Introduction
Enterococcus faecium has emerged as an
important cause of nosocomial infection (1). It is resistant to a variety of
antimicrobial agents, including β-lactam antibiotics. The
contribution of penicillin-binding protein 5 (PBP5) to β-lactam
resistance has been intensely investigated (2,3). The
existence of the low-affinity PBP5 has been found to be involved in
β-lactam resistance within laboratory mutants and clinical isolates
of E. faecium. Previously, it was demonstrated that
mutations in amino acids around the active site can alter the
affinities of PBP5 for β-lactams. Sequence data has revealed the
presence of amino acid mutations within the C-terminal domain of
PBP5 (PBP5-CD) (4). Notably,
certain mutations have been revealed to be significantly associated
with β-lactam resistance (5,6).
The crystal structures of PBP5 from Pseudomonas
aeru- ginosa, Escherichia coli and Haemophilus
influenzae have been previously determined (7–9).
These structures reveal the spatial arrangement of proteins, which
explain the reason for the observed substrate specificity of PBP5.
However, the crystal structure of PBP5 from E. faecium has
not been determined. The present study aimed to examine the PBP5-CD
variants of 13 clinical isolates with different levels of
penicillin resistance. The impacts of novel amino acid alterations
of E. faecium PBP5-CD on penicillin binding were analyzed
using homology modeling and molecular docking methods. The results
of the present study may assist in investigating the mechanisms
involved in β-lactam resistance in E. faecium. Additionally,
the binding patterns obtained in the present study may be useful in
the development of novel antibiotics against β-lactam resistant
E. faecium.
Materials and methods
Bacterial strains and identification
A total of 13 clinical isolates of E. faecium
were collected from local hospitals in Inner Mongolia, China.
Institutional ethical clearance was obtained. The specimens were
isolated from various clinical samples, as described previously
(10), including urine (9/13),
orotracheal fluid (1/13), vaginal/semen swab (1/13), pus (1/13) and
blood (1/13). A VITEK 2 Compact system (BioMérieux, Ltd., Marcy
l'Etoile, France) and polymerase chain reaction (PCR) analysis were
used to identify E. faecium. In brief, PCR amplification was
performed using 1 µl template DNA, 1 µl of each
primer (100 pmol), and 25 µl 2X PCR Master Mix (BioSci
Biotech, Hangzhou, China) in a total volume of 50 µl. A
C1000 Touch thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) was programmed with an initial denaturation step at 94°C for 3
min; 35 cycles of denaturation at 94°C for 30 sec, annealing at
54°C for 30 sec and elongation at 72°C for 1 min, followed by a
final extension at 72°C for 5 min. The PCR products were
analyzed using electrophoresis on a 1% agarose gel stained with
ethidium bromide (Sangon Biotech, Co., Ltd., Shanghai, China). The
PCR primers for the ddlE gene were forward
5′-TTGAGGCAGACCAGATTGACG-3′ and reverse 5′-TATGACAGCGACTCCGATTCC-3′
(11). The present study was
approved by the Ethics Comittee of Baotou Medical College, Baotou,
Chuna. All subjects provided written informed consent.
Susceptibility assessment
The antimicrobial susceptibility of the of E.
faecium isolates were determined using a disk diffusion
technique with the Kirby-Bauer (K-B) test method, according to the
standards and interpretive criteria described by the Clinical and
Laboratory Standards Institute (CLSI) (12). The drugs used for disc diffusion
assessment were obtained from Oxoid, Ltd. (Basingstoke, UK) in the
following concentrations: Chloramphenicol (30 µg),
gentamicin (120 µg), norfloxacin (30 µg),
ciprofloxacin (5 µg), ampicillin (10 µg),
tetracycline (30 µg), penicillin (10 IU), erythromycin (15
µg) and streptomycin (300 µg). E. faecalis
ATCC 29212 (susceptible) and E. fecalis ATCC 51299
(resistant) strains were used as the quality control strains for
the determination of antimicrobial susceptibility (12).
