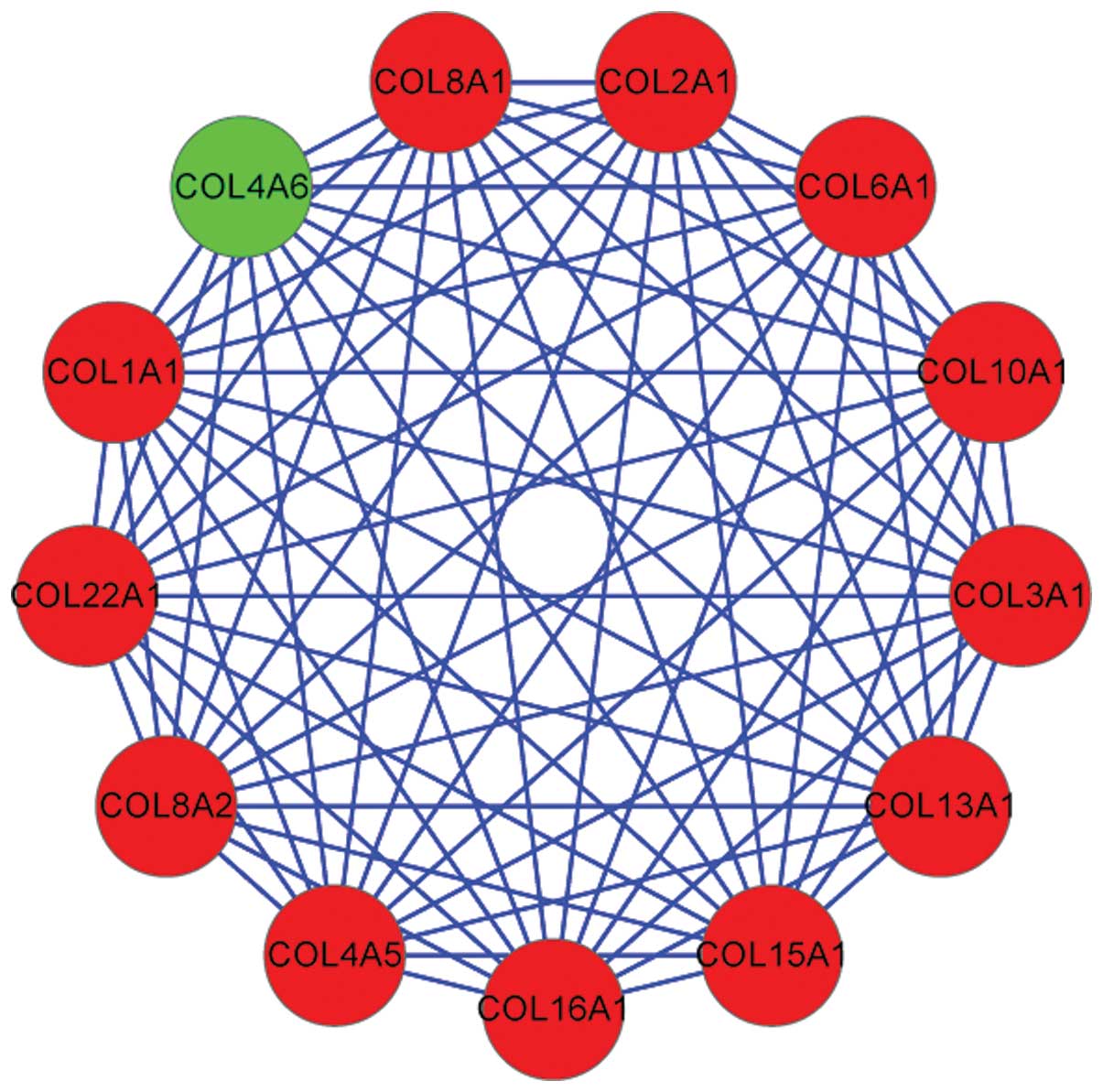
Two modules, including module 2 (Fig. 1) and module 5 (Fig. 2) were extracted from the
constructed PPI network. Protein domain enrichment analysis
revealed that the genes in module 5 were not significantly enriched
in any protein domain. A total of 10 protein domains were enriched
for the genes in module 2 (Table
V), including IPR001752: Kinesin, motor region (P=0.001518)
involving three genes (KIF2C, KIF11 and
KIF20A), IPR013212: Mad3/BUB1 homology region 1 (P=0.002759)
associated with two genes (BUB1 and BUB1B) and
IPR014400: Cyclin A/B/D/E (P=0.016448) involving two genes
(CCNB1 and CCNA2).
