Introduction
Osteoarthritis (OA) is a disabling and progressive
disease that affects multiple joints. Patients with OA constitute a
substantial medical, social and economic burden, therefore it is
necessary to conduct investigations to clarify the etiological and
predictive factors. There are two predominant types of OA; primary
OA, which is characterized by its late onset, and secondary OA,
which is differentiated from primary OA by its early onset with
established reasons, such as developmental abnormalities and
trauma. However, the causes for developing primary OA have not yet
been fully elucidated. It is hypothesized that the risk of primary
OA involves a variety of factors, including gender, age, behavior,
obesity and ethnicity, as well as genetic factors (1,2).
Results of linkage analysis and familial aggregation indicate that
complex genetic inheritance is critical in the underlying
pathogenesis of primary OA (3–6). The
interleukin-1 (IL-1) gene cluster on chromosome 2 (nucleotide
position, 113 531 492-113 891 593; 2q13-2q14.1) may serve as one of
the genetic loci that confer risk. IL-1 is a multifunctional
cytokine that predominantly impacts on inflammation and immunity.
In addition, IL-1 acts as a regulator of the homeostatic status of
the human body and is critical in the development of diseases
(7,8). Indeed, IL-1 is a family of three
associated proteins that are encoded by different genes. The family
comprises two major agonistic cytokines, interleukin-1α (IL-1A) and
interleukin-1β (IL-1B), as well as their endogenous receptor
antagonist, interleukin-1 receptor (IL-1R). It has been shown that
the potency of IL-1A, instead of IL-1B, in synovial fluid (SF) was
positively correlated with the occurrence of OA and that joint
tissues exposed to IL-1A in vitro were significantly
damaged, which triggered the development of OA in a rabbit
arthritis model (9,10). Evidence from numerous studies has
indicated that the polymorphisms or haplotypes in the IL-1A gene
were significantly associated with a risk for OA (11–13).
MicroRNAs (miRNAs) represent an abundant class of
short non-coding RNAs that act as post-transcriptional regulators.
To date, experiments have identified 600 human miRNAs, modulating
more than one-third of cellular messenger RNAs (mRNAs) (14). Increasing evidence has revealed
that dysregulated expression patterns of miRNAs were associated
with various diseases, including cancer, neurodegenerative and
cardiovascular diseases, and viral infections (14,15).
Binding of miRNA to mRNA is important in the regulation of mRNA
stability, as well as translation efficiency. However, this binding
may be compromised by the polymorphisms that are located within
miRNA target sites, which may either abrogate existent binding
sites or generate illegitimate binding sites (16–18),
and impact on the development of many diseases, such as various
types of cancer (19–21), Parkinson's disease (22), and other disorders (23-25).
A previous study by Gao et al revealed that the binding of
miR-122 and the regulation of IL-1A expression by the miRNA were
significantly affected by a 2-thiothiazolidine-4-carboxylic acid
(TTCA) insertion/deletion (Ins/Del) polymorphism (rs3783553) that
was residing in the IL-1A 3′-untranslated region (UTR), which
conferred a significantly increased risk for hepatocellular
carcinoma in a Chinese population (26).
Thus, the aim of the present study was to
demonstrate the functional influence of the rs3783553 polymorphism
on the interaction between miR-122 and IL-1A 3′-UTR. Furthermore,
the investigation of a Chinese Han population was conducted to
evaluate the association between the rs3783553 polymorphism and the
risk for OA, as well as the level of IL-1A in the SF of OA
patients.
Materials and methods
Subjects
A total of 931 Chinese Han OA patients with
clinically and radiologically confirmed diagnoses of OA were
recruited to the Department of Orthopedics, at Zhongshan Hospital
of Dalian University (Dalian, China). There were 317 females (34%)
and 614 males (66%; average age, 63.2±12.9 years). Informed consent
was obtained prior to the donation of blood for the purposes of
research. The radiographic criteria for OA were as follows: A
Kellgren-Lawrence (KL) grade >1 of at least one knee; and being
aged >38 years. The clinical criteria followed the guidelines of
the American College of Rheumatology (27,28)
in order to classify the type of OA, and the Western Ontario and
McMaster Universities Osteoarthritis Index (29) was used to evaluate pain and joint
stiffness, as well as physical function in the OA patients.
