Introduction
Major depressive disorder (MDD) is a considerable
public health concern, which affects patients worldwide. MDD is
associated with psychosocial impairment, poor quality of life, and
significant disability, morbidity and mortality (1). However, due to the poor understanding
regarding the pathogenic mechanisms associated with MDD, ~40% of
patients with MDD do not respond well to the currently available
treatments (2). A previous study
in patients with MDD detected neuronal atrophy, altered dendritic
morphology of hippocampal neurons and decreased hippocampal volume
(3).
Stress is a major factor in depression, which
impairs the structural and functional plasticity of the hippocampus
(4). Chronic and persistent stress
is able to induce dysfunction of the hypothalamic-pituitary-adrenal
axis, alter the immune system and induce other pathophysiological
effects detected in patients with depression (5). A previous study demonstrated that
chronic unpredictable mild stress is able to downregulate the
expression of brain-derived neurotrophic factor (BDNF), which has a
critical role in the etiology of depression; the downregulation of
which is reversed following classical antidepressant therapy
(6). Furthermore, emerging
evidence has demonstrated that microRNAs (miRNAs) are associated
with depression, particularly miRNAs involved with BDNF (7). miRNA alterations have been detected
in patients with MDD and rats exposed to chronic stress (8). miRNAs are a class of short (~22 nt)
non-coding RNAs that suppress the expression of various genes at
the post-transcriptional level by targeting the 3′ untranslated
region (3′UTR). miRNAs are essential for numerous cellular
processes, including survival, differentiation and apoptosis, in
humans and animals (9,10).
miRNAs are regulated by specific upstream molecules.
Epigenetic mechanisms are well-known mechanisms that regulate the
expression of miRNAs (11,12). Methyl-CpG-binding protein 2 (MeCP2)
is a transcription factor that binds to methylated cytosine
residues on CpG dinucleotides in DNA, resulting in the recruitment
of histone deacetylases and other transcriptional repressors that
silence target genes (13). A
previous study demonstrated that the loss of MeCP2 function causes
Rett syndrome (14). In addition,
it has been suggested that MeCP2 may have a critical role in
depression (15). Murgatroyd et
al (16) demonstrated that
neuronal activity controlled the ability of MeCP2 to regulate
activity-dependent transcription of the arginine vasopressin gene
and induced epigenetic marking. Hutchinson et al (15) demonstrated that MeCP2
phosphorylation was required for the beneficial effects of chronic
imipramine treatment on chronic social defeat stress-induced
depressive-like behaviors. In addition, miRNAs may decrease MeCP2
expression levels in human gastric carcinoma cell lines and in
cultured mouse cortical neurons (17,18).
Notably, MeCP2 is able to regulate cocaine intake through
controlling the effects of cocaine on striatal BDNF levels by
interacting with miR-212. (19).
Therefore, the present study hypothesized that interactions between
MeCP2 and miRNAs may have a role in depression.
The aim of the present study was to investigate the
regulatory association between BDNF, MeCP2 and miR-132 in
vitro and in vivo. The expression levels of BDNF, MeCP2
and miR-132 were detected in peripheral blood samples obtained from
patients with MDD (using ELISA, western blotting and reverse
transcription-quantitative polymerase chain reaction; RT-qPCR), and
in the hippocampi of depressed rats. In addition, the regulatory
association between BDNF, MeCP2 and miR-132 was determined using a
dual luciferase reporter gene system, as well as with gain- and
loss-function experiments.
Materials and methods
Blood samples
A total of 60 blood samples were collected from
patients with MDD (n=30) and age-matched normal subjects (n=30)
without other severe diseases, including diabetes, epilepsy and
dementia, from Nanfang Hospital, Southern Medical University
(Guangzhou, China). Written informed consent was obtained from the
patients. In addition, the present study was approved by the
ethical committee of the Southern Medical University. All samples
were collected according to the legislation and ethical boards of
Nanfang Hospital. All of the samples were stored at −80°C until
further use.
Animals
Male Sprague-Dawley rats, (age, 2 months; weight,
~220 g) obtained from Geneseed Biotech Co., Ltd. (Guangzhou, China)
were housed in groups of four per cage. The animals were allowed 1
week of habituation after arrival prior to stress exposure. All of
the rats were housed in standard conditions (12 h light/dark,
25±1°C, 50% humidity) with controlled access to food and water. All
of the animal procedures were performed in accordance with the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals, and the procedures were approved by the Local
Animal Use Committee of the Southern Medical University.
Cell culture and treatment
E18 rat fetuses (Geneseed Biotech Co., Ltd.) were
used to prepare primary hippocampal neurons. Hippocampi were
mechanically dissociated from the brains of the fetuses and treated
with trypsin (Sigma-Aldrich, St. Louis, MO, USA) in
phosphate-buffered saline (PBS) for 15 min at 37°C. The cell
suspensions were maintained in glial-conditioned medium (Invitrogen
Life Technologies, Carlsbad, CA, USA) in 100-mm dishes at 37°C in
an atmosphere containing 5% CO2 until further use.
