Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common type of cancer worldwide, with ~750,000 newly diagnosed
patients each year (1). Most HCCs
develop from either a viral hepatitis infection (hepatitis B or C)
or cirrhosis (2). Clinical
treatment of HCC has remained challenging due to of a lack of
effective chemotherapy and clearly defined end-points for clinical
protocols (3,4). The poor outcome of HCC patients has
largely been attributed to intrahepatic or distant metastasis. The
underlying mechanisms responsible for metastatic spread of HCC have
remained to be fully elucidated. Therefore, it is important to
explore the underlying mechanisms of HCC metastasis and develop
more effective treatments. RNA interference (RNAi) is a powerful
post-transcriptional gene-silencing mechanism in which homologous
RNA sequences are transfected into the cells by use of synthetic
small interfering RNA (siRNAs), or plasmid or viral vectors
encoding for short hairpin RNAs, inhibiting the expression of a
particular gene (5,6). RNAi-based approaches have recently
entered clinical trials (7).
Hence, RNAi has been regarded as a novel promising therapy for
HCC.
High-mobility group box 1 (HMGB1), a member of the
high-mobility group protein family, was originally characterized as
a non-histone, nuclear DNA-binding protein (8,9). It
has been implicated in several disease states, including sepsis,
arthritis, ischemia-re-perfusion injury and cancer (10,11).
It also functions as an extracellular signaling molecule during
inflammation, cell differentiation, cell migration and tumor
metastasis (12). Previous studies
have also reported that HMGB1 signaling proceeds via Toll-like
receptors (TLRs), including TLR-2 and TLR-4, and the receptor for
advanced glycation end-products (RAGE) to induce the nuclear
translocation of nuclear factor κB, resulting in an enhanced
production of pro-inflammatory cytokines, including tumor necrosis
factor α and interleukin 1b (13–15).
Blockade of the RAGE-HMGB1 interaction was shown to suppress tumor
growth and metastasis (16). In
addition, constant release of HMGB1 as a pro-inflammatory cytokine
from necrotic tumor cells can create a microenvironment similar to
that of chronic inflammation, a condition known to contribute to
the development of epithelial malignancies (17). Epithelial mesenchymal transition
(EMT) was shown to promote the migratory and invasive capacity of
HCC cells (18). EMT is a
critical, highly conserved process which controls cell
differentiation and embryonic development. Emerging evidence
revealed that EMT endows cancer cells with various malignant
characteristics, including increased mobility, invasiveness,
evasion of apoptosis and stem-like phenotypes (19). However, the association between
HMGB1 and EMT has remained elusive. Therefore, the present study
investigated whether inhibiting HMGB1 may attenuate EMT and thus
block the metastasis and invasion of HCC cells.
The present study evaluated HCC specimens with
regard to HMGB1 expression, and performed correlation analyses of
HMGB1 with clinicopathological parameters and survival rates in
order to determine the potential value of HMGB1 as an independent
prognostic factor in HCC. Furthermore, in vitro experiments
employing specific siRNAs targeting HMGB1 (HMGB1-siRNAs) were
performed in order to investigate the role of HMGB1 in cell
invasion and metastasis.
Materials and methods
Ethical review
All protocols were approved by the Xi'an Jiaotong
University Ethics Committee (Xi'an, China) according to the 1975
Declaration of Helsinki. Written informed consent was obtained from
each patient.
Clinical samples and cell lines
A total of 68 HCC patients, including 51 males and
17 females (range, 31–75 years; median, 54 years) who underwent
curative liver resection at the Department of Hepatobiliary
Surgery, First Affiliated Hospital of Medical College of Xi'an
Jiaotong University (Xi'an, China) from January 2010 to December
2010 were included in the present study. Patients had a median
follow-up time of 22.5 months. None of the patients received any
chemotherapy, radiotherapy or radiofrequency ablation prior to
surgery. HCC tissues and matched normal tumor-adjacent tissues
(>2 cm distance of the surgical margin) were collected and
immediately stored in 4% paraformaldehyde solution for
immunohistochemistry (IHC) or in liquid nitrogen for western blot
analysis. Data regarding the demographic and clinicopathological
characteristics were obtained from the medical records of the
patients. The human immortalized liver cell line LO2 and four HCC
cell lines, HepG2, Hep3B, Huh7 and MHCC97H, were obtained from the
Institute of Biochemistry and Cell Biology, Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in complete
Dulbecco's modified Eagle medium (DMEM; Mediatech, Manassas, VA,
USA) containing 10% fetal bovine serum (FBS; Gibco-BRL, Invitrogen
Life Technologies, Carlsbad, CA, USA) in a humidified 5%
CO2 incubator at 37°C. Cells in the logarithmic growth
phase were used in the in vitro assays.
IHC analysis
IHC was performed on paraformaldehyde-fixed paraffin
sections. Samples were subjected to immunostaining with HMGB1
polyclonal antibody (cat. no. 3935; dilution, 1:100) obtained from
Cell Signaling Technology, Inc., (Danvers, MA, USA) followed by
streptavidin peroxidase-conjugated secondary antibody (1:2,000;
cat. no. pv-9001; Zhongshan Goldenbridge Biotechnology Co., Ltd.,
Beijing, China). The staining for the HMGB1 protein was
semi-quantitatively evaluated by multiplying the staining intensity
with the percentage of positive liver cells. The staining intensity
was expressed as four grades: 0, None; 1, weak; 2, moderate; and 3,
strong. The percentage of HMGB1-positive liver cells was expressed
using the following grades: 0, <5%; 1, 6–25%; 2, 26–50%; 3,
51–75%; and 4, >75%. Ten independent high-magnification felds
(x400) were evaluated for each section using a laser scanning
confocal microscope (TCS2 SP5; Leica Microsystems, Wetzlar, G
ermany), and the average scores of ten fields represented the final
results. Sections with a total score of >1 were defined as
exhibiting positive staining for the HMGB1 protein.
