Introduction
The liver X receptor (LXR) is a nuclear receptor
which has an essential role in the regulation of metabolism and
inflammation through inducing and blocking target genes (1,2). Two
isoforms of LXR have been identified and are named as LXR-α and
LXR-β. LXR-β is expressed ubiquitously in all the tissues examined,
while LXR-α is more restricted, being most highly expressed in the
liver, adipose tissue, intestine, lung, macrophages, spleen and
kidney (3). A previous study has
shown that stimulating Kupffer cells (KCs) by lipopolysaccharide
(LPS) decreases the mRNA and protein expression of LXR-α (4). LXR-α has recently drawn attention due
to its anti-inflammatory properties; however, the underlying
molecular mechanism has remained elusive. The neuron-derived orphan
nuclear receptor-1 (NOR-1) functions as an early-response gene
which regulates key cellular processes, including proliferation,
differentiation and survival (5,6). It
has been shown to function in the regulation of important genes
involved in metabolic homeostasis in the liver and adipocytes
(7–9). It has been reported that the
overexpression of NOR-1 in macrophages reduced the synthesis of
inflammatory cytokines and chemokines (10). Kumar et al (8) discovered that the two nuclear
receptors LXR and NOR-1 have an interdependent regulatory
association in adipocytes and that LXR regulates gene transcription
of NOR-1 in adipocytes.
KCs, constituting 80–90% of the tissue-resident
macrophages present in the body, are increasingly recognized as
important modulators which control the liver's response to injury
and repair. They have a crucial role in liver homeostasis as well
as in initiation, maintenance and outcome of liver inflammation
(11). Upon activation, KCs
release various substances, including cytokines, nitric oxide and
reactive oxygen species. Therefore, KCs are intimately involved in
the liver's response to infection, toxins, ischemia, resection and
other stresses (12). KCs have
been implicated in the pathogenesis of various liver diseases,
including viral hepatitis, steatohepatitis, alcoholic liver
disease, intrahepatic cholestasis, activation or rejection of the
liver during liver transplantation and liver fibrosis (13).
However, to date, the association between the two
nuclear receptors in KCs during LPS-induced inflammation has
remained elusive, and the specific mechanisms are subject to
present research. It has been reported that T0901317 is the most
potent LXR-α ligand (2). The
present study hypothesized that upregulation of LXR-α by T0901317
may suppress LPS-induced inflammation by controlling the expression
of NOR-1 in KCs. The present study used T0901317 to activate LXR-α
as well as NOR-1 knockdown in order to identify a novel role of
LXR-α in regulating LPS-induced inflammation by affecting NOR-1
expression in KCs.
Materials and methods
Animals and experimental protocol
Male C57BL/6 mice (6–8 weeks) were obtained from the
laboratory animal research center of Chongqing Medical University
(Chongqing, China). All animals (n=60) were housed under specific
pathogen-free condition and allowed free access to sterile water
and food. The animals received humane care in compliance with the
institution's guidelines, as outlined in the guide for the care and
use of laboratory animals prepared by the National Academy of
Sciences (Beijing, China). The present study was approved by the
ethics committee of Chongqing Medical University (Chongqing,
China).
In vitro experiments
Primary KCs were isolated from mouse livers
according to a previously described procedure (14). Briefly, a three-step procedure was
applied to isolate KCs in sufficient number and purity from murine
liver, including enzymatic tissue treatment, gradient
centrifugation, and selective adherence. KCs were cultured in
24-well plates at a density of 1×106 cells in Dulbecco's
modified Eagle's medium (DMEM) (Hyclone, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Hyclone) and antibiotics
(100 U/ml penicillin G and 100 mg/ml streptomycin sulphate;
Beyotime Institute of Biotechnology, Haimen, China) at 37°C in the
presence of 5% CO2. The viability of KCs (>90%) was
determined by trypan blue exclusion (Beyotime Institute of
Biotechnology). KCs were observed under a light microscope
(TL-800C; Nikon Corporation, Tokyo, Japan). KCs were randomly
divided into six groups (six wells per group): Control (CON),
T0901317 (T09; Sigma-Aldrich, St. Louis, MO, USA), LPS (LPS; 10 mg;
Sigma-Aldrich), LPS + T0901317 (LPS + T09), LPS + T0901317 + NOR-1
small hairpin (sh) RNA (LPS + T09 + shRNA) and NOR-1 shRNA (shRNA)
groups.
Cultured KCs were transfected with a NOR-1 shRNA
plasmid (sc-38843-SH; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) using a transfection reagent [NOR-1 shRNA Plasmid (m); Santa
Cruz Biotechnology, Inc.] according to a previously described shRNA
transfection protocol (15), and
manufacturer's instructions. In brief, KCs were seeded into a
six-well tissue culture plate at a density of 1×106
cells/well and grown overnight in antibiotic-free normal growth
medium prior to transfection. Cells were washed twice with 2 ml
shRNA transfection medium (Santa Cruz Biotechnology, Inc.). 200
µl of the DNA/shRNA plasmid transfection reagent/shRNA (425
µg/ml) complexes were added to each well and the cells were
cultured at 37°C in a CO2 incubator for 5 h. Following
incubation, 1 ml growth medium containing twice the amount of serum
and antibiotics of normal medium was added to each well and the
cells were cultured for 24 h. The transfection efficiency of NOR-1
shRNA was measured by reverse transcription quantitative polymerase
chain reaction (RT-qPCR).