Ectopic cellular expression of miR-132 was achieved by transfection
with miR-132 mimics or inhibitors (cat. nos. miR10000838-1-5 and
miR20000838-1-5, respectively; Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) using Lipofectamine® 2000 (Invitrogen
Life Technologies) for 48 h, according to the manufacturer's
instructions. Following treatment with the demethylation drug
5-Aza-2′-deoxycytidine (Aza; 15.55 nM; Sigma-Aldrich) for 72 h, the
miR-132 expression in the primary hippocampal neurons was detected
using qPCR. For MeCP2 knockdown, a 70 nt short hairpin RNA (shRNA)
was designed by GenScript Co., Ltd. (Nanjing, China) and
pRNA-U6.1/Neo/CTL (cat. no. SD1801; GenScript Co., Ltd.) served as
the control shRNA. The shRNA was cloned into the pRNAT-U6.2/Lenti
expression vector by GenScript Co., Ltd. and the vector was
transfected into primary hippocampal neurons. Control vectors were
identical to the expression constructs without the gene insert.
CUS exposure
The rats were exposed to a variable sequence of mild
and unpredictable stressors for 28 days. The rats were exposed to
two random stressors per day, out of 10 various stressors. The
stressors included a forced cold (4°C) swim for 5 min, food
deprivation for 24 h, water deprivation for 24 h, light/dark cycle
reversal for 36 h, vibration for 1 h, cage tilting for 24 h, cold
(4°C) for 1 h, crowding for 24 h, soiled bedding for 24 h, and tail
clamp for 1 min.
Sucrose preference test
The sucrose preference test is used to detect
anhedonia. Prior to the experiment, the rats were trained to adapt
to a 1% sucrose solution (w/v) for 48 h. Following water
deprivation for 4 h, the rats were housed in individual cages for 4
h, which contained two identical bottles, one filled with 1%
sucrose solution and the other filled with water. At the end of the
4-h test, sucrose and water consumption (g) was measured. Sucrose
preference (%) was calculated using the following formula: Sucrose
preference (%) = sucrose consumption/(sucrose consumption + water
consumption). The less sucrose that was consumed, the more severe
the case of anhedonia.
Forced swim test (FST)
One day prior to the experiment, each rat was
individually placed into a plastic cylinder (diameter, 25 cm;
height, 55 cm) filled with water (23-25°C) to a depth of 45 cm for
15 min. The rats were subsequently removed from the water and
returned to their cages. After 24 h, a 5 min FST was performed. The
FST was performed as previously described (20). Immobility time (in sec) was
recorded by two independent observers. Floating was defined as the
minimum movement necessary to maintain the heads of the rats above
the water. The percentage of immobility time (%) was calculated
using the following formula: Immobility time (%) = immobility
time/total experimental time.
RT-qPCR
An Ultrapure RNA kit (Beijing CWBiotech Co., Ltd.,
Beijing, China) was used to extract total RNA from the primary
hippocampal neurons, which were collected by trypsinization,
according to the manufacturer's instructions. RNA was reverse
transcribed to cDNA using a RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher, Inc., Waltham, MA, USA) according to the
manufacturer's instruction. The cDNA (2 μg) was then used to
perform qPCR with a CFX96 Real-Time System (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and the following conditions were used:
95°C for 5 min; 40 cycles of 95°C for 30 sec, 58°C for 30 sec and
72°C for 30 sec; and 72°C for 10 min. A miScript SYBR-Green PCR kit
(Guangzhou RiboBio Co., Ltd., Guangzhou, China) was used to detect
the expression of miR-132. The specific primer sets for miRNA-132
and U6 were purchased from GeneCopoeia, Inc. (Rockville, MD, USA)
and miR-132 expression was normalized by U6. The 2−ΔΔCT
method was used to analyze the expression data.
ELISA determination of MeCP2, BDNF and
corticosterone
Human MeCP2, BDNF and corticosterone ELISA kits
(EpiQuik, Epigentek Group, Inc., Farmingdale, NY, USA) were used to
determine the levels in the blood samples. According to the
manufacturer's instructions, the supernatants (100 μl) of
the blood samples were used to measure the total protein of each
sample. The samples were stored overnight at 4°C prior to ELISA and
the supernatants were then collected. Briefly, the supernatants and
antibodies were mixed and incubated in 96-well plates at 37°C for 4
h. Subsequently, the plates were washed with PBS and incubated with
horseradish peroxidase-labeled anti-rabbit antibody for 30 min at
37°C. Stop solutions were then added to the wells in the dark, and
absorbance was measured at a wavelength of 450 nm using a Synergy™
Mx Microplate Reader (BioTek Instruments, Inc., Winooski, VT,
USA).