 | Table VEnriched protein domains for genes in
module 2 of the protein-protein interaction network (category,
Interpro). |
Table V
Enriched protein domains for genes in
module 2 of the protein-protein interaction network (category,
Interpro).
| Term | Name/function | Count | P-value | Genes |
|---|
| IPR001752 | Kinesin, motor
region | 3 | 0.001518 | KIF2C,
KIF11, KIF20A |
| IPR019821 | Kinesin, motor
region, conserved site | 3 | 0.001518 | KIF2C,
KIF11, KIF20A |
| IPR013212 | Mad3/BUB1 homology
region 1 | 2 | 0.002759 | BUB1,
BUB1B |
| IPR015661 | Mitotic checkpoint,
serine/threonine protein kinase, BUB1 | 2 | 0.002759 | BUB1,
BUB1B |
| IPR008271 | Serine/threonine
protein kinase, active site | 4 | 0.012294 | BUB1,
BUB1B, AURKA, AURKB |
| IPR014400 | Cyclin A/B/D/E | 2 | 0.016448 | CCNB1,
CCNA2 |
| IPR004367 | Cyclin,
C-terminus | 2 | 0.019164 | CCNB1,
CCNA2 |
| IPR017441 | Protein kinase, ATP
binding site | 4 | 0.023893 | BUB1,
BUB1B, AURKA, AURKB |
| IPR006671 | Cyclin,
N-terminus | 2 | 0.044611 | CCNB1,
CCNA2 |
| IPR013763 |
Cyclin-associated | 2 | 0.049887 | CCNB1,
CCNA2 |
Discussion
In the present study, the gene expression profiles
of the OA group and the non-OA group were analyzed and 1,377 DEGs
were identified, including 869 upregulated DEGs and 508
downregulated DEGs. The functional enrichment for the downregulated
DEGs revealed that the FGF19 gene was involved in the
response to stress as well as the defense response. According to
Tew et al (20), apoptosis
and proliferation are involved in the progression of cartilage
wounding during the development of OA. Costouros and Kim (21) reported that the use of programmed
cell death inhibitors can improve the results of cartilage repair
surgeries. fibroblast growth factor (FGF)2 is a mitogen that
induces chondrocyte proliferation in wounded cartilage (22). FGF18 induces the increase of
cartilage thickness of the tibial plateau and also increases
remodeling of the subchondral bone by repairing damaged cartilage
in OA rats (23). FGF2 and
FGF18 are involved in cartilage wounding, which may be
considered as one of the responses to stress. Therefore, the
present study hypothesized that FGF19 may be involved in the
repair of damaged cartilage in OA.
Functional enrichment analysis of the downregulated
DEGs further revealed that KIF11 and KIF2C were
involved in the response to stress, immune system process, response
to wounding and mitotic cell cycle. In addition, the protein domain
enrichment analysis of the genes in module 2 showed that
KIF2C, KIF11 and KIF20A, were significantly
enriched in the protein domain of IPR001752: Kinesin, motor region.
KIF2C, KIF11 and KIF20A belong to the Kinesin
family of proteins. Kinesins are important in cell division and at
least 12 kinesins are involved in mitosis and cytokinesis (24). KIF22 is a member of the
kinesin-like protein family and KIF22 mRNA was detected in human
bone, cartilage, joint capsules, ligaments, skin and primary
cultured chondrocytes (25).