24 h after transfection, the T09, LPS + T09 and LPS
+ T09 + shRNA groups were incubated with DMEM containing T0901317
(1 µM), and the other three groups were incubated in DMEM in
the absence of T0901317 for 6 h. Fresh medium was added to the CON,
T09 and shRNA groups, while fresh medium containing LPS (10 ng/ml)
was added to the LPS, LPS + T09 and LPS + T09 + shRNA groups,
followed by an additional 6-h incubation. The total incubation time
in each group was 30 h, which did not include the incubation time
with LPS/control (6 h).
F4/80 staining
KCs were identified by immunofluorescence using
fluorescein isothiocyanate-conjuugated monoclonal anti-F4/80
antibody [BM8] (Abcam, Cambridge, UK), according to the
manufacturer's instructions. Briefly, KCs were seeded in 24-well
chamber slides at a density of 2×104 cells/ml. The cells
were then fixed with 4% paraformaldehyde for 10 min and incubated
in 1% bovine serum albumin (Sigma-Aldrich)/10% normal goat serum
(Beyotime Institute of Biotechnology)/0.3M glycine (Beyotime
Institute of Biotechnology) in 0.1% phosphate-buffered saline
(PBS)-Tween for 1 h to permeabilise the cells and block
non-specific protein-protein interactions. The cells were then
incubated with the anti-F4/50 antibody (1µg/ml), overnight
at 4°C. Propidium iodide (Wuhan Boster Biological Technology Ltd.,
Wuhan, China) was used to stain the cell nuclei (red) at a
concentration of 1.43 µM. After washing with PBS, the cells
were covered with mounting medium (Wuhan Boster Biological
Technology Ltd.) and the slides were viewed by laser scanning
confocal microscopy (C2 Plus; Nikon Corporation).
RNA analysis
To investigate the association between LXR-α and
NOR-1 during inflammation, the mRNA expression of LXR-α and NOR-1
in KCs was analyzed by RT-qPCR. Briefly, total RNA was extracted
from cell samples of the experimental groups using
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer's instructions and
quantified via the 260 nm/280 nm absorption ratio of RNA samples
(Model 722; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each
total RNA sample was stored at −70°C. Equivalent amounts of
particle RNA extracted from each sample served as the template for
cDNA synthesis using the PrimeScript RT reagent kit with a gDNA
Eraser (cat no. DRR037A; Takara Bio Inc., Otsu, Japan). The
amplification of sample cDNA was monitored using the SsoFast
EvaGreen supermix (Bio-Rad Laboratories, Inc.) and the DNA
fluorescent dye SYBR Green (Molecular Probes, Eugene, OR, USA).
β-actin served as an endogenous normalization control (reference
gene). PCR cycling conditions were as follows: Initial denaturation
at 94°C for 2 min, 30 cycles at 94°C for 20 sec, 60°C for 30 sec
and 72°C for 30 sec. The thermal cycler was from Bio-Rad
Laboratories (S1000™ Thermal Cycler with 96-Well Fast Reaction
module). All PCR products were separated by electrophoresis on 2%
agarose gels. The DNA bands of LXR-α or NOR-1 were normalized to
the corresponding β-actin band using the Bio-Image analysis system
(Gel Doc2000; Bio-Rad Laboratories, Inc.), and the ratio of LXR-α
or NOR-1 to β-actin was used to assess the relative mRNA
expression. Bio-Rad CFX Manager software (version 2.0; Bio-Rad
Labotatories, Inc.) was used for data analysis, and the relative
mRNA expression levels were calculated using the Vandesompele
method (16). All values are
expressed as the ratio of the RNA concentration of the target
amplicon to the RNA concentration of the housekeeping gene
(β-actin), to take into account differences betweensample RNA
concentrations.
Sequences of specific LXR-α, NOR-1 and β-actin
primers were as follows: LXR-α forward,
5′-GAGACATCTCGGAGGTACAACCC-3′ and reverse,
5′-AGCAAGGCAAACTCGGCATC-3′; NOR-1 forward,
5′-TGTCTCAGTGTCGGGATGGTT-3′and reverse,
5′-TCCTGTTGTAGTGGGCTCTTTG-3′; and β-actin forward,
5′-TGCCCATCTACGAGGGCTAT-3′ and reverse,
5′-TGATGTCACGCACGATTTCC-3′.
