Introduction
Alzheimer's disease (AD) is a prototypical
age-associated neurodegenerative disorder and is the most common
progressive form of dementia. Although the pathogenesis of AD
remains to be clarified, missense mutations in three associated
genes coding for the proteins amyloid precursor protein (APP) and
the presenilins (PSs) have been identified as a causative factor in
early-onset familial AD (EOFAD) (1–3). Of
note, mutations within the PS gene are the most common cause of
EOFAD. PS1 mutations account for 18–50% of EOFAD cases (4), while mutations in PS2 are rare
(5). PS1 and PS2 are integral
membrane proteins with nine transmembrane domains (TMDs) similar in
amino acid composition (65%) and intracellular location (6). Of note, PS1 and PS2 exhibit a
different tissue-specific expression pattern in transcription
level. PS1 is uniformly expressed (7), while PS2 is restricted to the
heart, skeletal muscle and pancreas (8).
PS is the catalytic core of the γ-secretase complex,
containing nicastrin, PS enhancer 2 and anterior pharynx defective
1 (2,9,10).
γ-Secretases cleave various type I transmembrane proteins,
primarily APP, the Notch receptor, N-cadherin and E-cadherin, which
are associated with AD in vertebrates (11). APP, the most well-known substrate
of γ-secretase, generates a 37–49 amino acid peptide termed
amyloid-β (Aβ) following cleavage. These peptides, particularly
Aβ42, are abundantly released and aggregate into oligomeric
structures in the brain of patients with an APP or PS missense
mutation, and are considered to be the initiating factors of AD
(12). In addition, PS has also
been reported to regulate the kinase activity and control tau
phosphorylation, which is the causative factor of neurofibrillary
tangles in AD and other neurodegenerative disorders (13).
A number of invertebrate model organisms, including
Caenorhabditis (C.) elegans and
Drosophila (D) melanogaster, have been
employed to investigate AD, and have provided insight into AD
progression and the function of genes involved (14–17).
In C. elegans and D. melanogaster, homologues of
PSs have been identified. Sel-12, the homologous gene
of PS in C. elegans, has been confirmed to be involved in
the lin-12-mediated cell signaling pathway, which is homologous to
the Notch pathway in D. melanogaster and vertebrates
(18). Accumulating evidence has
demonstrated that the Notch receptor is the hydrolytic substrate of
Drosophila presenilin (DPS) in D. melanogaster, and
overexpression of wild-type DPS or DPS mutations in Drosophila
resulted in several phenotypes due to loss-of-function of Notch,
including wing margin loss, wing vein thickening and rough eye
(19–23), which are considered to be the
results of an apoptotic process of the developmental wing and eye
discs (24). Although Notch is
present in the adult brain and is required for long-term memory in
Drosophila (25), it
remains to be elucidated whether DPS regulates long-term memory via
Notch signaling. A DPS-null mutation has been reported to impair
learning and memory, which, however, cannot be exclusively
attributed to loss-of-function in the Notch pathway (26). In addition, DPS is also required
for calcium homeostasis, the disruption of which may lead to
cognitive deficits (27–29). Similarly to investigations in
invertebrates, Notch has also been confirmed to interact with PSs
and APP in mammals (30,31) and the expression of Notch 1 is
increased in the brains of patients with AD (32). Thus, the understanding of PS and
Notch signaling in AD may aid in the development of novel
strategies for the treatment of AD. Molecular and genetic studies
of PS homologues in invertebrates may also provide important
insight into the pathogenic mechanism of this gene family in
mammals.
The silkworm, Bombyx mori (B. mori),
is widely used in genetics and molecular biology and has numerous
beneficial traits as a model compared with mammals or other
insects. In particular, the whole genome of the silkworm has been
sequenced, which allows for genetic manipulation, and there are
already a number of ongoing research projects that utilize
transgenic and genetically modified silkworms (33,34).
Of note, a number of previous studies have demonstrated the
possibility of applying the results obtained in silkworms to
mammals (33,34).
To gain insight into the function of B. mori
presenilin (BmPS), the present study cloned and characterized the
BmPS gene and assessed its homology with PS from other
species. Furthermore, the expression of BmPS in various
B. mori tissues at the adult and larval/pupal stages was
assessed. The present study provided a basis for the understanding
of the function of PS, particularly with regards to the development
of AD.
Materials and methods
Reagents
The reagents and detection kits used in the present
study were as follows: LATaq, ExTaq, T4 Ligase, RNase-free DNase I,
pMD18 T-Vector, DL2000 DNA marker, DL5000 DNA marker, 5′-Full RACE
kit and 3′-Full RACE kit were purchased from Takara Bio, Inc.
(Otsu, Japan); ReverTra Ace® kit, KOD Plus ver.2, 100 bp
DNA Marker and SYBR® Green real-time PCR master mix were
purchased from Toyobo Co., Ltd. (Osaka, Japan); Agarose gel
extraction kit, Plasmid mini preparation kit and PCR clean-up kit
were purchased from Axygen (Union City, CA, USA); T7 polymerase and
PolyATtract® mRNA isolation system III were purchased
from Promega Corporation (Madison, WI, USA); TRIzol reagent was
purchased from Invitrogen Life Technologies (Carlsbad, CA, USA).
Unless stated otherwise, all other chemicals were purchased from
Sigma-Aldrich (St. Louis, MO, USA).
Animal model
The two B. mori strains, Dazao and R13Q, were
kindly provided by Professor Mu-wang Li and Professor An-yin Xu
(Department of Biotechnology, Jiangsu University of Science and
Technology, Zhenjiang, China). The larvae were reared on fresh
mulberries at 25°C under a 12 h-light/dark cycle. Eggs, pupae and
adults were maintained under the same light and temperature
conditions.
RNA extraction and reverse transcription
polymerase chain reaction (RT-PCR)
Total RNA from various tissues of B. mori was
extracted with TRIzol reagent. A total of 1 µg total RNA was
denatured at 65°C for 5 min and immediately chilled on ice.