Patients were excluded from the study as follows: If they had a
history of corticosteroid use; if they had undergone a bilateral
knee replacement; if they exhibited other types of arthritis,
cancer or other chronic diseases beyond hypertension or
hypercholesterolemia. The KL grade (0–4) was recorded after
radiographs had been obtained; a KL score of 1 or 2 grouped the
patient into 'mild OA' and a KL score of 3 or 4 grouped the patient
into 'severe OA'. A total of 952 healthy individuals (37% female;
mean age, 62.6±11.2 years) who exhibited no clinical manifestation
of OA were assigned to the control group. Written consent was
obtained from each participant, and the study was approved by the
medical ethics committees of Dalian University.
Collection of tissues and cell
culture
Sterile synovial tissues were obtained during
therapeutic open joint surgery from patients who had undergone a
synovectomy. The synovial tissue was obtained, digested and
cultured according to a previously reported protocol (30). Briefly, within 2 h after surgery,
the sample was washed three times, and the superficial layer of
synovium (~2 mm3) was cut and placed in Dulbecco's
modified Eagle's medium (DMEM; Gibco BRL, Eggenstein, Germany),
supplemented with penicillin and streptomycin (Gibco BRL) per
milliliter, and containing 4 mg/ml of clostridiopeptidase A
(Sigma-Aldrich, St. Louis, MO, USA). The tissue fragments were
sliced further with scissors and incubated for 5 h in 25 ml DMEM at
37°C in a humidified atmosphere of 5% carbon dioxide. Subsequently,
an equal volume of 0.05% trypsin and 0.02% EDTA in modified Puck's
Saline A (Gibco-BRL) was added and incubation continued for another
2 h under the same conditions. The digested cells were centrifuged
for 10 min at 400 × g at room temperature and the pellet (1–2 ml)
was suspended in DMEM (20–40 ml) supplemented with 10% fetal bovine
serum (Gibco-BRL). The cells were passaged after the treatment with
trypsin, split 1:3 and plated in 6-cm polystyrene petri dishes at
5×105 cells per dish, containing 2 ml medium.
Extraction and genotyping of DNA
Genomic DNA was extracted using a DNA extraction kit
(Sangong Biotech, Shanghai, China). A routine polymerase chain
reaction (PCR) using a PTC-200 cycler (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was performed to amplify the fragment containing
the TTCA Ins/Del polymorphism, rs3783553, with the following
primers: Forward, 5′-ATTGGTCCGATCTTT GACTC-3′ and reverse, 5′-TGA
TAA CAG TGG TCT CATGG-3′. The PCR products underwent genotyping
using a direct sequencing DNA sequencer 3730 (Applied Biosystems
Life Technologies, Foster City, CA, USA).
Measurement of IL-1A in SF
SF was obtained from 75 OA patients by either direct
aspiration (n=52) or lavage (n=23) and the samples were analyzed
blinded to the clinical information. The levels of cytokines
(ng/ml) were determined by multiplex bead assay (Bio-Rad
Laboratories, Inc.). The SF samples were spun down to remove cell
debris, and divided into equal quantities and stored at 80°C. The
concentrations of IL-1A in the SF samples were determined by
commercially available ELISA kits (MyBiosource LLC, San Diego, CA,
USA) and the assays were performed according to the manufacturer's
instructions.
Reverse transcription (RT) and
quantitation of miR-122 in SF by real-time PCR (qPCR)
The method was conducted as previously described
(31). Briefly, mRNA was reverse
transcribed into cDNA using a NCode VILO miRNA cDNA Synthesis kit
(Invitrogen Life Technologies, Carlsbad, CA, USA), and qPCR was
performed on an Applied Biosystems 7900HT Real-Time PCR system
(Applied Biosystems Life Technologies) with standard plasmids. The
preliminary qPCR products underwent TA cloning using a TA cloning
kit (Invitrogen Life Technologies) and the sequences of the
inserted nucleotides were verified. Plasmids with known copy
numbers were subject to qPCR, and standard curves for each of the
miRNAs were generated. Absolute copy numbers of the target miRNA in
the SF samples were obtained according to the generated standard
curves.
qPCR analysis of mRNA levels
Total RNA was isolated from the synovial cells
(obtained from patients of different genotypes) with or without
being transfected with miR-122 mimics using an Ambion RNA isolation
kit (Ambion Life Technologies). This was subsequently converted to
complementary DNA using an oligo (dT) 15 primer and SuperScript II
Reverse Transcriptase (Invitrogen Life Technologies).