Western blot analysis
The protein expression of LXR-α and NOR-1 in KCs was
detected by western blot analysis. Protein extracts were obtained
by homogenizing samples in a cell lysis buffer containing 20 mM
4-(2-hydroxyethyl)-1-piper-azineethanesulfonic acid, 0.42 mM NaCl,
15 mM MgCl2, 25% glycerol, 0.2 mM EDTA, 0.5 mM
phenylmethylsulfonyl fluoride and 0.5 mM dithiothreitol. The
protein concentration was determined using a Bradford Assay kit
(Bio-Rad Laboratories, Inc.). Equal amounts of protein samples were
each separated on 12% Tris-HCl gels (Bio-Rad Laboratories, Inc.) by
electrophoresis and transferred onto a polyvinylidene fluoride
membrane (Merck Millipore, Darmstadt, Germany). The membrane was
then blocked for 1 h with 5% nonfat dry milk and incubated with
goat anti-mouse LXR-α (1/1,000 dilution; SC1202; Santa Cruz
Biotechnology, Inc.) and rabbit anti-mouse NOR-1 (1/1,000 dilution;
SC133840; Santa Cruz Biotechnology, Inc.) polyclonal antibodies at
4°C overnight. The membrane was washed and incubated for 1 h at
room temperature with a 1/1,200-dilution of rabbit anti-goat (cat.
no. BA1006) or goat anti-rabbit (cat. no. BA1003) immunoglobulin G
(Wuhan Boster Biological Technology Ltd.) for 1 h. Finally, the
membrane was developed using an Enhanced Chemiluminescence
Detection kit (Pierce Biotechnology, Inc., Thermo Fisher
Scientific, Waltham, MA, USA) and exposed to an autoradiographic
film (Eastman Kodak, Rochester, NY, USA). The relative amounts of
LXR-α and NOR-1 protein were quantified via the relative density of
the protein bands using the Gel Doc 2000 image analysis system
(Bio-Rad Laboratories, Inc.).
ELISA analysis
ELISA was used to measure the protein levels of
tumor necrosis factor (TNF)-α (TNF-α Mouse ELISA kit; cat. no.
ab100747; Abcam) and interleukin (IL)-10 (IL-10 Mouse ELISA kit;
cat. no. ab46103; Abcam) in the supernatant of cultured cells
according to the manufacturer's instructions.
Statistical analysis
Values are expressed as the mean ± standard
deviation, and comparisons between values were performed by
analysis of variance using the statistical package SPSS version
18.0 (International Business Machines, Armonk, NY, USA). The
comparison mean values was performed using the Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference between values.
Results
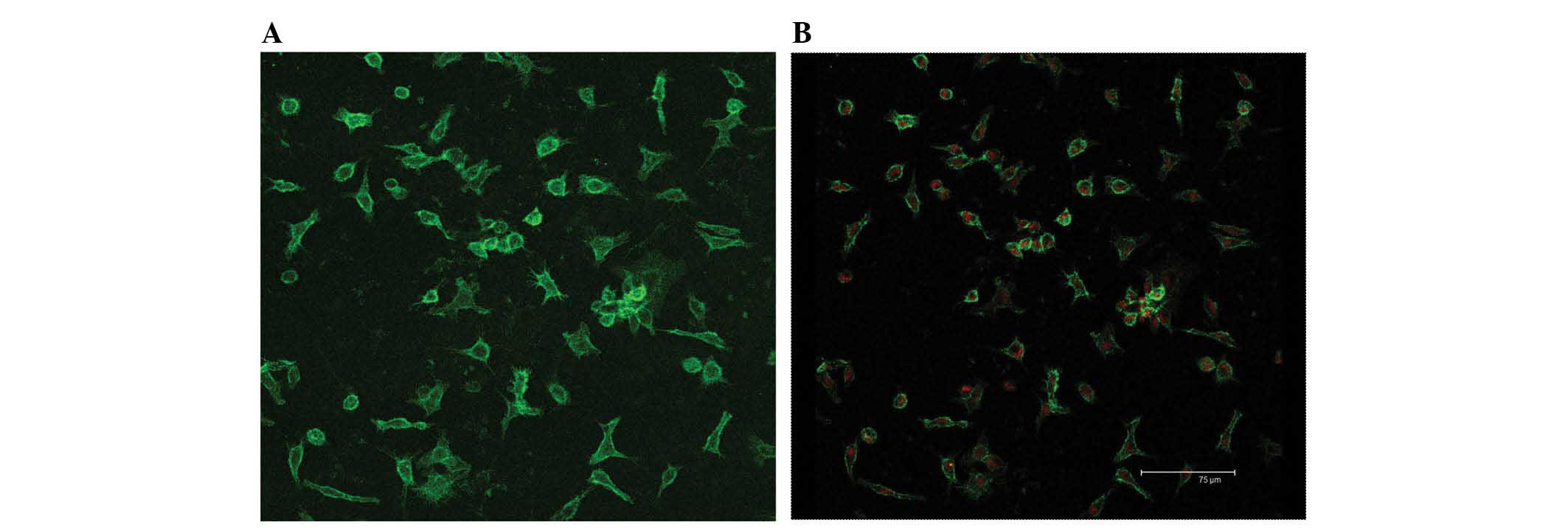
Identification of KCs
The viability of KCs determined by trypan blue
staining after isolation was ≥98%. The purity of KC fractions was
determined by morphological observation in combination with F4/80
staining (Fig. 1). The freshly
isolated cells had a round shape and were 8–10 µm in
diameter as indicated by light microscopy observation. 2 h later,
most of the cells adhered to the wall of plastic flask and
exhibited a fried-egg shape. With increasing culture time, the
cells became larger and more prominent, showing typical
morphological features of macrophages with irregular shape,
transparent cytoplasm and kidney-like nuclei (Fig. 2).