Western blotting
Total protein was extracted from the hippocampi of
the rats or the primary hippocampal neurons using cold
radioimmunoprecipitation lysis buffer (Wuhan Boster Biological
Technology., Ltd., Wuhan, China) Protein concentration was
determined by a Bicinchoninic Acid Protein Assay kit (Thermo
Fisher, Inc.). The protein samples were then separated by 10%
SDS-PAGE (Wuhan Boster Biological Technology., Ltd.) and
transferred to a nitrocellulose membrane (Wuhan Boster Biological
Technology., Ltd.). The membrane was blocked in 8% non-fat dried
milk in PBS for 4 h, and was then incubated with the following
primary antibodies: Rabbit monoclonal anti-MeCP2 [dilution, 1:2,000
(cat. no. 3456); Cell Signaling Technology, Inc., Danvers, MA,
USA], rabbit polyclonal anti-BDNF [dilution, 1:1,000 (cat. no.
ab6201); Abcam, Cambridge, UK), and mouse monoclonal anti-GAPDH
[dilution, 1:3,000 (cat. no. BM1985); Wuhan Boster Biological
Technology, Ltd.) overnight at 4°C. The membrane was washed with
Tris-buffered saline and Tween-20 (Wuhan Boster Biological
Technology., Ltd.) and incubated with a corresponding secondary
antibody for 1 h at 37°C. Enhanced chemiluminescence reagent (Wuhan
Boster Biological Technology, Ltd.) was used to detect the signal
on the membrane. The data were analyzed by densitometry using
Image-Pro Plus software 6.0 (Media Cybernetics, Inc., Rockville,
MD, USA), and normalized to internal control expression.
Dual luciferase reporter assay
Wild type (wt) and mutant (mut) 3′UTRs of MeCP2 and
BDNF were constructed into the XbaI site of the pGL3-control
vector (Promega Corporation, Madison, WI, USA), downstream of the
luciferase gene. All of the reporter vector construction and
site-directed mutagenesis were performed by GeneCopoeia, Inc.
(Guangzhou, China). For the luciferase assay, 1×105
cells were cultured in 24-well plates, until they had reached ~70%
confluence. Subsequently, the cells were co-transfected with
miR-132 mimic, and wt or mut 3′UTR of MeCP2 or BDNF dual luciferase
reporter vector, respectively. Following a 3 h incubation with the
trans-fection reagent/DNA complex, the medium was refreshed with
fresh medium supplemented with 10% fetal bovine serum (each from
Invitrogen Life Technologies). Post-transfection (48 h), a Dual
Luciferase Reporter Gene Assay kit (BioVision, Inc., Milpitas, CA,
USA) was used to determine the luciferase activities of each group
using a luminometer (Roche Diagnostics, Basel, Switzerland).
Renilla luciferase activity was normalized to firefly
luciferase activity.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). The data are
presented as the mean ± standard deviation. An unpaired two-tailed
Student's t-test was used to analyze the results. P<0.05 was
considered to indicate a statistically significant difference.
Results
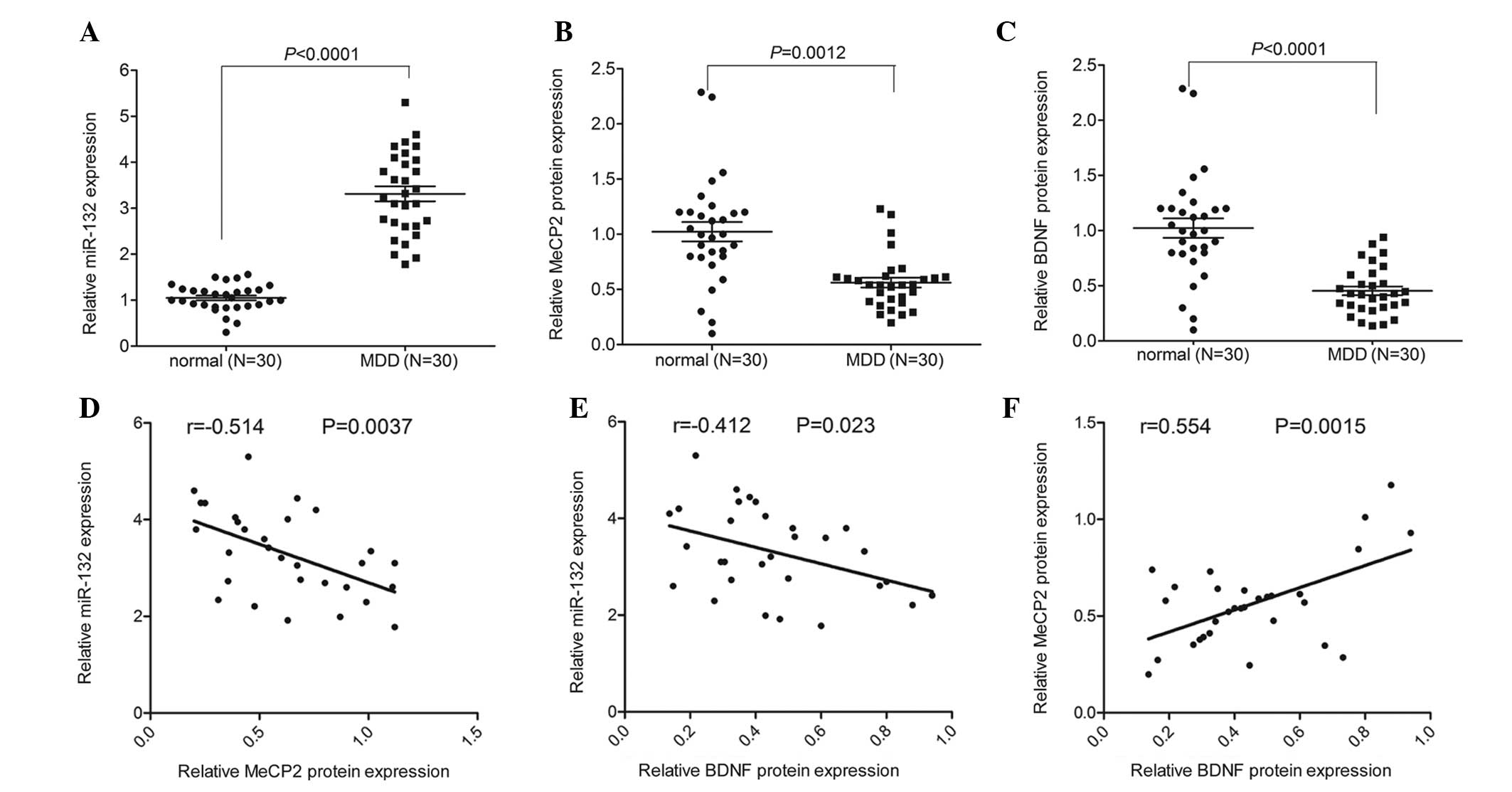
miR-132 expression is negatively
correlated with MeCP2 and BDNF protein expression in peripheral
blood samples of patients with MDD
The expression levels of miR-132 were detected by
RT-qPCR analysis. The large panel of samples included blood samples
from 30 patients with MDD and 30 normal controls. miR-132
expression was significantly increased in the peripheral blood of
patients with MDD, as compared with the paired normal controls
(Fig. 1A). In addition, an ELISA
assay was used to measure the protein expression of MeCP2. As shown
in Fig. 1B, the protein expression
of MeCP2 was significantly decreased in the peripheral blood of
patients with MDD, as compared with the normal controls.