siRNA transfection
The specific siRNA against HMGB1 (pre-designed
siRNA; Table I) and scramble siRNA
were synthesized by GenePharma Co., Ltd. (Shanghai, China). Huh7
cells were seeded in six-well plates at the concentration of
2×105/well and divided into two groups: The HMGB1 siRNA
group and the control siRNA group. Huh7 or MHCC97H cells were
transfected with 150 pmol siRNA (HMGB1 siRNA or control siRNA)
using 5 µl Lipofectamine 2000 (Invitrogen Life Technologies)
according to the manufacturer's instructions, and cultured in a
humidified 5% CO2 incubator at 37°C. Serum-free medium
was changed to complete medium 6 h after transfection. Cells were
harvested 48 h after transfection and used for further
experiments.
 | Table ISequences of HMGB1-specifc siRNAs. |
Table I
Sequences of HMGB1-specifc siRNAs.
| Name | Sequence (5′-3′) |
|---|
| HMGB1-siRNA-1 | S:
CCCGUUAUGAAAGAGAAAUTT |
| AS:
AUUUCUCUUUCAUAACGGGTT |
| HMGB1-siRNA-2 | S:
GGGAGGAGCAUAAGAAGAATT |
| AS:
UUCUUCUUAUGCUCCUCCCTT |
| HMGB1-siRNA-3 | S:
CUGGGAGAGAUGUGGAAUATT |
| AS:
UAUUCCACAUCUCUCCCAGTT |
| Scramble | S:
UUCUCCGAACGUGUCACGUTT |
| AS:
ACGUGACACGUUCGGAGAATT |
| β-actin | S:
GGGAAATCGTGCGTGACAT |
| AS:
CTGGAAGGTGGACAGCGAG |
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from HCC tissues and HCC
cells using TRIzol reagent (Invitrogen Life Technologies) according
to the manufacturer's instructions. The first-strand cDNA was
synthesized using the Revertid™ First Strand cDNA Synthesis kit
(Fermentas, Burlington, ON, Canada). cDNA (2 µl) obtained
from each sample was amplified and quantified by real-time PCR
using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus; Takara
Bio Inc., Shiga, Japan). PCR amplification was conducted using an
ABI Prism 7300 Sequence Detection system (Applied Biosystems Life
Technologies, Foster City, CA, USA) under the following conditions:
94°C for 3 min, followed by 35 cycles of 94°C for 30 sec and 60°C
for 30 sec. The human β-actin gene served as an internal control
gene to ensure that an equal amount of mRNA was analyzed from each
sample. The primers were synthesized by GenePharma Co., Ltd.
(Shanghai, China) and the sequences are shown in Table I. Relative gene expression was
calculated as 2−ΔCt[ΔCt=Ct(target gene) - Ct(β -actin). Three
independent experimental replicates were performed.
Western blot analysis
Western blotting was conducted as described in our
previous study (20). HMGB1 (cat.
no. SC-393423; 1:1,000) and β-actin (cat. no. SC-130301; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA; dilution, 1:5,000) antibodies
were incubated at 4°C overnight for western blot analysis.
Secondary horseradish peroxidase-conjugated goat anti-mouse
antibody (Bio-Rad Laboratories, Inc., Hercules, CA, USA) were used
at a 1:5,000 dilution at room temperature for 1 h, visualized using
an Enhanced Chemiluminescence Reagent (Millipore, Billerica, MA,
USA) and analyzed using Quantity One 1-D analysis software (Bio-Rad
Laboratories, Inc.).
Wound healing assay
The confluent cells were seeded in six-well plates
and allowed to form cell monolayers overnight. A sterile
200-µl plastic pipette tip was used to create a line-shaped
wound across the cell monolayer, and the detached cells were
removed by rinsing with phosphate-buffered saline (PBS). Cells were
cultured in serum-free DMEM in a humidified 5% CO2
incubator at 37°C for 48 h, and images were captured with a
phase-contrast microscope (COIC4365; Nikon, Tokyo, Japan).
Transwell assay
The 8-µM pore-size Transwell inserts (Nalge
Nunc, Penfield, NY, USA) were coated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA) at 1 mg/ml on the inner layer. Huh7 cells
were transfected with siRNA for 48 h and then seeded at
2.5×105 cells/ml in 200 µl into the upper
chambers of the Transwell inserts. 750 µl DMEM containing
10% FBS was added to the lower chambers, and the Transwell plates
were incubated for 24 h Cells were fixed in 4% paraformaldehyde for
2 min and then permeabilized with methanol for 20 min at room
temperature. The cells on the inner layer were gently removed with
a cotton swab and the adherent cells on the lower surface of the
insert were stained with 0.3% crystal violet dye (Bio-Rad, Inc.)
for 15 min. The filters were washed with PBS and images were
captured. Invaded cells on the lower surface were counted under a
light microscope.
Statistical analysis
Values are expressed as the mean ± standard
deviation. The SPSS statistical package for Windows version 13
(SPSS, Chicago, IL, USA) was used for Pearson's χ2 test
and the multivariate Cox regression analysis. Two-tailed Student's
t-test, Kaplan-Meier plots, log-rank test or analysis of
variance were used to evaluate statistical significance using
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference and all experiments were repeated
independently, three times.