Mutations in KIF22 may be the cause of
spondyloepimetaphyseal dysplasia with joint laxity, which is an
autosomal dominant skeletal disorder (26). Phosphocitrate is known to
downregulate multiple genes responsible for cell proliferation.
KIF23 and KIFC1 were shown to be differentially
expressed between phosphocitrate-treated and -untreated
telomerase-transduced OA 13A fibroblast-like synoviocytes (27). KIF2C was also identified as
a DEG in phosphocitrate-treated vs. untreated OA meniscal cells
(28). These results suggested
that KIF2C, KIF11 and KIF20A may be involved
in the mitotic cell cycle and contribute to the development of
OA.
In addition, functional enrichment analysis of the
down-regulated DEGs revealed that BUB1, BUB1B,
CCNB1 and CCNA2 were involved in the mitotic cell
cycle process. Furthermore, the protein domain enrichment analysis
of the genes in module 2, including BUB1 and BUB1B,
revealed that these genes were significantly enriched in the
protein domain of IPR013212: Mad3/BUB1 homology region (1), and the genes in module 2, including
CCNB1 and CCNA2, were significantly enriched in the
protein domain of IPR014400: Cyclin A/B/D/E. Several previous
studies have reported that BUB1, BUB1B, CCNB1
and CCNA2 had roles in the regulation of the cell cycle
(29–32). According to Tew et al
(20), apoptosis and proliferation
were involved in processes of cartilage wounding. Based on these
results, the present study hypothesized that BUB1,
BUB1B, CCNB1 and CCNA2 affected the
development of OA through the cell cycle.
In addition, upregulated DEGs, including
CD36, COL11A 2, COL1A1, COL2A1 and
COL3A1, were revealed to be involved in the ECM and
ECM-receptor interaction pathway. The structural matrix
macromolecules in the ECM and growth factors regulate the
chondrocyte function via specific membrane receptors (33). TSP-1 is involved in the
cell-matrix interactions of various tissues in cartilage. The
number of CD36-positive chondrocytes, which is considered to
be the receptor of TSP-1, is significantly increased in
severely osteoarthritic cartilage (34). The expression of CD36
patterning receptor has attenuating effects on the inflammatory,
pro-catabolic responses to S100A11 and tumor necrosis factor
α in chondrocytes (35). The α1(X)
collagen gene COL10A1 is a hypertrophic
chondrocytes-specific molecular marker and a transcriptional target
of RUNX2 during chondrogenesis (36,37).
RUNX2 also regulates the expression of COL10A1 in
hypertrophic chondrocytes and matrix metallopeptidase 13 in
terminal hypertrophic chondrocytes (38). The collagen genes (COL11A 2,
COL1A1 and COL2A1) are involved in the progression of
OA through the mediation of sensitivity to OA (39). The upregulated expression of
COL1A1 and COL2A1 reflects the metabolic activation
of OA chondrocytes, and the expression of COL11A2 is also
upregulated with the progression of OA (40). The results suggested that the DEGs
associated with the ECM are important in the regulation of
chondrocyte function.
In conclusion, in the present study, gene expression
analysis was performed and 1,377 DEGs were identified, among which
869 DEGs were upregulated and 508 DEGs were downregulated. The
identified DEGs may be involved in the pathogenesis of OA,
particularly FGF19, BUB1, KIF2C, CD36
and COL11A2, which are involved in the cellular stress
response, cell cycle and the ECM. However, the results of the
present study require confirmation by further studies.
References
|
1
|
Fransen M, Bridgett L, March L, Hoy D,
Penserga E and Brooks P: The epidemiology of osteoarthritis in
Asia. Int J Rheum Dis. 14:113–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahjoub M, Berenbaum F and Houard X: Why
subchondral bone in osteoarthritis? The importance of the cartilage
bone interface in osteoarthritis. Osteoporos Int. 23(Suppl 8):
S841–S846. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Henrotin Y, Pesesse L and Sanchez C:
Subchondral bone and osteoarthritis: Biological and cellular
aspects. Osteoporos Int. 23:S847–S851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chou CH, Wu CC, Song IW, Chuang HP, Lu LS,
Chang JH, Kuo SY, Lee CH, Wu JY, Chen YT, et al: Genome-wide
expression profiles of subchondral bone in osteoarthritis.