NOR-1 mRNA expression is partially
controlled by LXR-α under inflammatory and non-inflammatory
conditions
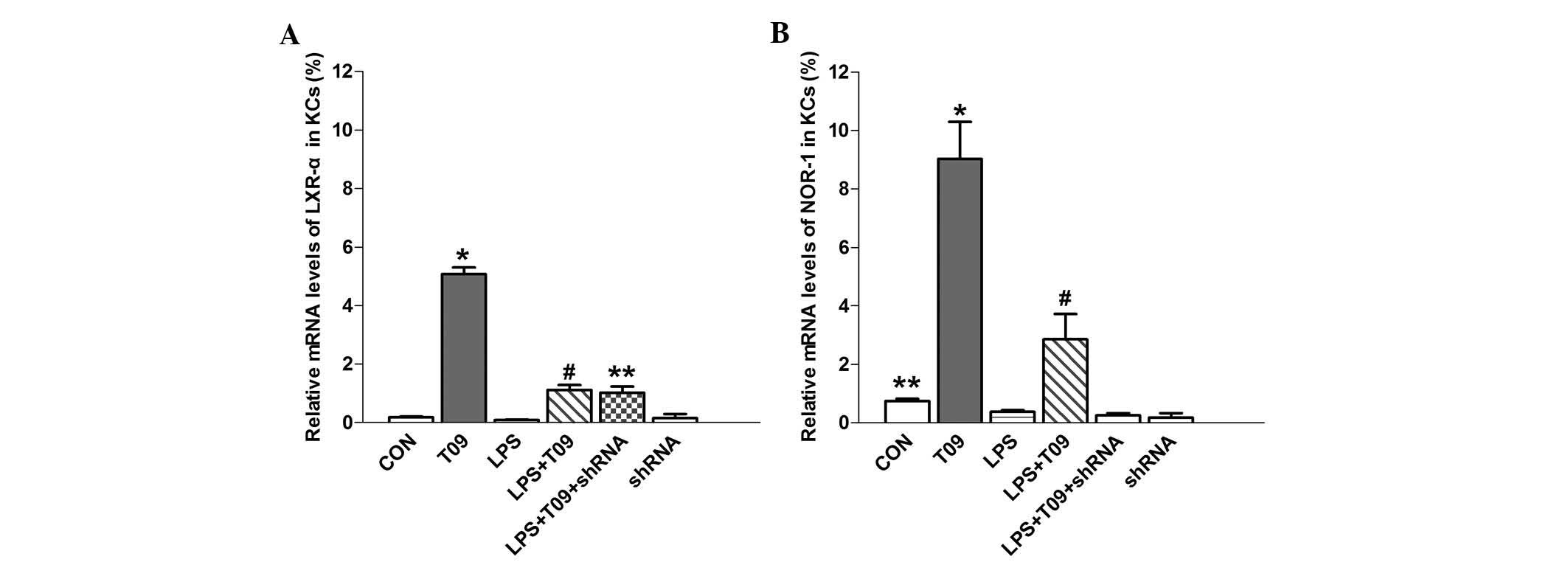
T0901317, a synthetic ligand of LXR-α, significantly
increased LXR-α mRNA expression in T09-KCs as compared with that in
the control group (P<0.05), and LXR-α expression in the LPS +
T09 group was higher than that in the LPS group (P<0.05)
(Fig. 3A). These results
encouraged the further exploration of the effect of NOR-1 shRNA on
LXR-α mRNA expression. However, the knockdown of NOR-1 using shRNA
had no effects on LXR-α mRNA expression, and there was no
significant difference in LXR-α expression between the LPS + T09
and LPS + T09 + shRNA groups (P>0.05) (Fig. 3A). The mRNA expression of NOR-1 in
the T09 group was significantly higher than that in the control
group (P<0.05), and the expression was also higher in the LPS +
T09 group than that in the LPS or LPS + T09 + shRNA group
(P<0.05) (Fig. 3B).
Furthermore, NOR-1 mRNA expression was reduced by shRNA compared
with that in the control cells (P<0.05), indicating that NOR-1
shRNA effectively suppressed the mRNA expression of NOR-1 (Fig 3B).
These results suggested that increased LXR-α
expression by its ligand can elevate NOR-1 mRNA expression under
normal and inflammatory conditions, and that there is a close
association between LXR-α and NOR-1. By contrast, shRNA targeting
NOR-1 suppressed NOR-1 mRNA expression but had no impact on LXR-α
mRNA levels (Fig. 3). These
findings indicated that the mRNA expression of NOR-1 is partially
controlled by LXR-α.