Results
Clinical significance of elevated HMGB1
expression in HCC tissues
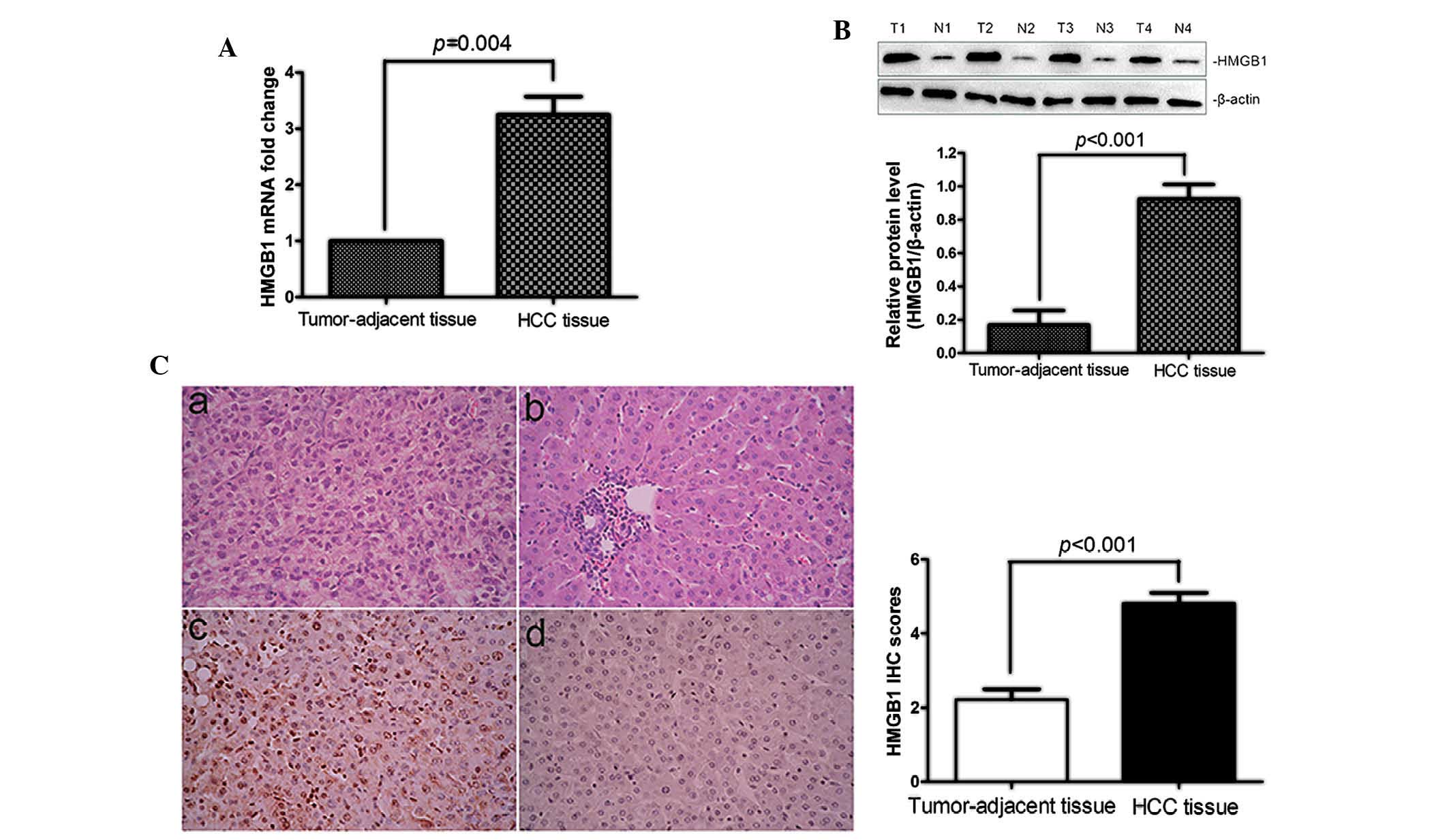
The expression of HMGB1 in HCC tissues and normal
adjacent tissues was at the mRNA and protein level by RT-qPCR, IHC
and western blot analysis. As shown in Fig. 1A and B, HMGB1 mRNA and protein
expression was significantly upregulated in HCC tissues compared
with that in their matched normal tumor-adjacent tissues (P=0.004
and P<0.001, respectively). IHC analysis revealed the presence
of HMGB1 in the cytoplasm and nuclei of HCC cells in the tissue
samples. The IHC results were further confirmed by western blot
analysis (P<0.001; Fig. 1C).
Clinical association analysis by the Pearson χ2 test
revealed that elevated HMGB1 expression in HCC tissues was
significantly associated with large tumor size (≥5 cm; P=0.001),
high histological grade (Edmondson-Steiner grades III and IV;
P=0.001) and advanced tumor stage [tumor-node-metastasis (TNM)
stages III and IV; P=0.003) (Table
II). These results demonstrated elevated expression of HMGB1 in
HCC and its correlation with poor clinicopathological features in
HCC.
 | Table IIClinical correlation of HMGB1
expression in HCC (n=68). |
Table II
Clinical correlation of HMGB1
expression in HCC (n=68).
| Clinical
parameter | Cases (n) | Expression status
| P-value |
|---|
| Positive (n=46) | Negative (n=22) |
|---|
| Age (years) | | | | 0.43 |
| <65 | 23 | 17 | 6 | |
| ≥65 | 45 | 29 | 16 | |