Furthermore, there was a significant decrease in BDNF expression
detected in the peripheral blood of patients with MDD, as compared
with the controls (Fig. 1C). The
expression of miR-132 was negatively correlated with the protein
expression levels of MeCP2 and BDNF, whereas MeCP2 expression was
positively correlated with BDNF expression in the peripheral blood
of patients with MDD (Fig.
1D–F).
MeCP2 and BDNF are downregulated, and
miR-132 is upregulated in rats exposed to CUS
The CUS model is a well-established animal model of
depression that mimics specific symptoms of human depression, such
as anhedonia and learned helplessness. Following exposure to CUS,
the rats exhibited high serum corticosterone levels and depressive
behaviors, including increased immobility time in the FST and
decreased sucrose consumption (Fig.
2A–C). Concordant with the results mentioned above, miR-132
expression was upregulated in the hippocampi of CUS-exposed rats,
whereas the protein expression levels of MeCP2 and BDNF were
significantly decreased (Fig.
2D–F). These results are concordant with the results from the
blood samples of human patients with MDD.
 | Figure 2Expression of miR-132, BDNF and MeCP2
in CUS-exposed rats. (A) FST demonstrated increased immobility time
in CUS-exposed rats, as compared with the control. (B) SPT
demonstrated decreased sucrose consumption in CUS-exposed rats, as
compared with the control. (C) Relative release of corticosterone
in CUS-exposed and control rats, as determined by ELISA. (D)
Relative expression levels of miR-132 in the hippocampi of
CUS-exposed and control rats. (E) Relative expression levels of
MeCP2 in the hippocampi of CUS-exposed and control rats. (F)
Relative expression levels of BDNF in the hippocampi of CUS-exposed
and control rats. Data are presented as the mean ± standard
deviation. *P<0.05, **P<0.01 and
***P<0.001 vs. the control rats. miR, microRNA; BDNF,
brain-derived neurotrophic factor; MeCP2, methyl-CpG-binding
protein 2; CUS, chronic unpredictable stress; SPT, sucrose
preference test; FST, forced swim test. |
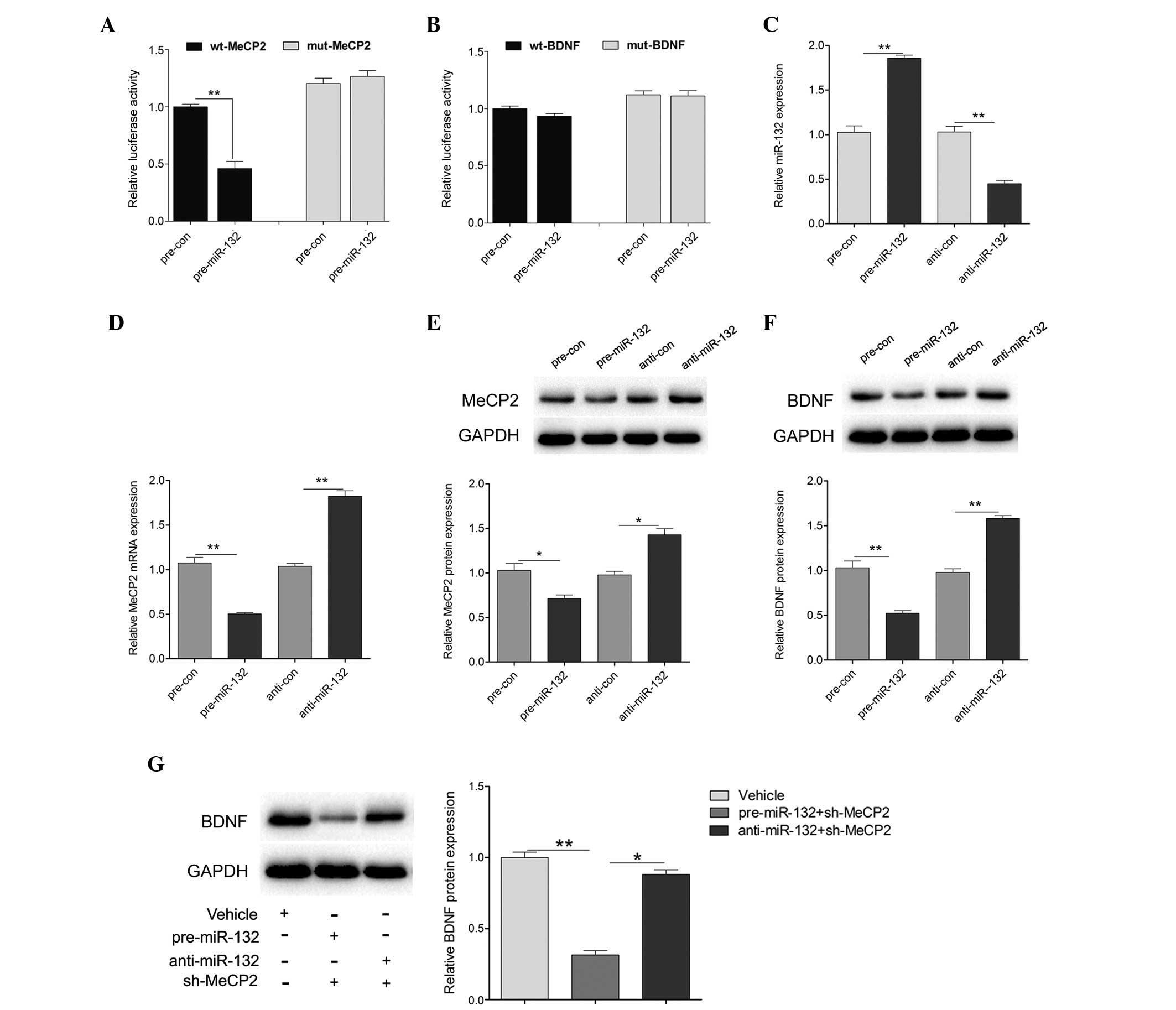
MeCP2 knockdown induces miR-132
expression and decreases BDNF expression
To determine the association between MeCP2, miR-132
and BDNF, the expression of MeCP2 was reduced by transfecting
sh-MeCP2 or a control vector, into primary hippocampal neurons.
Transfection with sh-MeCP2 efficiently decreased the expression