NOR-1 protein expression is partially
controlled by LXR-α under inflammatory and non-inflammatory
conditions
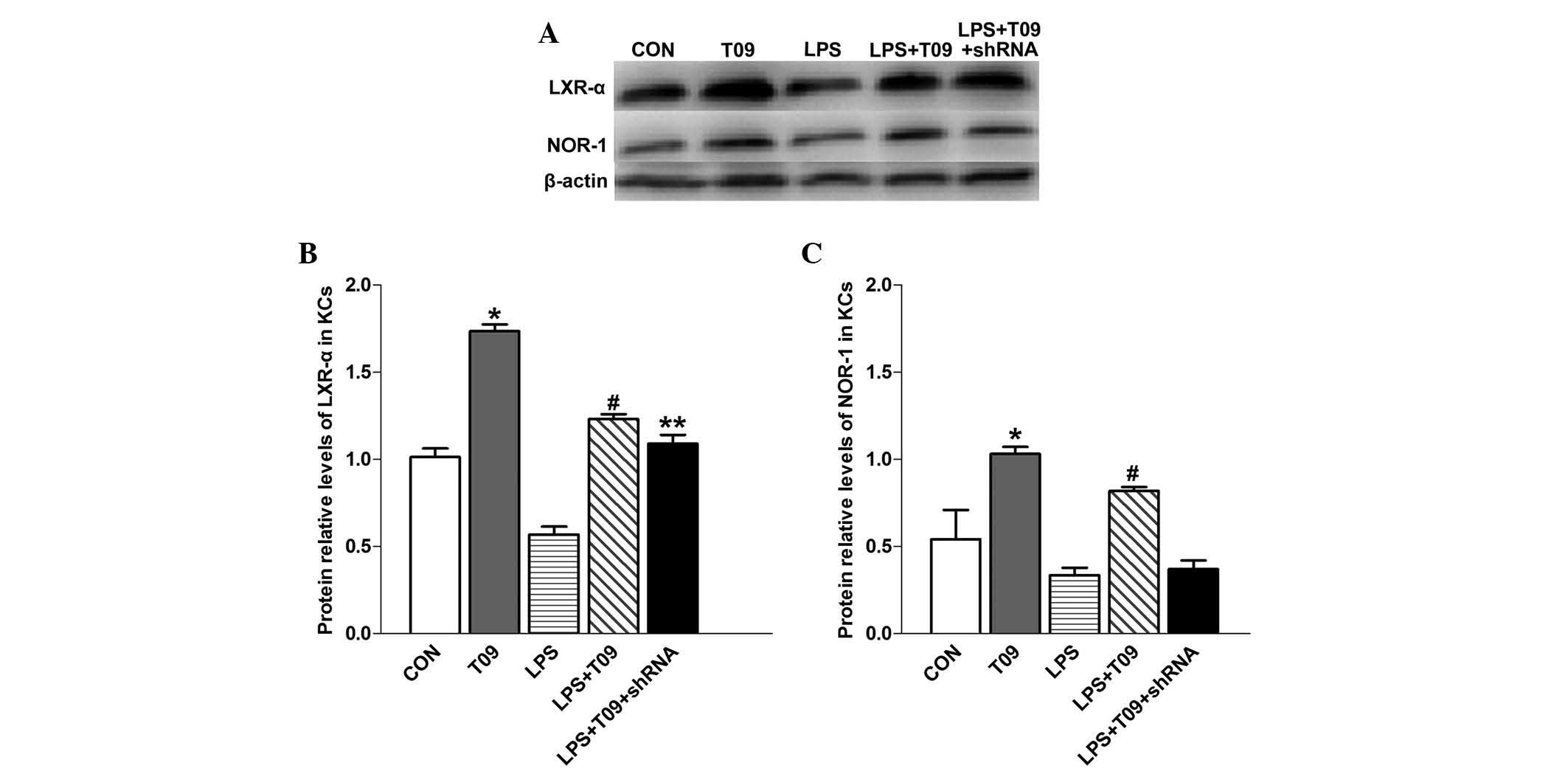
The protein expression of LXR-α was significantly
higher in T0901317 group than that in the control group
(P<0.05), and LXR-α was enhanced in the LPS + T09 group compared
with that in the LPS group (P<0.05) (Fig. 4A). However, there was no
significant difference in LXR-α levels between the LPS + T09 group
and the LPS + T09 + shRNA group (P>0.05). NOR-1 protein
expression was significantly higher in the T09 group than that in
the control group (P<0.05), and it was augmented in the LPS +
T09 group compared with that in the LPS + T09 + shRNA group
(P<0.05) (Fig. 4B). These
observations indicated that increased LXR-α expression enhances the
protein levels of NOR-1 under normal and inflammatory conditions.
The fact that NOR-1 shRNA had no effect on LXR-α expression in KCs
further confirms the interdependent association between LXR-α and
NOR-1.