First-strand cDNA was synthesized using ReverTra Ace®
according to the manufacturer's instructions, with Oligo
(dT)20 primers. Two specific primers F1 and R1 (as shown
in Table I) designed from the
flanks of the predicted BmPS coding sequence (CDS) present
in the expressed sequence tag (EST) sequences (BJ983375, BY936068,
CK563766, CK490866, CK488524, CK488055 and CK485294) from the
SilkBase (http://silkworm.genomics.org.cn/) were used for
amplification of the actual CDS. All PCR procedures were performed
using LA Taq with GC buffer (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) and consisted of 3 min of denaturation at 94°C followed by
35 cycles of 30 sec at 94°C, 30 sec at 58°C and 80 sec at 72°C, and
a final extension step at 72°C for 5 min.
 | Table IPrimer sequences designed for
Bombyx mori prese-nilin cloning. |
Table I
Primer sequences designed for
Bombyx mori prese-nilin cloning.
| Primer | Primer sequence
(5′–3′) | Positiona |
|---|
| F1 |
AGTGACTTAGACGCAACCGAGCA | Exon1 |
| R1 |
GACTTCTATCTACCTTACGCACA | Exon10 |
| F1-1 |
TTTGACTTTTATGGTTTTACC | Exon1 |
| R1-1 |
TTAGTTACGATTCGAATTTG | Exon10 |
| R2 |
GCTGGAACTCGAGGTTGCTGGG | Exon2 |
| R3 |
CCCCGATGGGCCACCTTCA | Exon2 |
| R4 |
CACCTAGACCAAGTTTTACGCCT | Exon9 |
| R5 |
ACCTTCGCCCTCGACAGTTC | Exon8 |
| F2 |
ATTTGCGGACGCTTTGGC | Exon10 |
| F3 |
GAACAGGTGTTCATTTAGC | Exon10 |
| F4 |
AACGCAATGAGCCTATATTTCCA | Exon5 |
| F5 |
ATGGACGGCCACATCGCAGAA | Exon1 |
The expression of BmPS alternative splice
sites was investigated by RT-PCR using total RNA extracted from
R13Q 3rd- and 5th instar larval tissues with the primers of F4 and
R5 (as shown in Table I). All PCRs
were performed using KOD Plus with 5% dimethyl sulfoxide and
consisted of 2 min of denaturation at 94°C followed by 28 cycles of
10 sec at 98°C, 30 sec at 58°C and 20 sec at 68°C.
The PCR products were purified by 1% agarose gel
electrophoresis, sub-cloned into the pMD18-T vector and sequenced
by Shanghai Invitrogen Biotechnology Co., Ltd. (Shanghai,
China).
Rapid amplification of cDNA ends
(RACE)
To clone the full-length cDNA of BmPS, 5′-
and 3′-RACE was performed using the 5′- and 3′-Full RACE kit
according to the manufacturer's instructions. For the 5′-RACE,
first-strand cDNA was generated by RT of poly(A)+ RNA
from the pupal brains of the R13Q strain using the 5′-cDNA
synthesis (CDS) primer provided in the kits. The cDNA was amplified
by PCR using gene-specific primers (GSPs) and R2/R3 (as shown in
Table I), which were synthesized
by Sangon Biotech Co., Ltd. (Shanghai, China). To determine the
BmPS transcripts with multiple transcription start sites and
alternative splice sites, 5′-RACE PCR was performed again with the
downstream primer R4 and the nest primer R5. In 3′-RACE,
first-strand cDNA was generated using the 3′-CDS primer. The cDNA
was amplified by PCR using GSPs and F2/F3 (as shown in Table I). The primers F1-1 and R1-1 were
used to assess whether the PCR products of 5′ and 3′ RACE belonged
to a single transcript.
RT-quantitative (q)PCR analysis
The brains of 4th instar larvae, 5th instar larvae,
pupae and adults were collected from the R13Q strain. The total
extracted RNA was treated with DNase to eliminate potential genomic
DNA contamination. The RT was performed using the ReverTra Ace kit
according to the manufacturer's instructions and was used in the
RT-qPCR (iQ5; Bio-Rad Laboratories, Inc.) assays. The negative
controls without the RT enzyme were prepared and analyzed in
parallel with the unknown samples during the RT-qPCR assay. RT-qPCR
was performed in a 20-µl volume using SYBR Green real-time PCR
master mix and the CFX96 system (Bio-Rad Laboratories, Inc.) with
template cDNA equivalent to 200 ng RNA and 0.4-mM primers. The
primer sequences for BmPS are shown in Table I, and the primers for the
BmActin3 gene, as the endogenous control, were as follows:
Forward, 5′-GCG CGG CTA CTC GTT CAC TACC-3′ and reverse, 5′-GGA TGT
CCA CGT CGC ACT TCA-3′. The two-step amplification protocol
consisted of 3 min at 95°C followed by target amplification via 40
cycles at 95°C for 15 sec, 63.5°C for 30 sec and 72°C for 30 sec.
The standard curves were generated by a 10-fold serial dilution of
template cDNA, and the reaction efficiency (E) was
determined by the equation E=10(−1/slope).
Quantification analyses of BmPS transcript expression
relative to BmActin3 were calculated according to the
2−ΔΔCt method using the CFX Manager software (version
1.6; Bio-Rad Laboratories, Inc.). Data from triplicate experiments
are expressed as the mean ± standard deviation.
Sequence analysis
To analyze the intron/exon of the BmPS, the
BmPS sequences were compared with silkworm genomic sequences
from the National Center for Biotechnology Information (NCBI)
GenBank database (http://www.ncbi.nlm.nih.gov/). The amino acid
sequences of PS homologues from various species were also
retrieved from the GenBank database using the basic local alignment
search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple
sequence alignments were performed using the DNAman, and
phylogenetic analysis was performed with MEGA software, version 5
(http://www.megasoftware.net/).
Statistical analysis of data
Student's one-tailed unpaired t-test was used to
perform statistical analysis. A minimum of three independent
replicates were used for each treatment and the error bars
represent the mean ± standard error.