| Gender | | | | 0.765 |
| Male | 51 | 34 | 17 | |
| Female | 17 | 12 | 5 | |
| Tumor size (cm) | | | | 0.001 |
| <5 | 22 | 9 | 13 | |
| ≥5 | 46 | 37 | 9 | |
| Tumor number | | | | 0.212 |
| Solitary | 39 | 24 | 15 | |
| Multiple | 29 | 22 | 7 | |
| Edmondson | | | | |
| I+II | 27 | 12 | 15 | 0.001a |
| III+IV | 41 | 34 | 7 | |
| TNM stage | | | | 0.003a |
| I–II | 38 | 20 | 18 | |
| III–IV | 30 | 26 | 4 | |
| Capsular
infltration | | | | 0.508 |
| Present | 49 | 32 | 17 | |
| Absent | 19 | 14 | 5 | |
| Vascular
invasion | | | | 0.120 |
| Present | 34 | 20 | 14 | |
| Absent | 34 | 26 | 8 | |
| AFP | | | | 0.988 |
| <400 ng/ml | 31 | 21 | 10 | |
| ≥400 ng/ml | 37 | 25 | 12 | |
| HBsAg | | | | 0.946 |
| Positive | 59 | 40 | 19 | |
| Negative | 9 | 6 | 3 | |
Positive expression of HMGB1 correlates
with a decreased three-year survival of HCC patients
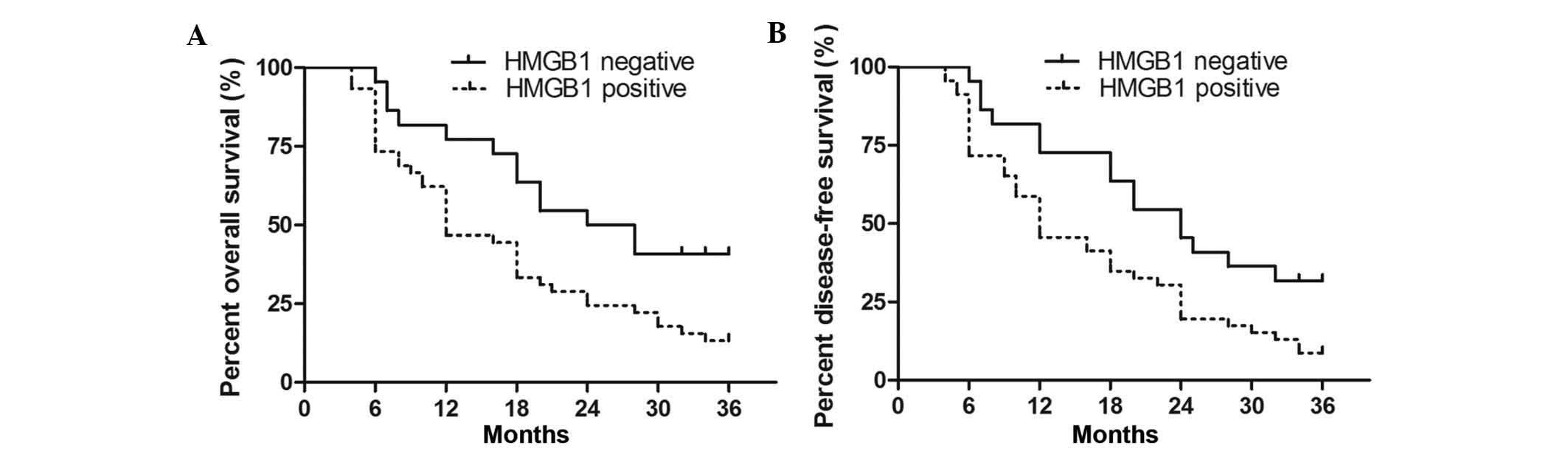
The present study next investigated whether HMGB1
was a prognostic factor for HCC patients. The overall survival (OS)
and disease-free survival (DFS) time of the 68 cases were
determined at the three-year follow-up, correlated with positive
and negative HMGB1 expression and depicted in Kaplan-Meier survival
curves. A significant correlation was detected between positive
expression of HMGB1 and shorter OS and DFS (P=0.004 and P=0.002,
respectively; Fig. 2). The OS and
DFS median survival time in the HMGB1-positive expression group was