LXR-α inhibits LPS-induced inflammation
in KCs through elevating NOR-1 expression
To further investigate the involvement of LXR-α and
NOR-1 in LPS-induced secretion of pro-inflammatory and
anti-inflammatory cytokines in KCs, TNF-α and IL-10 were quantified
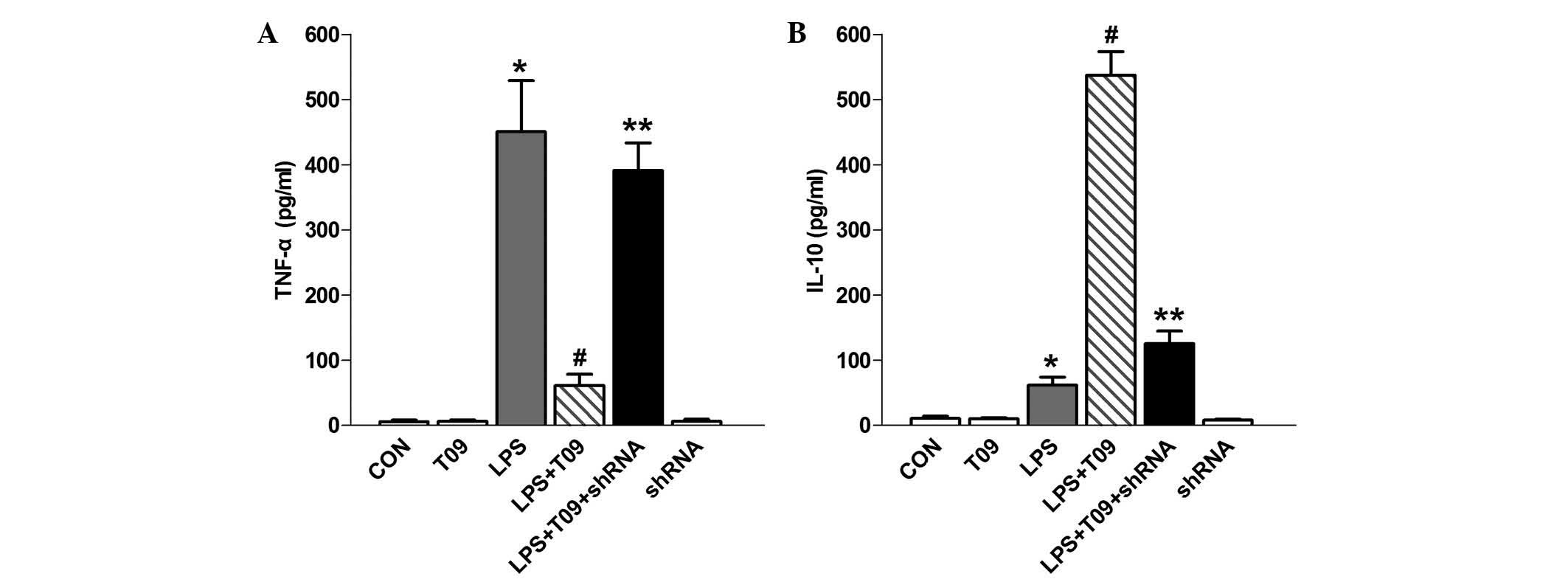
in each KC group by ELISA.
TNF-α levels in the LPS group were significantly
higher than those in the control group (P<0.05), whereas they
were obviously reduced in the LPS + T09 group compared with those
in the LPS group (P<0.05) (Fig
5A). The effect of NOR-1 shRNA on the production of TNF-α in
KCs was then explored. The production of TNF-α in KCs was
significantly enhanced in response to NOR-1 shRNA and T0901317
pre-treatment followed by LPS stimulation in comparison with that
in KCs treated with LPS and T0901317 (P<0.05). The levels of the
anti-inflammatory cytokine IL-10 in LPS-treated KCs was higher than
that in control cells (P<0.05), and of note, it was
significantly higher in the LPS + T09 group than that in the LPS
group (P<0.05) (Fig. 5B).
Furthermore, the IL-10 levels were decreased in the LPS + T09 +
shRNA group, which may be attributed to the suppression of NOR-1
(P<0.05). T0901317 blocked LPS-mediated TNF-α production and
induced IL-10 secretion. Of note, T0901317 or NOR-1 shRNA alone had
no effect on the expression levels of TNF-α or IL-10 (Fig. 5).
The ELISA results showed that LPS induced a marked
level of inflammation in KCs, which was partially suppressed
through ligand-induced upregulation of LXR-α expression.
Upregulated LXR-α inhibited LPS-induced inflammation through
elevating NOR-1 expression in KCs. These results suggested that
LPS-induced inflammation may be partially suppressed by LXR-α,
which may have an anti-inflammatory role by increasing NOR-1
expression and promoting ultimate secretion of the
anti-inflammatory cytokine IL-10 in KCs.
Discussion
LXR-α is a transcription factor belonging to the
nuclear receptor family, which has a central role in metabolic
homeostasis, being master regulators of key target genes in the
glucose and lipid pathways (17).
Previous studies have shown that LXR-α has a direct
anti-inflammatory effect. Musso et al (18) reported that agonists of LXR-α
improve cholesterol-induced hepatic inflammation and fibrosis by
reducing the activation of KCs and hepatic satellite cells. Spann
et al (19) provided
evidence that regulated accumulation of desmosterol effects
numerous homeostatic responses, including activation of LXR target
genes, inhibition of sterol regulatory element-binding protein
target genes, selective re-programming of fatty acid metabolism and
suppression of inflammatory-response genes, observed in macrophage
foam cells.