levels of MeCP2, as compared with the control vector (Fig. 3A). Furthermore, the expression
levels of miR-132 were detected. Increased expression levels of
miR-132 were detected in the primary hippocampal neurons
transfected with sh-MeCP2 (Fig.
3B). Furthermore, the expression of miR-132 was also induced
following treatment of the primary hippocampal neurons with the
demethylation agent Aza (Fig. 3C).
However, MeCP2 knockdown reduced BDNF expression (Fig. 3D). These results suggest that MeCP2
is able to regulate the expression of miR-132, which may be
involved in methylation.
miR-132 directly targets MeCP2 and
regulates BDNF expression
To investigate whether miR-132 targets the 3′UTR of
MeCP2 or BDNF, the wt 3′UTR of MeCP2 or BDNF (wt-MeCP2 or wt-BDNF)
was cloned downstream to a luciferase reporter gene. In addition, a
mutant 3′UTR (mut-MeCP2 or mut-BDNF) was constructed using binding
site mutagenesis. The wt-MeCP2 vector was co-transfected with
pre-miR-132 mimics or a scramble control into primary hippocampal
neurons. The luciferase activity of pre-miR-132 and wt-MeCP2
co-transfected cells was significantly reduced, as compared with
the scramble control cells (Fig.
4A). However, there were no significant changes in the
luciferase activity of pre-miR-132 and wt-BDNF co-transfected
cells, as compared with the scramble control cells (Fig. 4B). In addition, pre-miR-132 and
anti-miR-132 were transfected into primary hippocampal neurons. As
shown in Fig. 4C, the transfection
efficiency was satisfactory for further analysis. Overexpression of
miR-132 significantly reduced the expression levels of MeCP2, at
both the mRNA and protein level, whereas downregulation of miR-132
increased the mRNA and protein expression levels of MeCP2 (Fig. 4D and E). Furthermore, the protein
expression levels of BDNF were detected in the cells transfected
with pre-miR-132 or anti-miR-132. The protein expression levels of
BDNF were decreased following upregulation of miR-132, and
increased following downregulation of miR-132 (Fig. 4F). Reduced BDNF expression was
detected in the pre-miR-132 combined with sh-MeCP2 treatment group,
whereas treatment with anti-miR-132 combined with sh-MeCP2 had
little effect on BDNF expression (Fig.
4G). These results indicate that MeCP2 and miR-132 may regulate
hippocampal BDNF expression in an opposite manner, suggesting that
homeostatic interactions between these factors control hippocampal
BDNF expression.