shorter than that in the HMGB1-negative expression group (12 vs. 24
months and 9 vs. 21 months, respectively). Multivariate analysis of
all of the significant clinical factors for OS and DFS indicated
that HMGB1-positive expression (P=0.021 and P=0.034, respectively;
Table III) was an independent
prognostic factor for HCC patients. Thus, HMGB1 may be a potential
biomarker for predicting the clinical outcomes of HCC.
 | Table IIIMultivariate Cox regression analysis
of three-year overall survival and disease-free survival of 68
hepatocellular carcinoma patients. |
Table III
Multivariate Cox regression analysis
of three-year overall survival and disease-free survival of 68
hepatocellular carcinoma patients.
| Variable | Overall survival
| P-value | Disease-free
survival
| P-value |
|---|
| HR | 95% CI | HR | 95% CI |
|---|
| HMGB1 | 3.018 | 1.121–7.427 | 0.021a | 2.528 | 1.126–4.832 | 0.034a |
| Edmondson
grade | 1.643 | 0.753–3.548 | 0.032a | 1.645 | 0.583–4.629 | 0.018a |
| TNM stage | 4.116 | 1.101–9.623 | 0.004a | 7.314 | 2.018–23.69 | 0.002a |
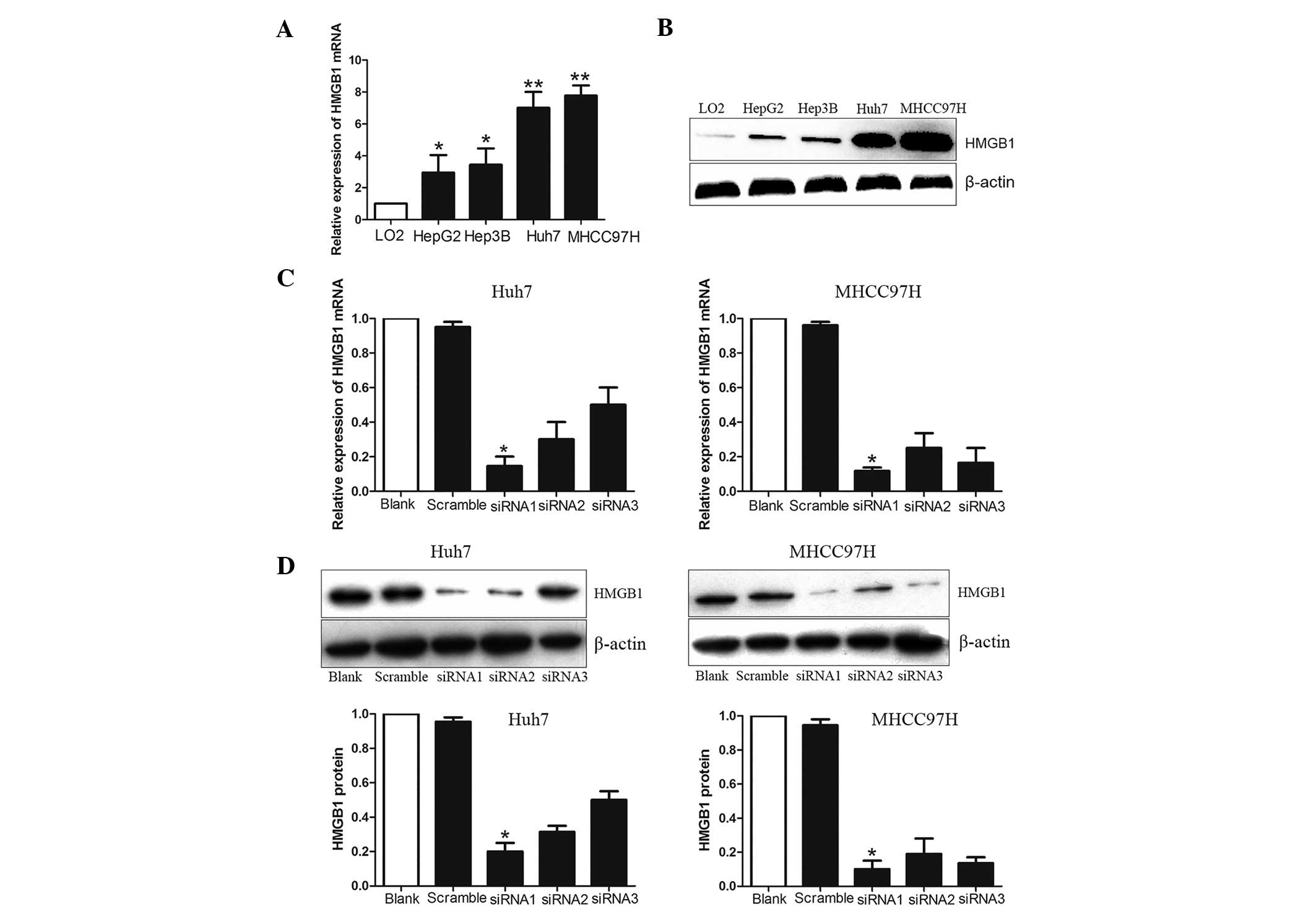
RNAi with HMGB1 expression in HCC
cells
To explore the effects of HMGB1 on the invasiveness
of HCC cells, the expression of HMGB1 was assessed in a panel of
HCC cell lines, which are known to exhibit an elevated invasive
behavior. The mRNA and protein expression levels of HMGB1 increased
progressively from healthy liver cells to HCC cells with low