Results
Cloning of the BmPS gene and its
variants
To characterize the B. mori presenilin gene
(BmPS), BmPS was cloned from 5th instar larval brains
of the R13Q strain. A 1,460-bp fragment (designated BmC-1) was
PCR-amplified using the specific primers F1 and R1 (as shown in
Table I), which were designed from
a conserved amino acid fragment of PSs from the silkworm EST
database. Based on the sequence of the original BmC-1 clone,
primers were designed for 5′- and 3′-RACE. A total of three
fragments (394 bp, 345 bp and 195 bp) were amplified by 5′-RACE
with the primer R2 and the nest primer R3 (as shown in Table I). Analysis of the sequence of
three fragments indicated that the 5′-region of the BmPS
transcripts contained at least three transcription start sites and
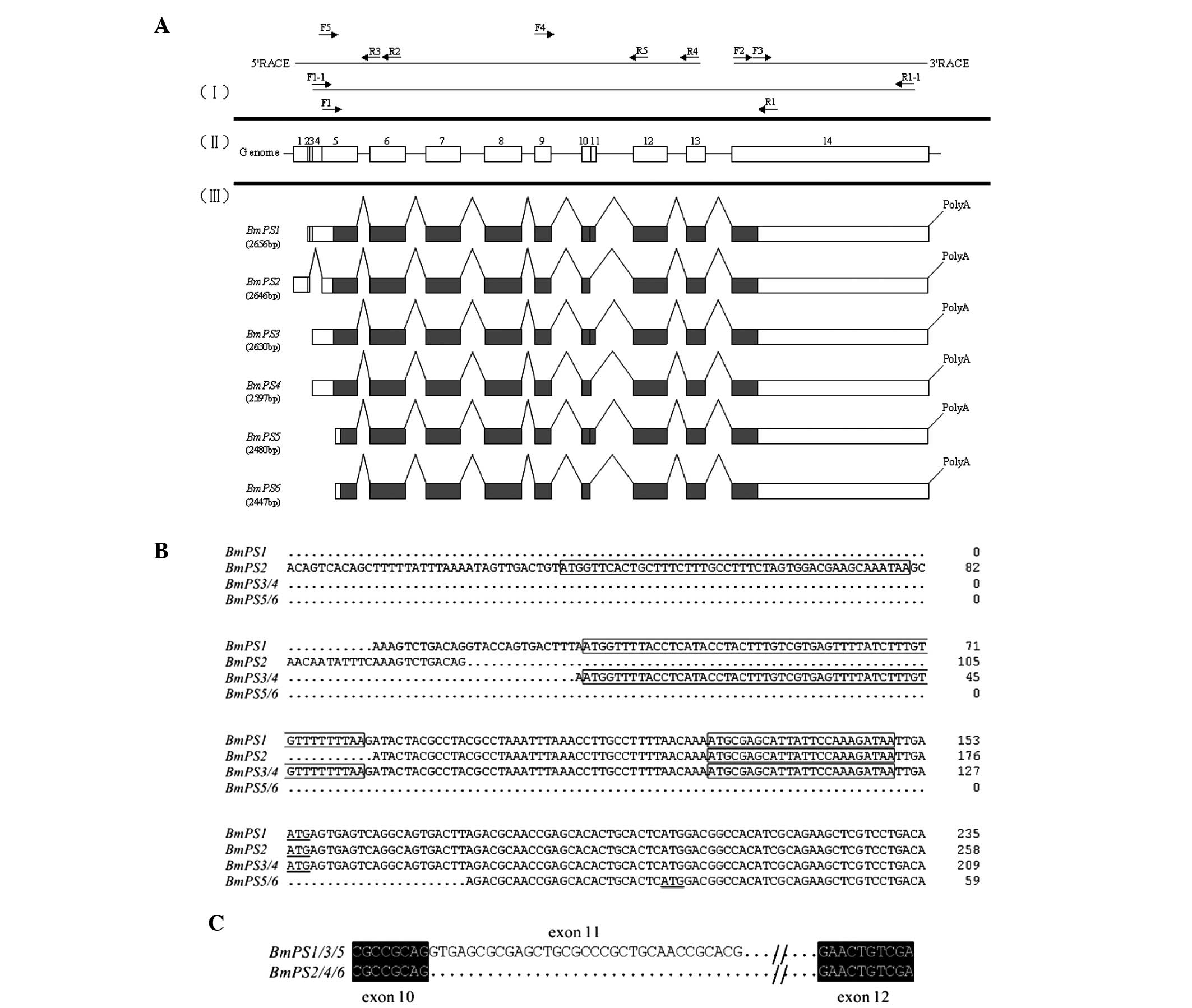
upstream open reading frames (uORFs; Figs. 1 and 2). Only a 1,113-bp fragment was
determined by 3′-RACE using the F2 and F3 primers (as shown in
Table I and Fig. 2), which exhibited a putative
polyadenylation signal (AATAAA) starting 15 nucleotides upstream of
the polyA tail (Fig. 1).
To confirm that 5′-RACE, BmC-1 and 3′-RACE belong to
the same transcript, the PCR amplification with primers F1-1 and
R1-1 (as shown in Table I and
Fig. 2) designed from the 5′-UTR
and 3′-UTR of BmPSs was performed. Only one fragment was
obtained and sequence analysis indicated that the PCR products of
5′-RACE, BmC-1 and 3′-RACE belonged to the same transcript. In
addition, a novel transcript was identified which had a 33
bp-deletion due to the alternative splicing (Fig. 1).
To determine the BmPS transcript combinations
with multiple transcription start sites and the 33 bp of
alternative splicing, 5′-RACE PCR was performed again with primers
R4/R5, which are downstream of the 33-bp deletion. The two bands
were amplified and sequence analysis of the 30 clones revealed that
the BmPS gene had six transcripts termed BmPS1 (2,656
bp), BmPS2 (2,646 bp), BmPS3 (2,630 bp), BmPS4
(2,597 bp), BmPS5 (2,480 bp) and BmPS6 (2,447 bp),
respectively (Fig. 2). The
nucleotide sequences of BmPS1 and BmPS2 have been
submitted to the GenBank/NCBI database with the accession numbers
JQ993471.1 and JQ993472.1, respectively.
Compared with the silkworm genome, BmPS was a
single gene in B. mori and there were at least six splice
variants, which were composed of 14 exons (Fig. 2A). In all the alternative splice
variants, the introns were in accordance with the GT-AG boundaries
rule (data not shown). BmPS1 (2,656 bp) was the longest
transcript, which comprised 13 exons (exon 2-14) and contained a
153 nt 5′-UTR, a 1,440 nt ORF encoding a putative protein of 479
amino acids and a 1,063 nt 3′-UTR (Fig. 1). In addition, three uORFs were
identified in the 5′UTRs of BmPS1-6. Among them, BmPS1 possessed
uORF1 and uORF3, BmPS2/3/4 possessed uORF2 and uORF3 and BmPS5/6
did not possess any uORFs (Figs. 1
and 2B). The alternative splicing
forms of BmPS2/4/6 has a 33 bp deletion (Fig. 2C).