Western blotting
Western blotting was performed to detect the
expression levels of PBP5 in the 13 clinical E. faecium
isolates, as described previously (13). Briefly, proteins were prepared from
3 ml cultures of the E. faecium sample in the stationary
growth phase. For western blot analysis, 15 µl of the
proteins were separated on SDS polyacrylamide gel and transferred
onto polyvinylidene fluoride (PVDF) membranes (Sangon Biotech, Co.,
Ltd.), blocked for 1 h at 37°C in PBS containing non-fat dry
milk (Sangon Biotech, Co., Ltd.) and incubated overnight at
4°C with the primary antibody (polyclonal anti-PBP5; Vital
River Laboratories, Beijing, China) diluted 1:1,000 in PBS.
Following washing in Tween 20 0.05% v/v in Tris-buffered saline,
the PVDF membranes were incubated for 1 h at room temperature with
peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no.
C006387; Sangon Biotech, Co., Ltd.) diluted 1:2,000 with PBS.
Signals were detected using enhanced chemiluminescence substrate
(Sangon Biotech, Co., Ltd.). Preparation of the anti-PBP5 antibody
was performed, as described previously (14).
PCR and DNA sequencing
The total DNA template was extracted from E.
faecium using a Bacteria Genomic DNA Extraction kit (BioSci
Biotech, Hangzhou, China), according to the manufacturer's
instructions. PCR was used to detect a 794 bp fragment from E.
faecium encoding PBP5 CD. The PCR conditions were as follows:
Initial denaturation at 94°C for 3 min; 40 cycles of
denaturation at 94°C for 30 sec, annealing at 52°C
for 1 min and elongation at 72°C for 2 min, followed by a
final extension at 72°C for 5 min. The PCR products were
analyzed using electrophoresis on agarose gel, as described above.
The sequence of the PCR primers were as follows: forward
5′-CGGGATCTCACAAGAAGAT-3′ and reverse 5′-TTATTGATAATTTTGGTT-3′. The
PCR products were purified using the DNA Agarose Gel Extraction Kit
(Sangon Biotech, Co., Ltd.) and were sequenced by Sangon Biotech
Co., Ltd. using a Primer-Walking method.
Homology modeling
The homology model of PBP5-CD from E. faecium
was constructed using the Modeller program (version 9.10) and the
structure of PBP2a from the 27r methicillin-resistant
Staphylococcus aureus strain (University of British
Columbia, British Columbia, Canada) was used as a template (PDB
code, 1VQQ). The model structure was subsequently refined by energy
minimization in the presence of explicit solvent model TIP3P water
using the CHARMm program (version c29b2). The obtained structure
was assessed using the Profiles-3D program within Insight II 2005
(Accelrys, San Diego, CA, USA).
Molecular docking
The Affinity program within Insight II 2005
(Accelrys) was used for docking experiments, as described
previously (15). The consistent
valence force field was selected prior to performing docking
calculations. Since the active site of PBP5-CD is conserved, the
crystal structure of covalent complexes of a β-lactam antibiotic
with E. coli PBP5 (PDB code, 3MZD) was used as a reference
structure, to identify the initial binding site of penicillin in
the E. faecium PBP5-CD.
Results and Discussion
Identification and susceptibility of E.
faecium
All 13 isolates were identified as E. faecium
using the VITEK 2 Compact system and PCR assay. The K-B method
revealed resistance to penicillin, with a zone of 32 mm for strain
7 and ≥64 mm for all other isolates. In the present study, the
resistance break-point was ≥16 mm, according to CLSI guidelines
(12). These results provided
clear evidence that strain 7 exhibited a lower level of penicillin
resistance, compared with the other isolates. Considering the
crucial role of PBP5 in β-lactam resistance, it was suggested that
the different levels of penicillin resistance between these E.
faecium isolates may have been due to the presence of
low-affinity PBP5 or the production of PBP5.
Expression of E. faecium pbp5 genes
In order to identify and understand the mechanisms
underlying penicillin resistance among the clinical E.
faecium isolates, the expression levels of pbp5 genes
were determined using western blotting (data was not shown). The
results demonstrated that the expression levels of the pbp5
genes were similar, indicating that the production of PBP5 did not
account for the different levels of penicillin resistance, which
were observed in the E. faecium isolates.