Arthritis Res Ther. 15:R1902013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chou CH, Wu CC, Song IW, Chuang HP, Lu LS,
Chang JH, Kuo SY, Lee CH, Wu JY and Chen YT: Genome-wide expression
profiles of subchondral bone in osteoarthritis. Arthritis Res Ther.
15:R1902013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhen G, Wen C, Jia X, Li Y, Crane JL,
Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, et al: Inhibition
of TGF-β signaling in mesenchymal stem cells of subchondral bone
attenuates osteoarthritis. Nat Med. 19:704–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Valverde-Franco G, Pelletier JP, Fahmi H,
Hum D, Matsuo K, Lussier B, Kapoor M and Martel-Pelletier J: In
vivo bone-specific EphB4 overexpression in mice protects both
subchondral bone and cartilage during osteoarthritis. Arthritis
Rheum. 64:3614–3625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karlsson C, Dehne T, Lindahl A, Brittberg
M, Pruss A, Sittinger M and Ringe J: Genome-wide expression
profiling reveals new candidate genes associated with
osteoarthritis. Osteoarthritis Cartilage. 18:581–592. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smyth G and Limma: Linear models for
microarray data Bioinformatics and Computational Biology Solutions
using R and Bioconductor 2005. Springer; New York: pp. 397–420.
2005
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. JR Stat Soc. 57:289–300. 1995.
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
15
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Detabase Issue): D808–D815. 2013. View Article : Google Scholar
|
|
16
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adamcsek B, Palla G, Farkas IJ, Derényi I
and Vicsek T: CFinder: Locating cliques and overlapping modules in
biological networks. Bioinformatics. 22:1021–1023. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Derényi I, Palla G and Vicsek T: Clique
percolation in random networks. Phys Rev Lett. 94:1602022005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Apweiler R, Attwood TK, Bairoch A, Bateman
A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD,
et al: The InterPro database, an integrated documentation resource
for protein families, domains and functional sites. Nucleic Acids
Res. 29:37–40. 2001. View Article : Google Scholar :
|
|
20
|
Tew SR, Kwan AP, Hann A, Thomson BM and
Archer CW: The reactions of articular cartilage to experimental
wounding: Role of apoptosis. Arthritis Rheum. 43:215–225. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Costouros JG and Kim HT: Preventing
chondrocyte programmed cell death caused by iatrogenic injury.
Knee. 14:107–111. 2007. View Article : Google Scholar
|
|
22
|
Khan IM, Palmer EA and Archer CW:
Fibroblast growth factor-2 induced chondrocyte cluster formation in
experimentally wounded articular cartilage is blocked by soluble
Jagged-1. Osteoarthritis Cartilage. 18:208–219. 2010. View Article : Google Scholar
|
|
23
|
Moore EE, Bendele AM, Thompson DL, Littau
A, Waggie KS, Reardon B and Ellsworth JL: Fibroblast growth
factor-18 stimulates chondrogenesis and cartilage repair in a rat
model of injury-induced osteoarthritis. Osteoarthritis Cartilage.
13:623–631. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu C, Zhao J, Bibikova M, Leverson JD,
Bossy-Wetzel E, Fan JB, Abraham RT and Jiang W: Functional analysis
of human microtubule-based motor proteins, the kinesins and
dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol
Cell. 16:3187–3199. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Min BJ, Kim N, Chung T, Kim OH, Nishimura
G, Chung CY, Song HR, Kim HW, Lee HR, Kim J, et al: Whole-exome
sequencing identifies mutations of KIF22 in spondyloepimetaphyseal
dysplasia with joint laxity, leptodactylic type. Am J Hum Genet.