Discussion
The symptoms of depression have been well
characterized, and include anhedonia and learned helplessness;
however, the underlying pathogenic mechanism remains unclear. The
present study demonstrated that miR-132 was significantly increased
in the peripheral blood of patients with MDD, as compared with the
normal subjects. The expression levels of miR-132 were shown to be
negatively correlated with the protein expression levels of MeCP2
and BDNF. In addition, in a rat model of CUS-induced depression,
miR-132 expression was upregulated in the hippocampi of CUS-exposed
rats, whereas the protein expression levels of MeCP2 and BDNF were
significantly decreased. Since it is difficult to determine BDNF
levels directly from the brain of patients, an increased effort has
been made regarding the measurement of BDNF levels in peripheral
blood, as the BDNF levels in the brain of patients with mood
disorders would be reliably reflected by levels in the peripheral
blood. It has previously been reported that serum BDNF levels were
decreased, whereas the levels of miR-132 were increased, in
patients with depression as compared with those of healthy controls
(7). Furthermore, serum BDNF
levels are increased following treatment with antidepressant agents
(21), thus suggesting that serum
BDNF levels may be used as a potential biomarker for measuring
depression status and the efficacy of antidepressant treatment. In
addition, previous studies have shown that BDNF gene polymorphisms
are associated with depression-related traits (22,23).
In an animal model of depression, decreased BDNF levels in the
brain and serum have been well demonstrated (24), suggesting that BDNF may have a
critical role as a susceptibility gene in depression. Therefore,
controlling BDNF levels may be an important factor for the
management of depression.
MeCP2 levels are closely correlated with BDNF
(25); however, the underlying
dynamics of this complex relationship in the brain are not fully
clear. Numerous studies have identified BDNF as a target that may
be regulated by MeCP2, which is relevant to the pathogenesis of MDD
(26–28). By binding to the BDNF promoter,
MeCP2 directly modulates BDNF expression in an activity-dependent
manner (29). Neuronal activity
rapidly induces the dissociation of MeCP2 from the BDNF promoter,
which enhances the expression of BDNF in response to neuronal
activity. Mecp2-deficient mice have been shown to exhibit reduced
BDNF mRNA and protein expression in various brain regions,
including the hippocampus (30).
In addition, reduced overall neuronal activity caused by MeCP2
deficiency is hypothesized to contribute to BDNF downregulation,
whereas BDNF overexpression rescued certain functional deficits
observed in MeCP2 mutants and extended their lifespan (31). Furthermore, MeCP2 overexpression in
cultured mouse cortical neurons results in increased BDNF
expression (18). A previous study
demonstrated that phosphorylation of MeCP2 at serine 421 has a
critical role in regulating the development of hippocampal
dendritic spines and the mature form of excitatory synapses
(32). Concordant with the
findings of previous studies, the present study demonstrated that
reduced BDNF and MeCP2 protein expression levels were detected in
CUS-exposed rats that exhibited depressive-like behavior.
Furthermore, knockdown of MeCP2 in primary hippocampal neurons
decreased BDNF protein expression levels. These results indicated
that hippocampal MeCP2 may directly regulate BDNF expression in
CUS-exposed rats, which may contribute to neuronal activity and
survival.
Knockdown of MeCP2 in primary hippocampal neurons
not only decreased BDNF protein expression levels, but also induced
miR-132 expression. In addition, treatment with Aza, a
demethylation agent, induced miR-132 expression in primary
hippocampal neurons. These results indicated that miR-132
expression may be regulated by MeCP2 via an epigenetic mechanism. A
previous study demonstrated that in patients with depression there
is a significant positive correlation between Self-Rating
Depression Scale scores and the expression of miR-132, and an
inverse correlation between serum BDNF levels and miR-132 levels in
depression (7). A previous study
reported on miRNAs associated with the sensitivity of human
lymphoblastoid cell lines to selective serotonin reuptake
inhibitors. It was demonstrated that miR-212 and miR-132 were
differentially expressed between two human lymphoblastoid cell
lines, which exhibited high or low sensitivities to paroxetine
(33). Furthermore, specific
overexpression of miR-132 in the perirhinal cortex impaired the
short-term recognition memory of rats (34). In addition, footshock stress and
predator scent stress induced a long-lasting hippocampal elevation
of miR-132 expression (35). These
results suggested a role of miR-132 in the neuronal mechanisms
underlying the symptoms of depression.
Furthermore, in the present study it was
demonstrated that miR-132 was able to regulate MeCP2 at the mRNA
and protein level by directly targeting its 3′UTR. However, it
could not directly target BDNF, although BDNF protein expression
levels were affected by miR-132. As mentioned above, MeCP2
expression was positively correlated, whereas miR-132 expression
was negatively correlated, with BDNF levels in patients with MDD.