Alternations in the E. faecium
PBP5-CD
The PBP5-CD variants from the isolates were further
investigated in the present study. The PBP5-CD genes of the
13 E. faecium isolates were amplified and sequenced. From
the primary structure alignment, a consensus sequence was observed
(Fig. 1A). As shown in Fig. 1B, an evolutionary path, as depicted
by the evolutionary tree, was observed. Unlike the PBP5 in the
majority of isolates, which was identified with the zone of ≥64 mm,
the PBP5 variant of strain 7, with the zone of 32 mm, had two novel
changes at positions 460 and 462. This is the first report, to the
best of our knowledge, to describe the Tyr460Phe and Ala462Thr or
Val462Thr PBP5-CD mutations in E. faecium isolates. These
substitutions are located in a loop, which forms the outer edge of
the active site, and are in close proximity to the inserted serine
at position 466. Therefore, the present study hypothesized that
these mutations may be involved in the binding of penicillin to
PBP5-CD, consequently resulting in a lower level of resistance
towards penicillin.
Homology model of E. faecium PBP5-CD
To further confirm the above hypothesis, homology
models of PBP5-CD were constructed. Strain 1 and strain 7, which
exhibited differences in the resistance to penicillin, were
included in this experiment. According to the sequence alignment
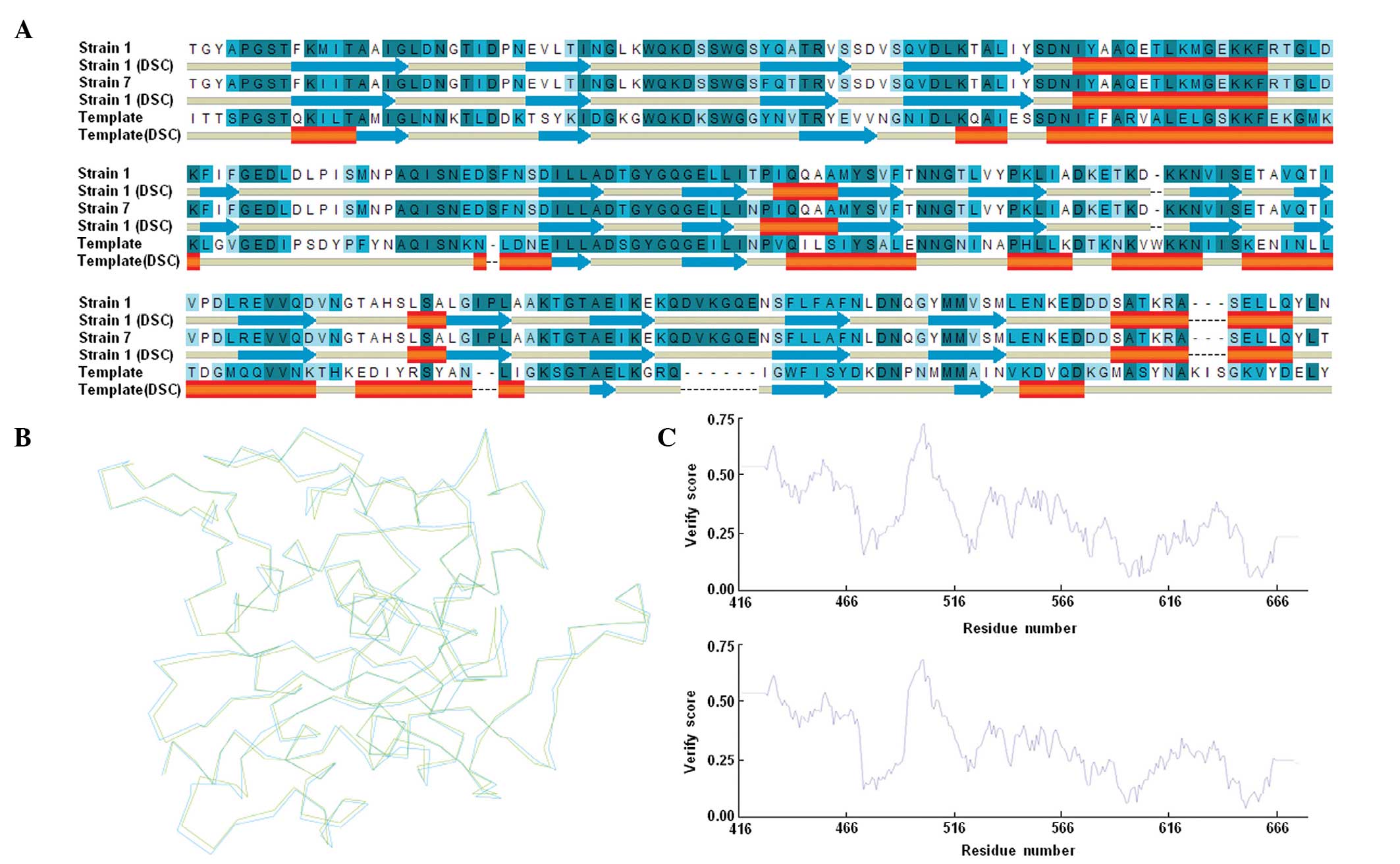
result, the percentages of identity of strain 1 and strain 7 with
the template were 36.7 and 37.1%, respectively. Sequence alignment