Sequence alignment and phylogenetic
analysis
The six transcripts encoded four BmPS isoforms,
termed BmPS-A (BmPS1/3), BmPS-B
(BmPS2/4), BmPS-C (BmPS5) and
BmPS-D (BmPS6), which consisted of 479, 468, 463 and
452 amino acid residues, respectively. Alignment of the amino acid
sequences of the BmPSs with those from mammals and insects
indicated a high level of conservation (Fig. 3A). BmPS-A exhibited ~44% sequence
identity to hPS1 and hPS2, ~51% to DPS and ~40% to sel-12. In the
TMDs, BmPS shared sequence identities of 70% with the human PSs,
including the two catalytic aspartate residues (Fig. 3A). Due to the sequence identities
in the functional regions of the PS proteins, it was hypothesized
that the BmPSs belong to the PS family. In addition, similarly to
DPS, one of the BmPS alternative splicing events occurs in the loop
region, which is located between TMD6 and TMD7 and had a markedly
low level of conservation between species (Fig. 3A). The primers BmPS F1 and BmPS R1
were used to amplify the BmPS in the brains of the Dazao strain,
and it was found that there was a 3-amino acid residue insertion in
the loop region compared with the R13Q strain (Fig. 3A).
 | Figure 3Amino acid sequence alignments and
phylogenetic analysis of BmPSs and their homologues. (A)
Full-length amino acid sequence comparison of PSs from Bombyx
mori (BmPS-A, BmPS-B, BmPS-C and
BmPS-D were from the R13Q strain, and Dazao-BmPS was
from the Dazao strain), Homo sapiens [accession nos.
NP_000012.1 (hPS1) and NP_000483.2 (hPS2)], Drosophila
melanogaster [accession no. NP_524184.1 (DPS)] and
Caenorhabditis elegans [accession no. NP_508175.1 (sel-12)].
Identical amino acid residues are indicated in black. The hPSs
transmembrane domains are marked by a single line and the two
conserved catalytic aspartate residues and 20 amino acid residues
of hPS1/hPS2 mutated in patients with familial Alzheimer's disease
are indicated by stars and dots, respectively. The difference in
amino acid sequences in BmPS between strains R13Q and Dazao
is indicated by a hollow box. (B) Phylogenetic tree of PSs, in
addition to the species presented in A, of Tribolium
castaneum (accession no. XP_967139.2), Apis mellifera
(accession no. XP_396620.4), Drosophila simulans (accession
no. XP_002085655.1), Anopheles gambiae (accession no.
XP_311942.3), Culex quinquefasciatus (accession no.
XP_001851522.1), Mus musculus (accession nos. NP_032969.1
and NP_001122077.1), Rattus norvegicus (accession nos.
NP_062036.2 and NP_112349.2), Gallus gallus (accession nos.
NP_989494.1 and NP_989633.1), Xenopus laevis (accession nos.
NP_00104023.1 and NP_001081211.1), Danio rerio (accession
nos. NP_571099.1 and NP_571589.2), Caenorhabditis elegans
(accession nos. NP_508175.1, NP_491328.1, CAP31133.1, NP_492095.1
and CAP30997.1), Schistosoma japonicum (accession no.
CAX74629.1) and Schistosoma mansoni (accession no.
ABS30416.1). The length of each branch represents the distance
between each protein and the common ancestor of that protein and
its neighbor. The phylogenic tree was constructed using MEGA5.0
software and the neighbor-joining method. BmPS, Bombyx
mori presenilin. PS, presenilin; TMD, transmembrane domain. |
A phylogenetic tree was constructed to analyze the
evolutionary associations of BmPS with PSs found in other insects,
nematodes, amphibians, reptiles, avian and mammalian/primate
species (Fig. 3B). The clustering
pattern confirmed that BmPS was grouped in the lineage of other