metastatic potential and, finally, to HCC cells with high
metastatic potential (Fig. 3A and
B). To further examine the effect of HMGB1 on the motility and
invasiveness, the knockdown effect of siRNA was first minimized by
using three specific HMGB1-siRNAs. siRNAs1-3 or scramble siRNA were
transfected into Huh7 and MHCC97H cells, which exhibited the
highest expression of HMGB1 among the cell lines tested, using
Lipofectamine 2000. HMGB1 expression was evaluated by RT-qPCR and
western blot analyses. The results indicated that all of the
HMGB1-siRNAs significantly inhibited the mRNA and protein
expression of HMGB1 in Huh7 and MHCC97H cells (Fig. 3C–E). Inhibition was highest
(80–90%) for HMGB1-siRNA-1, which was therefore selected to be used
in the subsequent knockdown experiments.
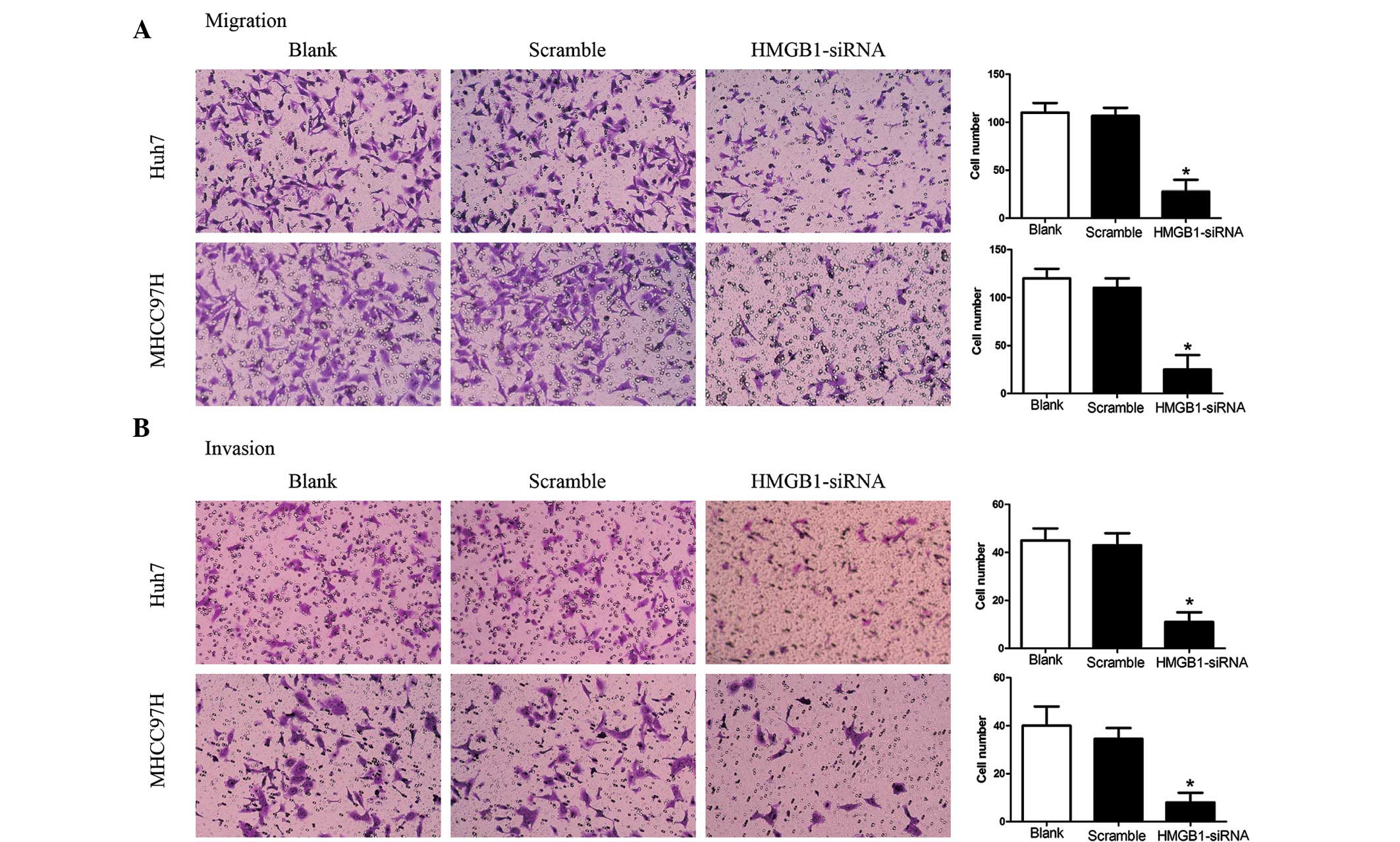
HMGB1-siRNA-1 inhibits the migration and
invasion of Huh7 and MHCC97H cells
As show in Fig. 4,
knockdown of HMGB1 in the highly invasive Huh7 and MHCC97H cells
decreased their migratory and invasive behaviors (Fig. 4A and B). A Transwell assay revealed
that the migration and invasion of cells in the
HMGB-siRNA-transfected group was inhibited compared with that in
the control groups (Huh7, P<0.01; MHCC97H, P<0.001). Cell
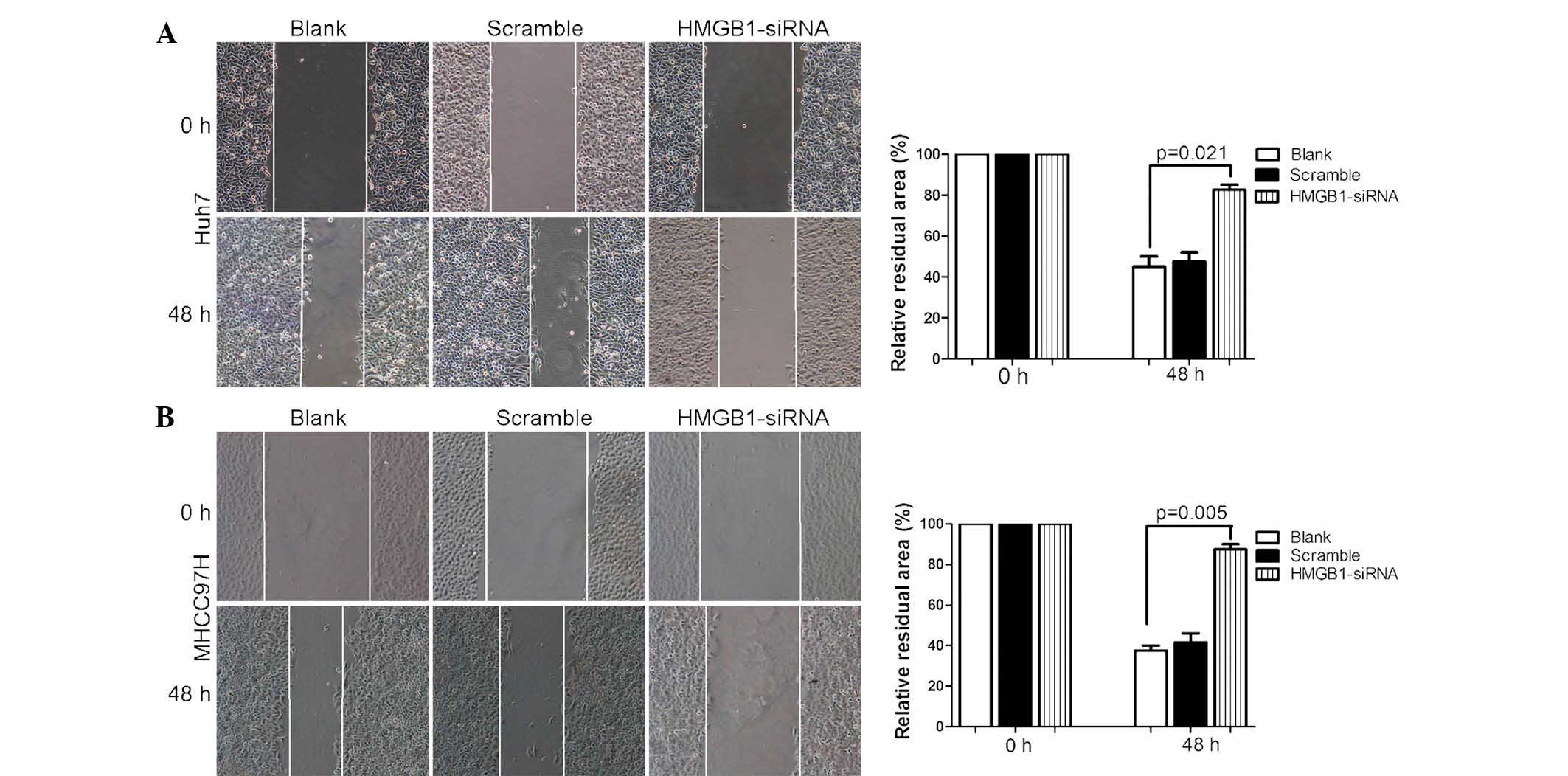
motility was also examined using a wound healing assay (Fig. 5). The results showed that HMGB1
knockdown significantly inhibited HCC cell migration and
invasion.