analysis also revealed the predicted secondary structures of
PBP5-CD, which indicated that these proteins were structurally
similar to one another (Fig. 2A).
The homology models of the PBP5-CD of strain 1 and strain 7 were
subsequently constructed, based on the structure of the template
(Fig. 2B), and the quality of the
modeled structures was determined. All residues were in sterically
allowed regions of the Ramachandran plot. In addition, the overall
self-compatibility scores for the predicted structures were all
above the expected values (Fig.
2C). These results indicated that the predicted models were
reliable. Subsequently, the two predicted structures of PBP5-CD
were compared, to understand the effects of the substitutions on
the spatial arrangement. The superposition of Cα atoms from the
PBP5-CD of strain 1 onto the corresponding atoms of strain 7
provided a root mean square deviation of 0.529 Å (Fig. 2B), which indicated that certain
substitutions were able to produce minor conformational changes,
which may have the effect of altering the orientation of residues
in the binding sites.
Detection of binding between penicillin
and PBP5-CD
In order to obtain additional evidence to support
the hypothesis that Phe460 and Thr462 are involved in penicillin
binding, molecular docking was performed. However, calculation of
covalent bonds is challenging due to the principle limitation of
molecular simulation methods, particularly for PBP5, and no docking
methods have been previously reported. In order to set up a