insects (red flour beetle, honey bee, fruit fly and mosquito), with
a high bootstrap value, the highest similarity with the PS of
Tribolium castaneum, whereas avian species (chicken),
amphibians (frog), teleosts (zebrafish) and mammals (human and rat)
grouped into distinct lineages.
Expression patterns of the B. mori PS
gene
In order to determine whether the alternative
splicing event in the loop region exhibited tissue-specific
expression, the RT-PCR experiments with primers F4 and R5,
detecting exon 11 in the ORF, were examined in different tissues of
5th instar silkworm larvae. The two bands corresponding to the
alternative splicing of BmPS1/3/5 and
BmPS2/4/6 were detected in the silkworm
tissues. The expression of BmPS2/4/6 was more
abundant than that of BmPS1/3/5; however, the
ratio of BmPS1/3/5 to
BmPS2/4/6 was 2.2-2.4 in the brain, fat body,
silk gland and testes, and 2.7-3.0 in the prothoracic gland, the
midgut, malpi-ghian tubules and the ovaries (Fig. 4). This result indicated that
BmPS and its alternative splicing are expressed in all of
the tissues assessed in the present study.
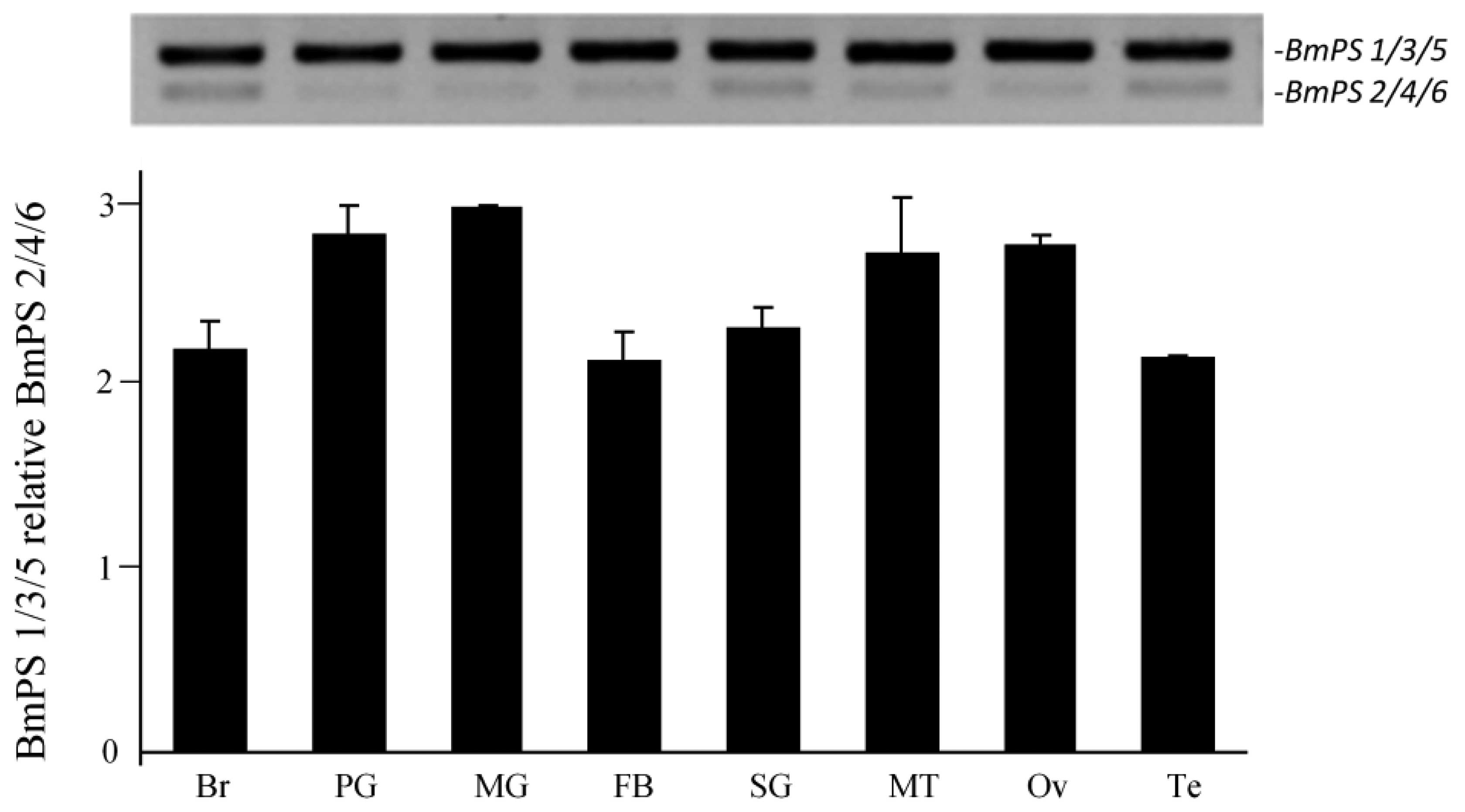 | Figure 4RT-PCR analysis of BmPS splice
variants in tissues of 3rd- and 5th-day instar larvae of Bombyx
mori (R13Q strain). BmPS1/3/5 and
BmPS2/4/6 were detected in the Br, PG, MG, FB,
SG, MT, Ov and Te, and the quantification of the RT-PCR results
produced by gel imaging system is shown. Data are presented as the
mean ± standard error; 3 independent experiments. L, larval; P,
pupal; A, adult; BmPS, Bombyx mori presenilin;
RT-PCR, reverse transcription quantitative polymerase chain
reaction; Br, brain; PG, prothoracic gland; MG, midgut; FB, fat
body; SG, silk gland; MT, malpighian tubules; Ov, ovaries; TE,
testis. |
As PS is important in neurodegenerative disorders in
mammals and the developmental processes in Drosophila, the
temporal expression of BmPS in the brain was investigated in
the present study using RT-qPCR from the larval, pupal and adult
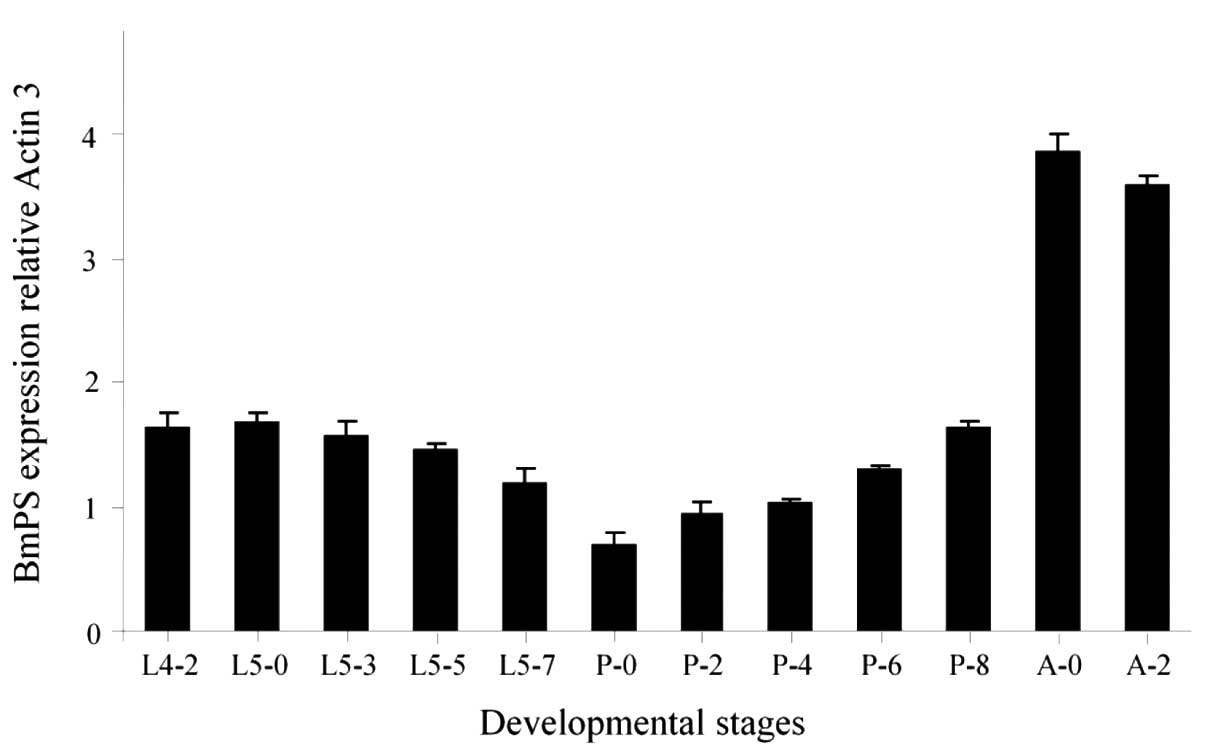
stages (Fig. 5). The expression
levels of BmPS were constant in the 4th instar and the
initial stage of the 5th instar, and slowly declined between day 0
of the 5th instar and day 0 of the pupal stage. However, during the
development of the pupal stage, the BmPS expression levels
persistently increased and suddenly markedly improved in the adult
stage. This dynamic expression pattern in the brain suggested that
BmPS may have roles during B. mori development,
particularly in the adult stages.
Discussion
PSs are highly conserved transmembrane proteins,
which are aspartyl proteases with catalytic residues localized
inside the lipid bilayer on TMD6 and TMD7 (2). There are several motifs conserved
evolutionarily across numerous species in the two amino acid
sequences. The most important motifs include YD and GxGD motifs,
which contain catalytic aspartyl residues or an endoplasmic
reticulum-retention sequence (35–37).
The topology of PSs has established a view of PS as containing nine
TMDs (38–40). In metazoans, PS is synthesized as a
holoprotein and subsequently undergoes autocatalytic
endoproteolysis between TMD6 and TMD7. The conserved two catalytic
aspartate residues (Asp257 in TM6 and Asp385
in TM7 of hPS1) are essential for the released amino- and
carboxy-terminal fragments (NTF and CTF, respectively), which are
associated with conformational changes in the complex and function
together as a heterodimer (41–43).