HMGB1 induces EM T in HCC cells
EMT has an impor tant role in metastasis, as it
induces tumor-associated epithelial cells to obtain mesenchymal
features, which leads to reduced cell-cell contact and increased
migratory ability (16). Thus, the
present study next investigated how HMGB1 regulates the migratory
and invasive phenotypes of HCC cells. The expression of EMT markers
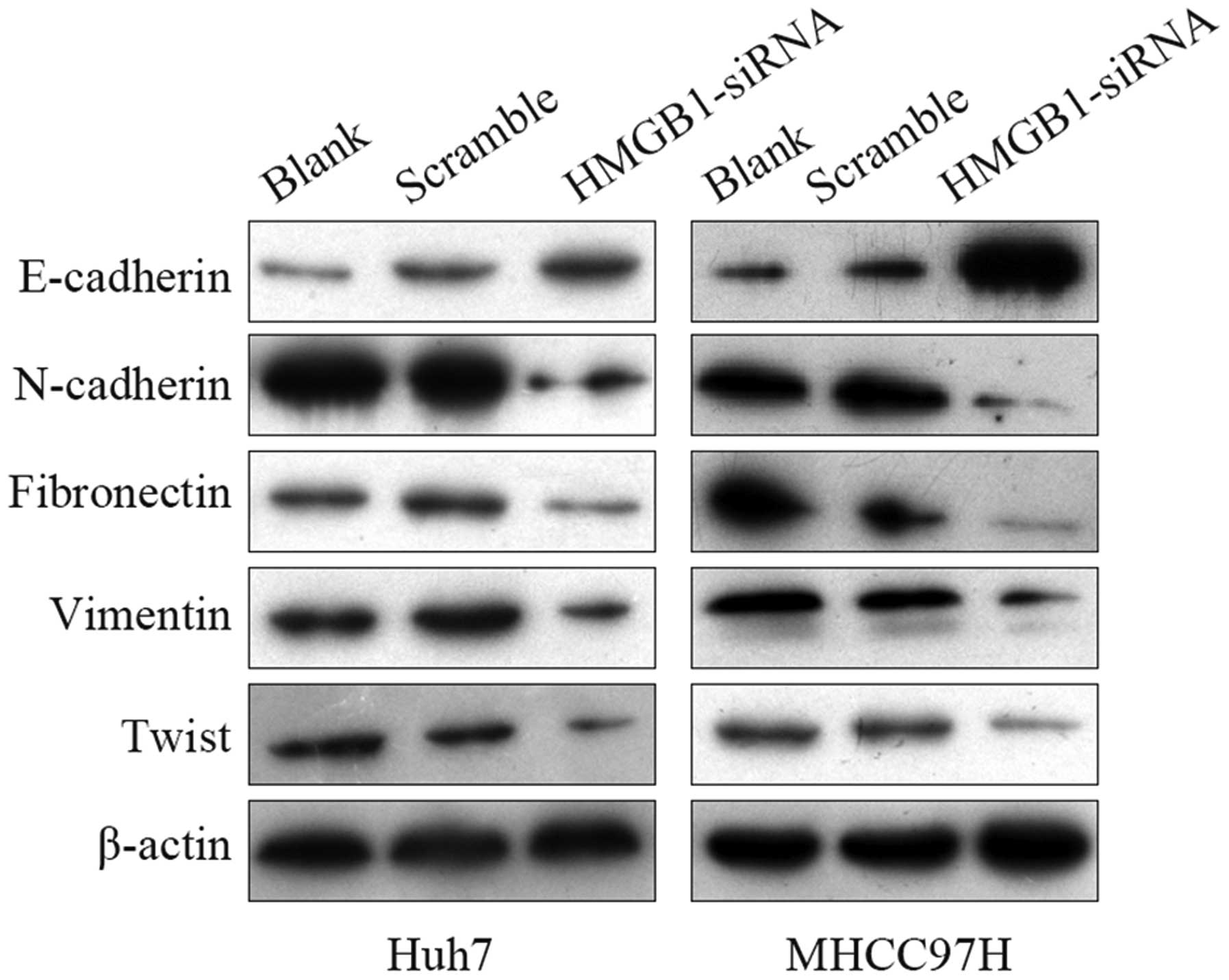
was evaluated by western blot analysis (Fig. 6). The results showed that knockdown
of HMGB1 in Huh7 and MHCC97H cells led to a marked increase in the
expression of epithelial marker E-cadherin and marked decreases in
the expression of mesenchymal markers N-cadherin, fibronectin,
vimentin and twist. These findings suggested that HMGB1 induced EMT
in HCC cells.
Discussion
HCC is one of the leading causes of
cancer-associated mortality worldwide (17). Surgical treatment, including
hepatic resection and liver transplantation at the early stage, is
the only curative therapy for HCC patients. Unfortunately, due to
the high frequency of intrahepatic and extrahepatic metastasis,
most HCC patients are diagnosed at the advanced stage and are not
eligible for hepatic resection, which leads to a poor prognosis for
HCC. Hence, it is urgently required to explore the molecular
mechanism of HCC progression and discover novel HCC markers and
targeting agents.
The present study demonstrated that HMGB1 was
signifcantly upregulated in HCC tissues compared with that in
tumor-adjacent tissues. Furthermore, elevated HMGB1 expression was
significantly associated with large tumor size (>5 cm), high
Edmonson-Steiner classification (stages III and IV) and advanced
TNM stage (III and IV). Kaplan-Meier analysis revealed that HMGB1
expression was correlated with a poorer survival of HCC patients
after hepatectomy. Furthermore, multivariate Cox regression
analysis showed that HMGB1 was an independent factor for predicting
the three-year overall and disease-free survival in HCC patients.
These results showed that the status of HMGB1 was critical for the
prognosis of HCC patients.
Furthermore, the present study determined the
functional signifcance of HMGB1 overexpression in HCC cell lines.
The HMGB1 content was low in normal hepatocytes, increased in
non-invasive and primary HCC cells, and was highest in invasive HCC
cell lines. The expression of HMGB1 was in parallel with the
invasiveness and motility of the respective cell lines, indicating
that HMGB1 may be used as a potential marker for HCC. The in
vitro experiments demonstrated, for the first time, to the best
of our knowledge, that HMGB1 knockdown by a specifc siRNA inhibited
cell migration and invasion of Huh7 and MHCC97H cells. As migration
and invasion represent the cytological basis of tumor metastasis
and the specifc inhibition of HMGB1 expression reduced HCC
migration and invasion in vitro, HMGB1 may be utilized as a
therapeutic target.
Metastasis is regarded as the main cause of
mortality in cancer patients (21). For patients with HCC, the poor
prognosis is mainly attributed to intrahepatic and distant
metastasis. The metastatic process includes a series of
interdependent events, including cancer cell proliferation,
migration and invasion (22). As a
nuclear protein, HMGB1 has dual roles as a chromatin structural
protein and a cytokine (23). It
is involved in a variety of processes of cancer progression,
including cell proliferation, angiogenesis, invasion and metastasis
(11,24). The present study demonstrated that
knockdown of HMGB1 inhibited the migration and invasion of Huh7 and
MHCC97H cells, which was in agreement with previous studies on
other malignancies (11,25), indicating that HMGB1 is involved in
tumor metastasis.
In the present study, wester n blot a nalysis of
EMT-associate d proteins showed that the EMT was reduced after
knockdown of HMGB1. This indicated that HMGB1 mediated metastasis
of HCC through regulating the EMT. Therefore, HMGB1 may be an ideal
therapeutic target for metastatic HCC.