reliable theoretical method to fully understand the binding mode
between PBP5-CD and penicillin, the active site of PBP5-CD was
obtained by using the crystal structure of E. coli PBP5 as
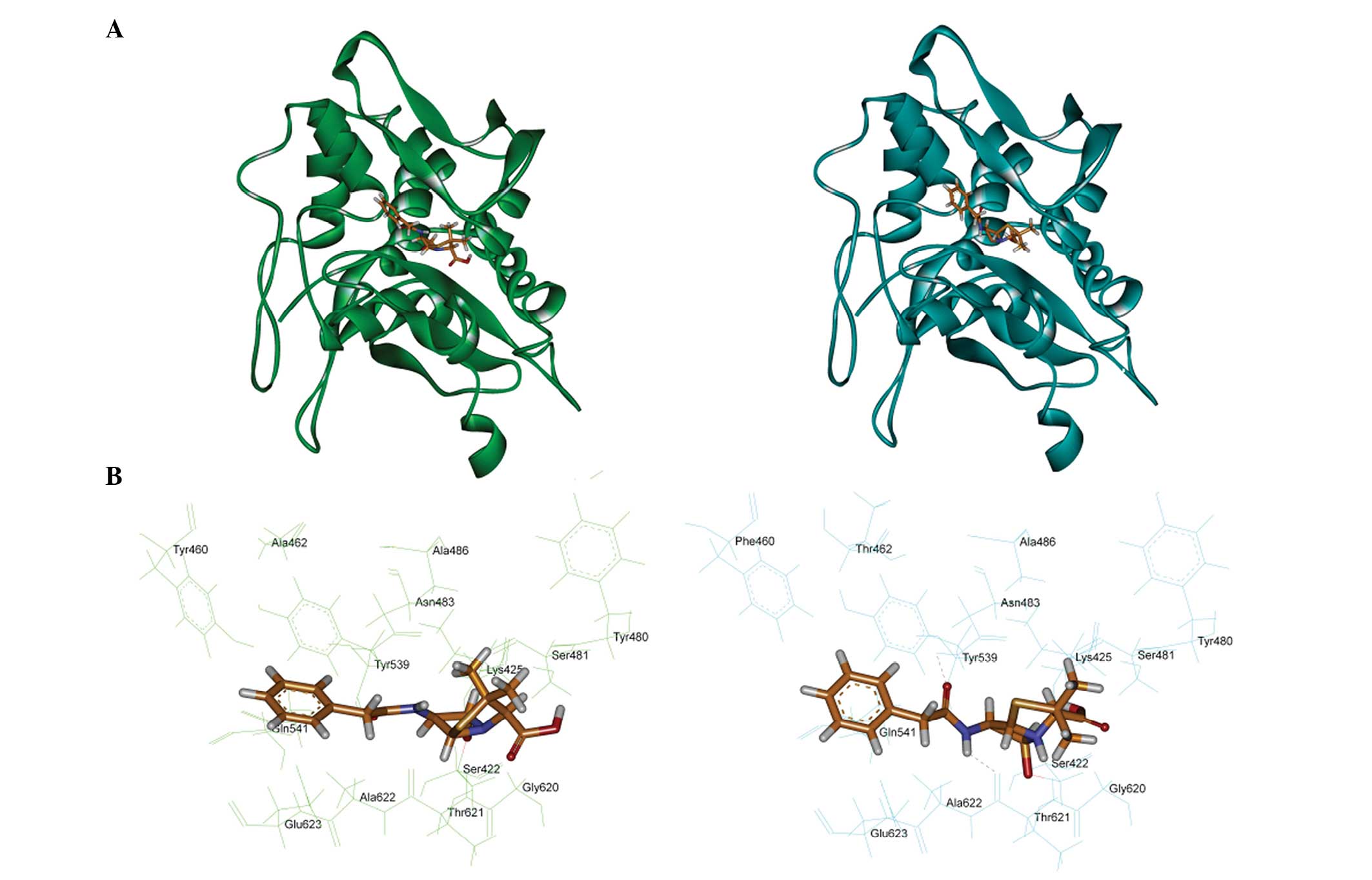
the template. Fig. 3 shows the
covalent binding mode of penicillin with PBP5-CD. The penicillin
covalently interacted with the serine at position 422 of the
active-site, which was consistent with the results of a previous
study (4). For the PBP5-CD of
strain 7, the results demonstrated that hydrophobic groups were
positioned in lipophilic pockets, which were created by Tyr539,
Phe460 and Thr 462. The theoretical calculation also indicated that
penicillin formed two hydrogen bonds with Asn483 and Thr621.
However, no hydrogen bonding was observed in strain 1, and the
replacement of Phe460 and Thr462 with Tyr460 and Ala462 resulted in
a reduction in hydrophobic interactions.
In conclusion, Phe460 and Thr462 in strain 7 may
increase the ability of PBP5 to bind to penicillin and affect the
level of penicillin resistance of E. faecium. This is the
first report, to the best of our knowledge, demonstrating that the
two residues, Phe460 and Thr462, were responsible for the lower
penicillin resistance of E. faecium observed in
vitro.
Acknowledgments
The present study was supported by the Natural
Science Foundation of Inner Mongolia Autonomous Region of China
(grant. nos. 2014JQ04 and 2012MS1159) and the National Natural
Science Foundation of China (grant. no. 81460049).