NOR-1 (also known as NR4A3) together with Nurr77
(NR4A1) and Nurr1 (NR4A2) form a superfamily named NR4As. The NR4A
proteins are among the most evolutionarily conserved nuclear
hormone receptor superfamilies (20). Members of the NR4A sub-family,
including NOR-1, lack a classical ligand-binding domain, function
as constitutively active transcription factors and respond to
stimuli such as immediate early response genes (21). Although initially proved to control
certain key processes, including differentiation, proliferation and
apoptosis, and associated with central nervous system disorders
(22), recent observations
identified roles of NR4As in controlling lipid metabolism and
inflammation (23). Zhao et
al (24) discussed how NR4As
control hepatic and skeletal muscle glucose metabolism, plasma and
hepatic lipid metabolism, as well as differentiation and function
of white and brown adipocytes.
It has been shown that NR4A receptors in murine
macrophages can be activated by LPS, and NOR-1 is potently and
transiently induced at an early time-point (21); however, the exact underlying
mechanism has remained elusive. Based on the anti-inflammatory
action of NOR-1 and the regulation of NOR-1 by activated LXR-α, the
present study hypothesized that LXR-α may inhibit inflammation by
regulating NOR-1 expression. The present study reported several
findings supporting a novel role of LXR-α and NOR-1 in controlling
LPS-induced inflammation. KCs were treated with LPS, LXR-α agonist
and NOR-1 shRNA to examine their effects. First, it was found that
LPS and/or LXR-α agonist led to profound changes in LXR-α
expression, which were paralleled by similar alterations in NOR-1
expression. Furthermore, it was revealed that LXR-α agonist
resulted in upregulation of NOR-1 at the mRNA and protein level;
however, NOR-1 shRNA had no effect on LXR-α expression, indicating
that LXR-α regulated NOR-1 expression. Finally, the levels of TNF-α
and IL-10, which are classic pro-inflammatory and anti-inflammatory
factors, respectively, were all elevated in KCs treated with LPS.
Of note, TNF-α was decreased but IL-10 was elevated in KCs treated
with LPS + T09. However, T0901317 alone affected neither TNF-α nor
IL-10 levels. NOR-1 knockdown in KCs treated with LPS + T09
increased the levels of TNF-α to a similar extent to that in the
LPS group, while the levels of IL-10 were markedly decreased. These
results indicated that enhancement of LXR-α expression regulates
inflammation in KCs by reducing the secretion of pro-inflammatory
factor TNF-α and elevating the secretion of anti-inflammatory
factor IL-10, which is closely associated with the expression of
NOR-1.
The results of the present study suggested that
during inflammation, NOR-1 has an anti-inflammatory role by
increasing the expression of the anti-inflammatory cytokine IL-10
in KCs. This phenomenon underscores the fact that the cross-talk
and expression modulation between the nuclear receptor families
has, at large, remained elusive. Further study is required to
confirm that LXR-α regulates LPS-mediated inflammation through
controlling NOR-1 expression. An enhanced understanding of the
complex association between LXR-α and NOR-1 during KC growth is
important for revealing the role of adaptive immunity in
inflammation, which may further benefit the application of adaptive
immunity in clinical treatment of inflammation.
In conclusion, the present study discovered that
promoting LXR-α expression by its ligand T0901317 elevated NOR-1
expression in KCs, which in turn upregulated cytokine IL-10 to
resist LPS-induced inflammation. The association between LXR-α and
NOR-1 in regulating metabolism, inflammation and immunity provides
novel insight into the feasibility of using LXR-α to regulate
inflammation; therefore, the present study suggested LXR-α as a
novel target to treat inflammatory diseases.
Acknowledgments
The present study was supported by the National
Nature Science Foundation of China (grant nos. 31370753, 81301656
and 81400614).
References
|
1
|
Xia X, Jung D, Webb P, Zhang A, Zhang B,
Li L, Ayers SD, Gabbi C, Ueno Y, Gustafsson JÅ, et al: Liver X
receptor β and peroxisome proliferator-activated receptor δ
regulate cholesterol transport in murine cholangiocytes.
Hepatology. 56:2288–2296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calkin AC and Tontonoz P: Liver X receptor
signaling pathways and atherosclerosis. Arterioscler Thromb Vasc
Biol. 30:1513–1518. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rébé C, Filomenko R, Raveneau M, Chevriaux
A, Ishibashi M, Lagrost L, Junien JL, Gambert P and Masson D:
Identification of biological markers of liver X receptor (LXR)
activation at the cell surface of human monocytes. PLoS One.