In the present study, a B. mori homologue of the PS gene was
cloned and characterized. The deduced amino acid sequence of BmPS
is equally homologous to DPS (~51% sequence identity), hPS1/hPS2
(~44% sequence identity), the highest similarity with insect PS,
particularly T. castaneum PS. A total of nine TMDs and the
hydrophilic domains at the beginning of the TM6-TM7 loop domain
exhibited >70% identity to DPS and hPSs. Two highly-conserved
aspartate residues of PS, Asp257 in TM6 and
Asp385 in TM7 of hPS1, corresponded to Asp265
and Asp397 in BmPS-A. Furthermore, the mutations in
hPS1/hPS2, present in human familial AD, have been reported (for a
list of the mutations, see molgen.ua.ac.be/ADMutations) (4,44,45).
Of note, among the 20 amino acid residues of hPS1/hPS2 mutated in
patients with familial AD, 17 are identical and three represent
conserved substitutions in BmPS, indicating that the conserved
mutations may be important for functions and/or structures of PS.
This result suggested that the BmPS cloned in the present study is
a member of the PS family and may be relevant to the pathogenesis
of AD.
Human PSs have two homologues, hPS1 and hPS2,
encoded by two genes on chromsome 14 and chromosome 1,
respectively. However, invertebrate PSs, such as Drosophila
PS, derived from a single gene, exhibit several splice variants
(46,47). In B. mori, PS has at least
six alternative splice sites corresponding to four isoforms, which
belong to a single gene. Of note, three uORFs were identified in
the 5′-UTR of BmPS. uORFs in the 5′-UTR of an mRNA
transcript are able to modulate the translational efficiency of the
major ORF (mORF), and disruptions of a functional uORF are
associated with human genetic diseases (48–51).
In Drosophila, ~40% of transcripts contain uORFs (52). DPS genes also have three
uORFs in the 5′-UTR of mRNA (data from FlyBase http://flybase.org/). An upstream ORF has been
observed to be involved in Sex lethal to inhibit msl-2
translation during Drosophila gender determination (53). Although uORF-mediated translational
control has been rarely reported in Bombyx mori, the
translational efficiency of the mORF of BmPS may be
modulated by the uORFs, highlighting the potential physiological
implications of BmPS.
Although PSs share extensive homology throughout
their length, the N-terminal domain and the large hydrophilic loop
facing the cytosolic compartment between TM6 and TM7 are highly
divergent between PS1 and PS2, and are poorly conserved across
species. However, numerous PS-linked mutations have been mapped to
specific amino acid residues within the loop region, which are
highly conserved between species (54,55),
the filamin (an actin-binding protein) and the methyltransferases
(an evolutionary conserved protein in a variety of species) in
Drosophlia interact with the loop region of either human or
Drosophila PSs (56,57),
suggesting the functional importance of the loop. Analyzing the
BmPS splice variants, the alternative splicing occurs only in the
large hydrophilic loop region between TMD6 and TMD7, as exon 11.
Similarly, the alternative splice variants of DPS predominantly
occur in this region (58). Of
note, comparison of BmPS between two silkworm strains R13Q and
Dazao showed that the three amino acid residues were inserted in
this region in Dazao. It has been established that the strains R13Q
and Dazao exhibit morphological and physiological differences,
including body stripe, size, voltinism and life span. Although it
remains to be elucidated whether the alternative splicing in the
TM6-TM7 loop of BmPS has an important function, there remains
reason to hypothesize that BmPS may be associated with certain
physiological processes in B. mori.
PS has been implicated in numerous molecular
processes, including Notch signaling, the metabolism of β-catenin
and calcium homeostasis (2,19,59–62).
mRNAs of PSs are ubiquitously detected in a number of human
and mouse tissues, including the brain, heart, kidney and muscle
(63). In Drosophila, DPS
is expressed at different developmental stages, exhibiting higher
levels of expression in adults than larvae, and is predominantly
expressed in the central nervous system (46,47,58).
In the present study, BmPS and its splice variants were
observed to be expressed in the silkworm tissues, including the
brain, fat body, silk gland, testis, prothoracic gland, midgut,
malpighian tubules and ovaries, and the expression of BmPS
in the developmental brain was higher in the adult stage. This
dynamic expression pattern of BmPS suggested that it may
have multiple roles during Bombyx development.
While vertebrate models provide a closer match to
humans from an evolutionary perspective, invertebrates, including
the nematode worm C. elegans and the fruit fly D.
melanogaster, have been used extensively in transgenic
experiments and classical genetic analyses to investigate the
function of AD-relevant genes and to analyze the underlying
mechanisms of AD (16,17,64).
The Notch signaling pathway was initially linked to AD in C.
elegans and D. melanogaster, contributing greatly to the
current understanding of the PS-mediated pathogenesis of AD.
Although the silkworm B. mori, as a model organism, has not
been previously utilized in the study of AD, the present study on
B. mori PSs may provide valuable insight into the mechanisms
of silkworm development regulation and the pathogenesis of AD.
Acknowledgments
The authors would like to thank Professor Yong-hua
Ji for encouragement and helpful advice on the manuscript. This
study was supported by grants from the National Science Foundation
of China (grant no. 31172147) and the Shanghai Committee of Science
and Technology (grant nos. 10JC14162002 and 10DZ2271800).
References
|
1
|
Tanzi RE: A brief history of Alzheimer's
disease gene discovery. J Alzheimers Dis. 33(Suppl 1): S5–S13.
2013.
|
|
2
|
Zhang S, Zhang M, Cai F and Song W:
Biological function of Presenilin and its role in AD pathogenesis.
Transl Neurodegener. 2:152013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guerreiro RJ, Baquero M, Blesa R, Boada M,
Bras JM, Bullido MJ, Calado A, Crook R, Ferreira C, Frank A, et al:
Genetic screening of Alzheimer's disease genes in Iberian and
African samples yields novel mutations in presenilins and APP.
Neurobiol Aging. 31:725–731. 2010. View Article : Google Scholar :
|
|
4
|
Theuns J, Del-Favero J, Dermaut B, van
Duijn CM, Backhovens H, Van den Broeck MV, Serneels S, Corsmit E,
Broeckhoven CV and Cruts M: Genetic variability in the regulatory
region of presenilin1 associated with risk for Alzheimer's disease
and variable expression. Hum Mol Genet. 9:325–331. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bekris LM, Yu CE, Bird TD and Tsuang DW:
Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol.