In addition, in CUS-exposed rats, hippocampal MeCP2 and BDNF
expression was decreased, whereas hippocampal miR-132 expression
was increased. Furthermore, downregulated expression of BDNF,
induced by MeCP2 knockdown, was enhanced by miR-132 mimics and
rescued by miR-132 inhibitors. These results demonstrated that
MeCP2-miR-132 homeostatic interactions may control the hippocampal
BDNF levels, which are associated with depression. BDNF can be
synthesized locally in the hippocampus in an activity-dependent
manner, and unpredictable and persistent stress can reduce local
BDNF production. The results of the present study suggested that
unpredictable and persistent stress was able to decrease the de
novo production of BDNF in the hippocampus. Furthermore,
downregulation of MeCP2 may result in loss of the inhibitory
capacity of the repressors of BDNF transcription, such as RE1
silencing transcription factor (36). In the same manner, miR-132 may
decrease hippocampal BDNF levels by silencing MeCP2 expression. A
recent study demonstrated that MeCP2 serves as a necessary
co-activator of cAMP response element-binding protein activity at
the BDNF promoter (37). In this
way, MeCP2 levels may determine the stimulatory effects of CREB
signaling on BDNF production in the hippocampus. The present study
demonstrated that MeCP2 and miR-132 have opposite effects on
hippocampal BDNF levels, suggesting that homeostatic interactions
between MeCP2 and miR-132 may have a key role in depression.
In conclusion, the present study highlights the
interaction between MeCP2 and miR-132 in regulating hippocampal
BDNF levels, suggesting that miR-132 may be a key factor in
controlling stress-induced hippocampal neuroplasticity and neuronal
survival in depression.
References
|
1
|
Dwivedi Y: Emerging role of microRNAs in
major depressive disorder: Diagnosis and therapeutic implications.
Dialogues Clin Neurosci. 16:43–61. 2014.PubMed/NCBI
|
|
2
|
Mouillet-Richard S, Baudry A, Launay JM
and Kellermann O: MicroRNAs and depression. Neurobiol Dis.
46:272–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sala M, Perez J, Soloff P, Ucelli di Nemi
S, Caverzasi E, Soares JC and Brambilla P: Stress and hippocampal
abnormalities in psychiatric disorders. Eur Neuropsychopharmacol.
14:393–405. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li M, Fu Q, Li Y, Li S, Xue J and Ma S:
Emodin opposes chronic unpredictable mild stress induced
depressive-like behavior in mice by upregulating the levels of
hippocampal glucocorticoid receptor and brain-derived neurotrophic
factor. Fitoterapia. 98:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maripuu M, Wikgren M, Karling P, Adolfsson
R and Norrback KF: Relative hypo- and hypercortisolism are both
associated with depression and lower quality of life in bipolar
disorder: A cross-sectional study. PLoS One. 9:e986822014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Réus GZ, Abelaira HM, Stringari RB, Fries
GR, Kapczinski F and Quevedo J: Memantine treatment reverses
anhedonia, normalizes corticosterone levels and increases BDNF
levels in the prefrontal cortex induced by chronic mild stress in
rats. Metab Brain Dis. 27:175–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang
YX, Li XX, Zhang C, Xie SY and Wang PY: Alterations of serum levels
of BDNF-related miRNAs in patients with depression. PLoS One.
8:e636482013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai M, Zhu X, Zhang Y, Zhang S, Zhang L,
Xue L, Yi J, Yao S and Zhang X: Abnormal hippocampal BDNF and
miR-16 expression is associated with depression-like behaviors
induced by stress during early life. PLoS One. 7:e469212012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bahi A, Chandrasekar V and Dreyer JL:
Selective lentiviral-mediated suppression of microRNA124a in the
hippocampus evokes antidepressants-like effects in rats.
Psychoneuroendocrinology. 46:78–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Connor RM, Grenham S, Dinan TG and Cryan
JF: microRNAs as novel antidepressant targets: Converging effects
of ketamine and electroconvulsive shock therapy in the rat
hippocampus. Int J Neuropsychopharmacol. 16:1885–1892. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dell'Osso B, D'Addario C, Carlotta Palazzo
M, Benatti B, Camuri G, Galimberti D, Fenoglio C, Scarpini E, Di
Francesco A, Maccarrone M and Altamura AC: Epigenetic modulation of
BDNF gene: Differences in DNA methylation between unipolar and
bipolar patients. J Affect Disord. 166:330–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zucchi FC, Yao Y, Ward ID, Ilnytskyy Y,
Olson DM, Benzies K, Kovalchuk I, Kovalchuk O and Metz GA: Maternal
stress induces epigenetic signatures of psychiatric and
neurological diseases in the offspring. PLoS One. 8:e569672013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murgatroyd C, Wu Y, Bockmühl Y and
Spengler D: Genes learn from stress: How infantile trauma programs
us for depression. Epigenetics. 5:194–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Forlani G, Giarda E, Ala U, Di Cunto F,
Salani M, Tupler R, Kilstrup-Nielsen C and Landsberger N: The
MeCP2/YY1 interaction regulates ANT1 expression at 4q35: Novel
hints for Rett syndrome pathogenesis. Hum Mol Genet. 19:3114–3123.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hutchinson AN, Deng JV, Cohen S and West
AE: Phosphorylation of MeCP2 at Ser421 contributes to chronic
antidepressant action. J Neurosci. 32:14355–14363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murgatroyd C, Patchev AV, Wu Y, Micale V,
Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF and
Spengler D: Dynamic DNA methylation programs persistent adverse
effects of early-life stress. Nat Neurosci. 12:1559–1566. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wada R, Akiyama Y, Hashimoto Y, Fukamachi
H and Yuasa Y: miR-212 is downregulated and suppresses
methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J
Cancer. 127:1106–1114. 2010. View Article : Google Scholar
|
|
18
|
Klein ME, Lioy DT, Ma L, Impey S, Mandel G
and Goodman RH: Homeostatic regulation of MeCP2 expression by a
CREB-induced microRNA. Nat Neurosci. 10:1513–1514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Im HI, Hollander JA, Bali P and Kenny PJ:
MeCP2 controls BDNF expression and cocaine intake through
homeostatic interactions with microRNA-212. Nat Neurosci.