89:760–766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boyden ED, Campos-Xavier AB, Kalamajski S,
Cameron TL, Suarez P, Tanackovic G, Andria G, Ballhausen D, Briggs
MD, Hartley C, et al: Recurrent dominant mutations affecting two
adjacent residues in the motor domain of the monomeric kinesin
KIF22 result in skeletal dysplasia and joint laxity. Am J Hum
Genet. 89:767–772. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun Y, Mauerhan DR, Franklin AM, Norton J,
Hanley EN Jr and Gruber HE: Phosphocitrate is potentially a
disease-modifying drug for noncrystal-associated osteoarthritis.
Biomed Res Int. 2013:3262672013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Y, Roberts A, Mauerhan DR, Sun AR,
Norton HJ and Hanley EN Jr: Biological activities of
phosphocitrate: A potential meniscal protective agent. Biomed Res
Int. 2013:7265812013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taylor SS and Mckeon F: Kinetochore
localization of murine Bub1 is required for normal mitotic timing
and checkpoint response to spindle damage. Cell. 89:727–735. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davenport JW, Fernandes ER, Harris LD,
Neale GA and Goorha R: The mouse mitotic checkpoint gene bub1b, a
novel bub1 family member, is expressed in a cell cycle-dependent
manner. Genomics. 55:113–117. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sartor H, Ehlert F, Grzeschik KH, Müller R
and Adolph S: Assignment of two human cell cycle genes, CDC25C and
CCNB1, to 5q31 and 5q12, respectively. Genomics. 13:911–912. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang C, Wang L, Wu D, Chen H, Chen Z,
Thomas-Ahner JM, Zynger DL, Eeckhoute J, Yu J, Luo J, et al:
Defnition of a FoxA1 Cistrome that is crucial for G1 to S-phase
cell-cycle transit in castration-resistant prostate cancer. Cancer
Res. 71:6738–6748. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Van Der Kraan PM, Buma P, Van Kuppevelt T
and Van Den Berg WB: Interaction of chondrocytes, extracellular
matrix and growth factors: Relevance for articular cartilage tissue
engineering. Osteoarthritis Cartilage. 10:631–637. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pfander D, Cramer T, Deuerling D, Weseloh
G and Swoboda B: Expression of thrombospondin-1 and its receptor
CD36 in human osteoarthritic cartilage. Ann Rheum Dis. 59:448–454.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cecil DL, Appleton CT, Polewski MD, Mort
JS, Schmidt AM, Bendele A, Beier F and Terkeltaub R: The pattern
recognition receptor CD36 is a chondrocyte hypertrophy marker
associated with suppression of catabolic responses and promotion of
repair responses to inflammatory stimuli. J Immunol. 182:5024–5031.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng Q, Zhou G, Morello R, Chen Y,
Garcia-Rojas X and Lee B: Type X collagen gene regulation by Runx2
contributes directly to its hypertrophic chondrocyte-specific
expression in vivo. J Cell Biol. 162:833–842. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin AC, Seeto BL, Bartoszko JM, Khoury MA,
Whetstone H, Ho L, Hsu C, Ali SA and Alman BA: Modulating hedgehog
signaling can attenuate the severity of osteoarthritis. Nat Med.
15:1421–1425. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Komori T: Regulation of bone development
and extracellular matrix protein genes by RUNX2. Cell Tissue Res.
339:189–195. 2010. View Article : Google Scholar
|
|
39
|
Jin SY, Hong SJ, Yang HI, Park SD, Yoo MC,
Lee HJ, Hong MS, Park HJ, Yoon SH, Kim BS, et al: Estrogen
receptor-alpha gene haplotype is associated with primary knee
osteoarthritis in Korean population. Arthritis Res Ther.
6:R415–R421. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lamas JR, Rodríguez-Rodríguez L, Vigo AG,
Alvarez-Lafuente R, López-Romero P, Marco F, Camafeita E, Dopazo A,
Callejas S, Villafuertes E, et al: Large-scale gene expression in
bone marrow mesenchymal stem cells: A putative role for COL10A1 in
osteoarthritis. Ann Rheum Dis. 69:1880–1885. 2010. View Article : Google Scholar : PubMed/NCBI
|