23:213–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schon EA and Area-Gomez E: Is Alzheimer's
disease a disorder of mitochondria-associated membranes? J
Alzheimers Dis. 20(Suppl 2): S281–S292. 2010.PubMed/NCBI
|
|
7
|
Sherrington R, Rogaev EI, Liang Y, Rogaeva
EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al:
Cloning of a gene bearing missense mutations in early onset
familial Alzheimer's disease. Nature. 375:754–760. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rogaev EI, Sherrington R, Rogaeva EA,
Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et
al: Familial Alzheimer's disease in kindreds with missense
mutations in a novel gene on chromosome 1 related to the
Alzheimer's Disease type 3 gene. Nature. 376:775–778. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dries DR and Yu G: Assembly, maturation
and trafficking of the gamma-secretase complex in Alzheimer's
disease. Curr Alzheimer Res. 5:132–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De Strooper B: Aph-1, pen-2 and nicastrin
with presenilin generate an active gamma-secretase complex. Neuron.
38:9–12. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bergmans BA and De Strooper B:
Gamma-secretases: From cell biology to therapeutic strategies.
Lancet Neurol. 9:215–226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Querfurth HW and LaFerla FM: Alzheimer's
Disease. N Engl J Med. 362:329–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baki L, Shioi J, Wen P, Shao Z, Schwarzman
A, Gama-Sosa M, Neve R and Robakis NK: PS1 activates PI3K thus
inhibiting GSK-3 activity and tau over phosphorylation: effects of
FAD mutations. EMBO J. 23:2586–2596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mhatre SD, Paddock BE, Saunders AJ and
Marenda DR: Invertebrate models of Alzheimer's disease. J
Alzheimers Dis. 33:3–16. 2013.
|
|
15
|
Bonner JM and Boulianne GL: Drosophila as
a model to study age-related neurodegenerative disorders:
Alzheimer's disease. Exp Gerontol. 46:335–339. 2011. View Article : Google Scholar
|
|
16
|
Moloney A, Sattelle DB, Lomas DA and
Crowther DC: Alzheimer's disease: Insights from Drosophila
melanogaster models. Trends Biochem Sci. 35:228–235. 2010.
View Article : Google Scholar :
|
|
17
|
Link CD: Invertebrate models of
Alzheimer's disease. Genes Brain Behav. 4:147–156. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Levitan D and Greenwald I: Facilitation of
lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182
Alzheimer's disease gene. Nature. 377:351–354. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Struhl G and Greenwald I: Presenilin is
required for activity and nuclear access of notch in Drosophila.
Nature. 398:522–525. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ye YH, Lukinova N and Fortini ME:
Neurogenic phenotypes and altered notch processing in Drosophila
Presenilin mutants. Nature. 398:525–529. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Y, Livne-Bar I, Zhou L and Boulianne
GL: Drosophila presenilin is required for neuronal differentiation
and affects notch subcellular localization and signaling. J
Neurosci. 19:8435–8442. 1999.PubMed/NCBI
|
|
22
|
Mahoney MB, Parks AL, Ruddy DA, Tiong SY,
Esengil H, Phan AC, Philandrinos P, Winter CG, Chatterjee R,
Huppert K, et al: Presenilin-based genetic screens in Drosophila
melano- gaster identify novel notch pathway modifiers. Genetics.
172:2309–2324. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van de Hoef DL, Hughes J, Livne-Bar I,
Garza D, Konsolaki M and Boulianne GL: Identifying genes that
interact with Drosophila presenilin and amyloid precursor protein.
Genesis. 47:246–260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye YH and Fortini ME: Apoptotic activities
of wild-type and Alzheimer's disease-related mutant presenilins in
Drosophila melanogaster. J Cell Biol. 146:1351–1364. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Presente A, Boyles RS, Serway CN, de Belle
JS and Andres AJ: Notch is required for long-term memory in
Drosophila. Proc Natl Acad Sci USA. 101:1764–1768. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Knight D, Iliadi K, Charlton MP, Atwood HL
and Boulianne GL: Presynaptic plasticity and associative learning
are impaired in a Drosophila presenilin null mutant. Dev Neurobiol.
67:1598–1613. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu Y, Lv Y, Ye Y, Wang Y, Hong Y, Fortini
ME, Zhong Y and Xie Z: A role for presenilin in post-stress
regulation: Effects of presenilin mutations on Ca2+ currents in
Drosophila. FASEB J. 21:2368–2378. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Michno K, Knight D, Campussano JM, van de
Hoef D and Boulianne GL: Intracellular calcium deficits in
Drosophila cholinergic neurons expressing wild type or FAD-mutant
presenilin. PLoS One. 4:e69042009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McBride SM, Choi CH, Schoenfeld BP, Bell
AJ, Liebelt DA, Ferreiro D, Choi RJ, Hinchey P, Kollaros M,
Terlizzi AM, et al: Pharmacological and genetic reversal of
age-dependent cognitive deficits attributable to decreased
presenilin function. J Neurosci. 30:9510–9522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ray WJ, Yao M, Nowotny P, Mumm J, Zhang
WJ, Wu JY, Kopan R and Goate AM: Evidence for a physical
interaction between presenilin and Notch. Proc Natl Acad Sci USA.
96:3263–3268. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Roncarati R, Sestan N, Scheinfeld MH,
Berechid BE, Lopez PA, Meucci O, McGlade JC, Rakic P and D'Adamio
L: The gamma-secretase-generated intracellular domain of
beta-amyloid precursor protein binds Numb and inhibits Notch
signaling. Proc Natl Acad Sci USA. 99:7102–7107. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Woo HN, Park JS, Gwon AR, Arumugam TV and
Jo DG: Alzheimer's disease and notch signaling. Biochem Biophys Res
Commun. 390:1093–1097. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsumoto Y, Sumiya E, Sugita T and
Sekimizu K: An invertebrate hyperglycemic model for the
identification of anti-diabetic drugs. PLoS One. 6:e182922011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sekimizu N, Paudel A and Hamamoto H:
Animal welfare and use of silkworm as a model animal. Drug Discov
Ther. 6:226–229. 2012.PubMed/NCBI
|
|
35
|
Kaether C, Capell A, Edbauer D, Winkler E,
Novak B, Steiner H and Haass C: The presenilin C-terminus is
required for ER- retention, nicastrin-binding and gamma-secretase
activity. EMBO J. 23:4738–4748. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wolfe MS: Gamma-secretase in biology and
medicine. Semin Cell Dev Biol. 20:219–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fassler M, Li X and Kaether C: Polar
transmembrane-based amino acids in presenilin 1 are involved in
endoplasmic reticulum localization, Pen2 protein binding and
γ-secretase complex stabilization. J Biol Chem. 286:38390–38396.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sato C, Takagi S, Tomita T and Iwatsubo T:
The C-terminal PAL motif and transmembrane domain 9 of presenilin 1
are involved in the formation of the catalytic pore of the
gamma-secretase. J Neurosci. 28:6264–6271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tolia A, Horré K and De Strooper B:
Transmembrane domain 9 of presenilin determines the dynamic
conformation of the catalytic site of gamma-secretase. J Biol Chem.