13:1120–1127. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo YW, Xu Y, Cao WY, Zhong XL, Duan J,
Wang XQ, Hu ZL, Li F, Zhang JY, Zhou M, Dai RP and Li CQ:
Insulin-like growth factor 2 mitigates depressive behavior in a rat
model of chronic stress. Neuropharmacology. 89:318–324. 2015.
View Article : Google Scholar
|
|
21
|
Munno D, Sterpone S, Fania S, Cappellin F,
Mengozzi G, Saroldi M, Bechon E and Zullo G: Plasma brain derived
neurotrophic factor levels and neuropsychological aspects of
depressed patients treated with paroxetine. Panminerva Med.
55:377–384. 2013.
|
|
22
|
Yu H, Wang DD, Wang Y, Liu T, Lee FS and
Chen ZY: Variant brain-derived neurotrophic factor Val66Met
polymorphism alters vulnerability to stress and response to
antidepressants. J Neurosci. 32:4092–4101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carballedo A, Amico F, Ugwu I, Fagan AJ,
Fahey C, Morris D, Meaney JF, Leemans A and Frodl T: Reduced
fractional anisotropy in the uncinate fasciculus in patients with
major depression carrying the met-allele of the Val66Met
brain-derived neurotrophic factor genotype. Am J Med Genet B
Neuropsychiatr Genet. 159B:537–548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Valvassori SS, Budni J, Varela RB and
Quevedo J: Contributions of animal models to the study of mood
disorders. Rev Bras Psiquiatr. 35(Suppl 2): S121–S131. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu X, Kozikowski AP and Pozzo-Miller L: A
selective histone deacetylase-6 inhibitor improves BDNF trafficking
in hippocampal neurons from Mecp2 knockout mice: Implications for
Rett syndrome. Front Cell Neurosci. 8:682014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khoshnan A and Patterson PH: Elevated IKKα
accelerates the differentiation of human neuronal progenitor cells
and induces MeCP2-dependent BDNF expression. PLoS One.
7:e417942012. View Article : Google Scholar
|
|
27
|
Cortés-Mendoza J, Díaz de León-Guerrero S,
Pedraza-Alva G and Pérez-Martínez L: Shaping synaptic plasticity:
the role of activity-mediated epigenetic regulation on gene
transcription. Int J Dev Neurosci. 31:359–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv J, Xin Y, Zhou W and Qiu Z: The
epigenetic switches for neural development and psychiatric
disorders. J Genet Genomics. 40:339–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Zhong X, Chau KF, Williams EC and
Chang Q: Loss of activity-induced phosphorylation of MeCP2 enhances
synaptogenesis, LTP and spatial memory. Nat Neurosci. 14:1001–1008.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li W, Calfa G, Larimore J and Pozzo-Miller
L: Activity-dependent BDNF release and TRPC signaling is impaired
in hippocampal neurons of Mecp2 mutant mice. Proc Natl Acad Sci
USA. 109:17087–17092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang Q, Khare G, Dani V, Nelson S and
Jaenisch R: The disease progression of Mecp2 mutant mice is
affected by the level of BDNF expression. Neuron. 49:341–348. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY,
Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al:
Brain-specific phosphorylation of MeCP2 regulates
activity-dependent Bdnf transcription, dendritic growth, and spine
maturation. Neuron. 52:255–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oved K, Morag A, Pasmanik-Chor M,
Oron-Karni V, Shomron N, Rehavi M, Stingl JC and Gurwitz D:
Genome-wide miRNA expression profiling of human lymphoblastoid cell
lines identifies tentative SSRI antidepressant response biomarkers.
Pharmacogenomics. 13:1129–1139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scott HL, Tamagnini F, Narduzzo KE,
Howarth JL, Lee YB, Wong LF, Brown MW, Warburton EC, Bashir ZI and
Uney JB: MicroRNA-132 regulates recognition memory and synaptic
plasticity in the perirhinal cortex. Eur J Neurosci. 36:2941–2948.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shaltiel G, Hanan M, Wolf Y, Barbash S,
Kovalev E, Shoham S and Soreq H: Hippocampal microRNA-132 mediates
stress-inducible cognitive deficits through its
acetylcholinesterase target. Brain Struct Funct. 218:59–72. 2013.
View Article : Google Scholar :
|
|
36
|
Abuhatzira L, Makedonski K, Kaufman Y,
Razin A and Shemer R: MeCP2 deficiency in the brain decreases BDNF
levels by REST/CoREST-mediated repression and increases TRKB
production. Epigenetics. 2:214–222. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chahrour M, Jung SY, Shaw C, Zhou X, Wong
ST, Qin J and Zoghbi HY: MeCP2, a key contributor to neurological
disease, activates and represses transcription. Science.
320:1224–1229. 2008. View Article : Google Scholar : PubMed/NCBI
|