In conclusion, the results of the present study
revealed that RNAi-mediated knockdown of HMGB1 effectively
inhibited the migration and invasion of Huh7 and MHCC97H cells. The
expression of EMT markers was inhibited by HMGB1-siRNA-1,
indicating that HMGB1 is involved in the development of metastasis.
Therefore, HMGB1 may serve as a potential target for the treatment
of metastatic HCC. Furthermore, HMGB1 was demonstrated to be an
independent prognostic factor for HCC. Further study is required to
clarify the mechanisms by which HMGB1 is involved in the initiation
and progression of HCC as well as the formation of metastasis via
stimulating EMT.
Acknowledgments
This study was supported by grants from the National
Natural Scientifc Foundation of China (nos. 81272645 and 81072052
to Qingguang Liu), Key Science and Technology Fund of Shaanxi
Province (no. 2010K01-131 to Tao Song) and the Funds for Creative
Research sponsored by The First Affliated Hospital of Xi'an
Jiaotong University (14YB10 to Zhikui Liu).
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith MW, Yue ZN, Korth MJ, Do HA, Boix L,
Fausto N, Bruix J, Carithers RL Jr and Katze MG: Hepatitis C virus
and liver disease: Global transcriptional profling and
identifcation of potential markers. Hepatology. 38:1458–1467. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Di Bisceglie AM, Bruix J,
Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M,
Talwalkar J, et al: Design and endpoints of clinical trials in
hepatocellular carcinoma. J Natl Cancer Inst. 100:698–711. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Talwalkar JA and Gores GJ: Diagnosis and
staging of hepatocellular carcinoma. Gastroenterology. 127(5 Suppl
1): S126–S132. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim DH and Rossi JJ: Strategies for
silencing human disease using RNA interference. Nat Rev Genet.
8:173–184. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gartel AL and Kandel ES: RNA interference
in cancer. Biomol Eng. 23:17–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castanotto D and Rossi JJ: The promises
and pitfalls of RNA-interference-based therapeutics. Nature.
457:426–433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Javaherian K, Liu JF and Wang JC:
Nonhistone proteins HMG1 and HMG2 change the DNA helical structure.
Science. 199:1345–1346. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lotze MT and Tracey KJ: High-mobility
group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal.
Nat Rev Immunol. 5:331–42. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sims GP, Rowe DC, Rietdijk ST, Herbst R
and Coyle AJ: HMGB1 and RAGE in infammation and cancer. Annu Rev
Immunol. 28:367–88. 2010. View Article : Google Scholar
|
|
11
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1 and cancer. Biochim Biophys Acta.
1799:131–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kostova N, Zlateva S, Ugrinova I and
Pasheva E: The expression of HMGB1 protein and its receptor RAGE in
human malignant tumors. Mol Cell Biochem. 337:251–58. 2010.
View Article : Google Scholar
|
|
13
|
Park JS, Gamboni-Robertson F, He Q,
Svetkauskaite D, Kim JY, Strassheim D, Sohn JW, Yamada S, Maruyama
I, Banerjee A, et al: High mobility group box 1 protein interacts
with multiple Toll-like receptors. Am J Physiol Cell Physiol.
290:C917–924. 2006. View Article : Google Scholar
|
|
14
|
Yu M, Wang H, Ding A, Golenbock DT, Latz
E, Czura CJ, Fenton MJ, Tracey KJ and Yang H: HMGB1 signals through
toll-like receptor (TLR) 4 and TLR2. Shock. 26:174–79. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wolfson RK, Chiang ET and Garcia JG: HMGB1
induces human lung endothelial cell cytoskeletal rearrangement and
barrier disruption. Microvasc Res. 81:189–97. 2011. View Article : Google Scholar
|
|
16
|
Larocca C, Cohen JR, Fernando RI, Huang B,
Hamilton DH and Palena C: An autocrine loop between TGF-β1 and the
transcription factor brachyury controls the transition of human
carcinoma cells into a mesenchymal phenotype. Mol Cancer Ther.
12:1805–815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang JD, Harmsen WS, Slettedahl SW,
Chaiteerakij R, Enders FT, Therneau TM, Orsini L, Kim WR and
Roberts LR: Factors that affect risk for hepatocellular carcinoma
and effects of surveillance. Clin Gastroenterol Hepatol.
9:617–623.e1. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
El-Nahas AM: Plasticity of kidney cells:
Role in kidney remodeling and scarring. Kidney Int. 64:1553–563.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sato M, Shames DS and Hasegawa Y: Emerging
evidence of epithelial-to-mesenchymal transition in lung
carcinogenesis. Respirology. 17:1048–059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Dou C, Jia Y, Li Q, Zheng X, Yai Y,
Liu Q and Song T: RIG-I suppresses the migration and invasion of
hepatocellular carcinoma cells by regulating MMP9. Int J Oncol.
46:1710–720. 2015.PubMed/NCBI
|
|
21
|
Rudmik LR and Magliocco AM: Molecular
mechanisms of hepatic metastasis in colorectal cancer. J Surg
Oncol. 92:347–59. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumar S and Weaver VM: Mechanics,
malignancy and metastasis: The force journey of a tumor cell.
Cancer Metastasis Rev. 28:113–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Naghavi MH, Nowak P, Andersson J,
Sönnerborg A, Yang H, Tracey KJ and Vahlne A: Intracellular high
mobility group B1 protein (HMGB1) represses HIV-1 LTR-directed
transcription in a promoter-and cell-specifc manner. Virology.
314:179–189. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ueda M, Takahashi Y, Shinden Y, Sakimura
S, Hirata H, Uchi R, Takano Y, Kurashige J, Iguchi T, Eguchi H, et
al: Prognostic signifcance of high mobility group box 1 (HMGB1)
expression in patients with colorectal cancer. Anticancer Res.
34:5357–5362. 2014.PubMed/NCBI
|
|
25
|
Li LC, Gao J and Li J: Emerging role of
HMGB1 in fbrotic diseases. J Cell Mol Med. 18:2331–2339. 2014.
View Article : Google Scholar : PubMed/NCBI
|