283:19793–19803. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sobhanifar S, Schneider B, Löhr F,
Gottstein D, Ikeya T, Mlynarczyk K, Pulawski W, Ghoshdastider U,
Kolinski M, Filipek S, et al: Structural investigation of the
C-terminal catalytic fragment of presenilin 1. Proc Natl Acad Sci
USA. 107:9644–9649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shirotani K, Edbauer D, Capell A, Schmitz
J, Steiner H and Haass C: Gamma-secretase activity is associated
with a conformational change of nicastrin. J Biol Chem.
278:16474–16477. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Deng Y, Tarassishin L, Kallhoff V,
Peethumnongsin E, Wu L, Li YM and Zheng H: Deletion of presenilin 1
hydrophilic loop sequence leads to impaired gamma-secretase
activity and exacerbated amyloid pathology. J Neurosci.
26:3845–3854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McCarthy JV, Twomey C and Wujek P:
Presenilin-dependent regulated intramembrane proteolysis and
gamma-secretase activity. Cell Mol Life Sci. 66:1534–1555. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Boteva K, Vitek M, Mitsuda H, de Silva H,
Xu PT, Small G and Gilbert JR: Mutation analysis of presenillin 1
gene in Alzheimer's disease. Lancet. 347:130–131. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chapman J, Asherov A, Wang N, Treves TA,
Korczyn AD and Goldfarb LG: Familial Alzheimer's disease associated
with S182 codon 286 mutation. Lancet. 346:10401995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Boulianne GL, Livne-Bar I, Humphreys JM,
Liang Y, Lin C, Rogaev E and St George-Hyslop P: Cloning and
characterization of the Drosophila presenilin homologue.
Neuroreport. 8:1025–1029. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hong CS and Koo EH: Isolation and
characterization of Drosophila presenilin homolog. Neuroreport.
8:665–668. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Calvo SE, Pagliarini DJ and Mootha VK:
Upstream open reading frames cause widespread reduction of protein
expression and are polymorphic among humans. Proc Natl Acad Sci
USA. 106:7507–7512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Morris DR and Geballe AP: Upstream open
reading frames as regulators of mRNA translation. Mol Cell Biol.
20:8635–8642. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Scheper GC, van der Knaap MS and Proud CG:
Translation matters: Protein synthesis defects in inherited
disease. Nat Rev Genet. 8:711–723. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wen Y, Liu Y, Xu Y, Zhao Y, Hua R, Wang K,
Sun M, Li Y, Yang S, Zhang XJ, et al: Loss-of-function mutations of
an inhibitory upstream ORF in the human hairless transcript cause
Marie Unna hereditary hypotrichosis. Nat Genet. 41:228–233. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hayden CA and Bosco G: Comparative genomic
analysis of novel conserved peptide upstream open reading frames in
Drosophila melanogaster and other dipteran species. BMC Genomics.
9:612008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yao P and Fox PL: Sex lethal and upstream
ORFs: A bait-and-trap system for ribosomes. Genome Biol.
12:1212011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tolia A, Chávez-Gutiérrez L and De
Strooper B: Contribution of presenilin transmembrane domains 6 and
7 to a water-containing cavity in the gamma-secretase complex. J
Biol Chem. 281:27633–27642. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wanngren J, Frånberg J, Svensson AI,
Laudon H, Olsson F, Winblad B, Liu F, Näslund J, Lundkvist J and
Karlström H: The large hydrophilic loop of presenilin 1 is
important for regulating gamma-secretase complex assembly and
dictating the amyloid beta peptide (Abeta) Profile without
affecting Notch processing. J Biol Chem. 285:8527–8536. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Guo YQ, Zhang SX, Sokol N, Cooley L and
Boulianne GL: Physical and genetic interaction of filamin with
presenilin in Drosophila. J Cell Sci. 113:3499–3508.
2000.PubMed/NCBI
|
|
57
|
Zhang SX, Gua Y and Boulianne GL:
Identification of a novel family of putative methyltransferases
that interact with human and Drosophila presenilins. Gene.
280:135–144. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nowotny P, Gorski SM, Han SW, Philips K,
Ray WJ, Nowotny V, Jones CJ, Clark RF, Cagan RL and Goate AM:
Posttranslational modification and plasma membrane localization of
the Drosophila melanogaster presenilin. Mol Cell Neurosci.
15:88–98. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Song W, Nadeau P, Yuan M, Yang X, Shen J
and Yankner BA: Proteolytic release and nuclear translocation of
Notch-1 are induced by presenilin-1 and impaired by pathogenic
presenilin-1 mutations. Proc Natl Acad Sci USA. 96:6959–6963. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kang DE, Soriano S, Frosch MP, Collins T,
Naruse S, Sisodia SS, Leibowitz G, Levine F and Koo EH: Presenilin
1 facilitates the constitutive turnover of β-catenin: Differential
activity of Alzheimer's disease-linked PS1 mutants in
β-catenin-signaling pathway. J Neurosci. 19:4229–4237.
1999.PubMed/NCBI
|
|
61
|
Stutzmann GE, Caccamo A, LaFerla FM and
Parker I: Dysregulated IP3 signaling in cortical neurons of
knock-in mice expressing an Alzheimer's-linked mutation in
presenilin1 results in exaggerated Ca2+ signals and altered
membrane excitability. J Neurosci. 24:508–513. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Soriano S, Kang DE, Fu M, Pestell R,
Chevallier N, Zheng H and Koo EH: Presenilin 1 negatively regulates
beta-catenin/T cell factor/lymphoid enhancer factor-1 signaling
independently of beta-amyloid precursor protein and notch
processing. J Cell Biol. 152:785–794. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee MK, Slunt HH, Martin LJ, Thinakaran G,
Kim G, Gandy SE, Seeger M, Koo E, Price DL and Sisodia SS:
Expression of presenilin 1 and 2 (PS1 and PS2) in human and murine
tissues. J Neurosci. 16:7513–7525. 1996.PubMed/NCBI
|
|
64
|
Dillin A and Cohen E: Ageing and protein
aggregation-mediated disorders: from invertebrates to mammals.
Philos Trans R Soc Lond B Biol Sci. 366:94–98. 2011. View Article : Google Scholar :
|