7:e487382012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang YY, Dahle MK, Steffensen KR, Reinholt
FP, Collins JL, Thiemermann C, Aasen AO, Gustafsson JA and Wang JE:
Liver X receptor agonist GW3965 dose-dependently regulates
lpsmediated liver injury and modulates posttranscriptional
TNF-alpha production and p38 mitogen-activated protein kinase
activation in liver macrophages. Shock. 32:548–553. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nomiyama T, Zhao Y, Gizard F, Findeisen
HM, Heywood EB, Jones KL, Conneely OM and Bruemmer D: Deficiency of
the NR4A neuron-derived orphan receptor-1 attenuates neointima
formation after vascular injury. Circulation. 119:577–586. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rodríguez-Calvo R, Guadall A, Calvayrac O,
Navarro MA, Alonso J, Ferrán B, de Diego A, Muniesa P, Osada J,
Rodríguez C, et al: Over-expression of neuron-derived orphan
receptor-1 (NOR-1) exacerbates neointimal hyperplasia after
vascular injury. Hum Mol Genet. 22:1949–1959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vacca M, Murzilli S, Salvatore L, Di
Tullio G, D'Orazio A, Lo Sasso G, Graziano G, Pinzani M, Chieppa M,
Mariani-Costantini R, et al: Neuron-derived orphan receptor 1
promotes proliferation of quiescent hepatocytes. Gastroenterology.
144:1518–1529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kumar N, Wang H, Liu D and Collins S:
Liver X receptor is a regulator of orphan nuclear receptor NOR-1
gene transcription in adipocytes. Int J Obes (Lond). 33:519–524.
2009. View Article : Google Scholar
|
|
9
|
Pearen MA, Eriksson NA, Fitzsimmons RL,
Goode JM, Martel N, Andrikopoulos S and Muscat GE: The nuclear
receptor, Nor-1, markedly increases type II oxidative muscle fibers
and resistance to fatigue. Mol Endocrinol. 26:372–384. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Van Tiel CM and De Vries CJ: NR4All in the
vessel wall. J Steroid Biochem Mol Biol. 130:186–193. 2012.
View Article : Google Scholar
|
|
11
|
Wynn TA, Chawla A and Pollard JW:
Macrophage biology in development, homeostasis and disease. Nature.
2013.496:445–455. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baffy G: Kupffer cells in non-alcoholic
fatty liver disease: The emerging view. J Hepatol. 51:212–223.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jenne CN and Kubes P: Immune surveillance
by the liver. Nat Immunol. 14:996–1006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li PZ, Li JZ, Li M, Gong JP and He K: An
efficient method to isolate and culture mouse Kupffer cells.
Immunol Lett. 158:52–56. 2014. View Article : Google Scholar
|
|
15
|
Ohkura N, Ito M, Tsukada T, Sasaki K,
Yamaguchi K and Miki K: Structure, mapping and expression of a
human NOR-1 gene, the third member of the Nur77/NGFI-B family.
Biochim Biophys Acta. 1308:205–214. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hellemans J and Vandesompele J: Selection
of reliable reference genes for RT-qPCR analysis. Methods Mol Biol.
1160:19–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Archer A, Laurencikiene J, Ahmed O,
Steffensen KR, Parini P, Gustafsson JÅ and Korach-André M: Skeletal
muscle as a target of LXR agonist after long-term treatment: Focus
on lipid homeostasis. Am J Physiol Endocrinol Metab. 306:E494–E502.
2014. View Article : Google Scholar
|
|
18
|
Musso G, Gambino R and Cassader M:
Cholesterol metabolism and the pathogenesis of non-alcoholic
steatohepatitis. Prog Lipid Res. 52:175–191. 2013. View Article : Google Scholar
|
|
19
|
Spann NJ, Garmire LX, McDonald JG, Myers
DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D,
et al: Regulated accumulation of desmosterol integrates macrophage
lipid metabolism and inflammatory responses. Cell. 151:138–152.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Briand O, Helleboid-Chapman A, Ploton M,
Hennuyer N, Carpentier R, Pattou F, Vandewalle B, Moerman E, Gmyr
V, Kerr-Conte J, et al: The nuclear orphan receptor Nur77 is a
lipotoxicity sensor regulating glucose-induced insulin secretion in
pancreatic β-cells. Mol Endocrinol. 26:399–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McMorrow JP and Murphy EP: Inflammation: A
role for NR4A orphan nuclear receptors? Biochem Soc Trans.
39:688–693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Papac-Milicevic N, Breuss JM, Zaujec J,
Ryban L, Plyushch T, Wagner GA, Fenzl S, Dremsek P, Cabaravdic M,
Steiner M, et al: The interferon stimulated gene 12 inactivates
vasculoprotective functions of NR4A nuclear receptors. Circ Res.
110:e50–e63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Veum VL, Dankel SN, Gjerde J, Nielsen HJ,
Solsvik MH, Haugen C, Christensen BJ, Hoang T, Fadnes DJ, Busch C,
et al: The nuclear receptors NUR77, NURR1 and NOR1 in obesity and
during fat loss. Int J Obes (Lond). 36:1195–1202. 2012. View Article : Google Scholar
|
|
24
|
Zhao Y and Bruemmer D: NR4A orphan nuclear
receptors: Transcriptional regulators of gene expression in
metabolism and vascular biology. Arterioscler Thromb Vasc Bio.
30:1535–1541. 2010. View Article : Google Scholar
|