References
|
1
|
Murray BE: The life and times of the
Enterococcus. Clin Microbiol Rev. 3:46–65. 1990.PubMed/NCBI
|
|
2
|
Rice LB, Carias LL, Hutton-Thomas R,
Sifaoui F, Gutmann L and Rudin SD: Penicillin-binding protein 5 and
expression of ampicillin resistance in Enterococcus faecium.
Antimicrob Agents Chemother. 45:1480–1486. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rybkine T, Mainardi JL, Sougakoff W,
Collatz E and Gutmann L: Penicillin-binding protein 5 sequence
alterations in clinical isolates of Enterococcus faecium with
different levels of β-lactam resistance. J Infect Dis. 178:159–163.
1998. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rice LB, Bellais S, Carias LL,
Hutton-Thomas R, Bonomo RA, Caspers P, Page MG and Gutmann L:
Impact of specific pbp5 mutations on expression of beta-lactam
resistance in Enterococcus faecium. Antimicrob Agents Chemother.
48:3028–3032. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh SE, Hsu LL, Hsu WH, Chen CY, Chen HJ
and Liao CT: Importance of amino acid alterations and expression of
penicillin-binding protein 5 to ampicillin resistance of
Enterococcus faecium in Taiwan. Int J Antimicrob Agents.
28:514–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poeta P, Costa D, Igrejas G, Sáenz Y,
Zarazaga M, Rodrigues J and Torres C: Polymorphisms of the pbp5
gene and correlation with ampicillin resistance in Enterococcus
faecium isolates of animal origin. J Med Microbiol. 56:236–240.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith JD, Kumarasiri M, Zhang W, Hesek D,
Lee M, Toth M, Vakulenko S, Fisher JF, Mobashery S and Chen Y:
Structural analysis of the role of Pseudomonas aeruginosa
penicillin-binding protein 5 in β-lactam resistance. Antimicrob
Agents Chemother. 57:3137–3146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nicola G, Tomberg J, Pratt RF, Nicholas RA
and Davies C: Crystal structures of covalent complexes of β-lactam
antibiotics with Escherichia coli penicillin-binding protein 5:
Toward an understanding of antibiotic specificity. Biochemistry.
49:8094–8104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kawai F, Clarke TB, Roper DI, Han GJ,
Hwang KY, Unzai S, Obayashi E, Park SY and Tame JR: Crystal
structures of penicillin-binding proteins 4 and 5 from Haemophilus
influenzae. J Mol Biol. 396:634–645. 2010. View Article : Google Scholar
|
|
10
|
Li W, Li J, Wei Q, Hu Q, Lin X, Chen M, Ye
R and Lv H: Characterization of aminoglycoside resistance and
virulence genes among Enterococcus spp. isolated from a hospital in
China. Int J Environ Res Public Health. 12:3014–3025. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Macovei L and Zurek L: Ecology of
antibiotic resistance genes: characterization of enterococci from
houseflies collected in food settings. Appl Environ Microbiol.
72:4028–4035. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clinical and Laboratory Standards
Institute: Performance Standards for Antimicrobial Susceptibility
Testing: Twentieth Informational Supplement. Wayne, PA: Clinical
and Laboratory Standards Institute; pp. M100–S23. 2013
|
|
13
|
Rybkine T, Mainardi JL, Sougakoff W,
Collatz E and Gutmann L: Penicillin-binding protein 5 sequence
alterations in clinical isolates of Enterococcus faecium with
different levels of beta-lactam resistance. J Infect Dis.
178:159–163. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsieh SE, Hsu LL, Hsu WH, Chen CY, Chen HJ
and Liao CT: Importance of amino acid alterations and expression of
penicillin-binding protein 5 to ampicillin resistance of
Enterococcus faecium in Taiwan. Int J Antimicrob Agents.
28:514–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lei M, Zhao X, Wang Z and Zhu Y:
Pharmacophore modeling, docking studies and synthesis of novel
dipeptide proteasome inhibitors containing boron atoms. J Chem Inf
Model. 49:2092–2100. 2009. View Article : Google Scholar : PubMed/NCBI
|