TaqMan® Gene Expression assays (Applied Biosystems Life
Technologies) were conducted using an ABI 7900HT Real-Time PCR
system (Applied Biosystems Life Technologies) to quantify the
relative IL-1A and miR-122 expression in the samples. U6 (RNU6-6P
RNA) served as the internal control. All reactions were run in
triplicate to reduce confounding variance.
Western blot analysis
Equal quantities of cell lysates were loaded onto
SDS-polyacrylamide gel for electrophoresis. The separated proteins
were subsequently transferred onto a polyvinylidene difluoride
(PVDF) membrane, followed by blocking at room temperature for 1 h
with Tris-buffered saline and Tween-20 [10 mM Tris-Cl (pH 8.0), 150
mM NaCl and 0.05% Tween-20) containing 5% nonfat dry milk powder.
The PVDF membrane was incubated with the rabbit anti-IL-1A
polyclonal antibody (cat. no. AV54322; Sigma-Aldrich; dilution,
1:2,000) at 4°C overnight, followed by incubation with horseradish
peroxidase-conjugated anti-rabbit secondary antibody (cat. no.
A0545; Sigma-Aldrich; dilution, 1:10,000) at room temperature for 2
h. The chemical fluorescence signal was detected using an enhanced
chemiluminescence kit (GE Healthcare Life Sciences, Chalfont, UK)
according to the manufacturer's instructions. A band of β-actin
served to normalize the result of the target gene.
IL-1A 3′-UTR luciferase assay
The full length of human IL-1A 3′-UTR containing the
gene locus (rs3783553) was PCR amplified using the following primer
sequence obtained from a genomic DNA sample that was isolated from
a wild-type subject: Forward,
5′-GATCTCTAGAGTCTGGAGTCTCACTTGTCTCACTTGTG-3′ and reverse,
5′-CATGGATCCGTCAGAGAATTTTGTTGCAAGCTTTATTTAG-3′. The PCR product was
cloned using a TA cloning kit (Invitrogen Life Technologies), and
the accuracy of the insert was established by Sanger sequencing.
Subsequently, a QuickChange XL Site-Directed Mutagenesis kit
(Agilent Technologies, Inc., Santa Clara, CA, USA) was used to
introduce another allele of rs3783553 using the following primer
sequence to delete TTCA: Forward,
5′-TTACCTGGGCATTCTTGTATTCCACCTGCAATCAAG-3′; and complementary
reverse, 5′-CTTGATTGCAGGTGGAATACAAGAATGCCCAGGTAA-3′. Finally, the
3′-UTR of Renilla luciferase in the pRL-SV40 vector (Promega
Corporation, Madison, WI, USA) was substituted with the generated
wild-type and mutant 3′-UTR of IL-1A. A luciferase assay was
performed in the human synovial cells as previously described
(26). Briefly, cells were seeded
at 1×105 cells per well in 24-well plates. After 12 h,
the cells were transfected using Lipofectamine 2000 (Invitrogen
Life Technologies) according to the manufacturer's instructions.
Each well comprised the wild-type or mutant construct (500 ng) and
pGL3 control vector (50 ng) that were co-transfected with 100 pmol
pre-miR-122 or the 22-nt oligonucleotide negative control (Ambion
Life Technologies, Austin, TX, USA). Following transfection
(duration, 24 h), the cells were harvested by the addition of a
passive lysis buffer (Promega Corporation). Luciferase activity in
the cell lysate was determined using the Dual-Luciferase Reporter
assay system (Promega Corporation) in a TD-20/20 Luminometer
(Turner Biosystems Inc., Sunnyvale, CA, USA).
Statistical analysis
Fisher's exact and χ2 tests, as well as
Student's t-test were used to compare the frequency distribution of
age, gender, body mass index (BMI) and smoking status between the
OA cases and control subjects. The Hardy-Weinberg equilibrium (HWE)
of the genotype frequencies of the controls was assessed using the
χ2 test. The association between the rs3783553
polymorphism and OA risk was estimated by computing odds ratios
(ORs), and their 95% confidence intervals (CIs) from unconditional
logistic regression analysis with the adjustment for possible
confounders. Student's t-test was used to evaluate the association
between the rs3783553 genotype and SF concentration of miR122 and
IL-1A. P<0.05 was considered to indicate a statistically
significant difference, and all analyses were performed using IBM
SPSS 19.0 software (Armonk, NY, USA).
Results
rs3783553 genotypes affect IL-1A
expression by regulating the miR-122 binding in human synovial
cells
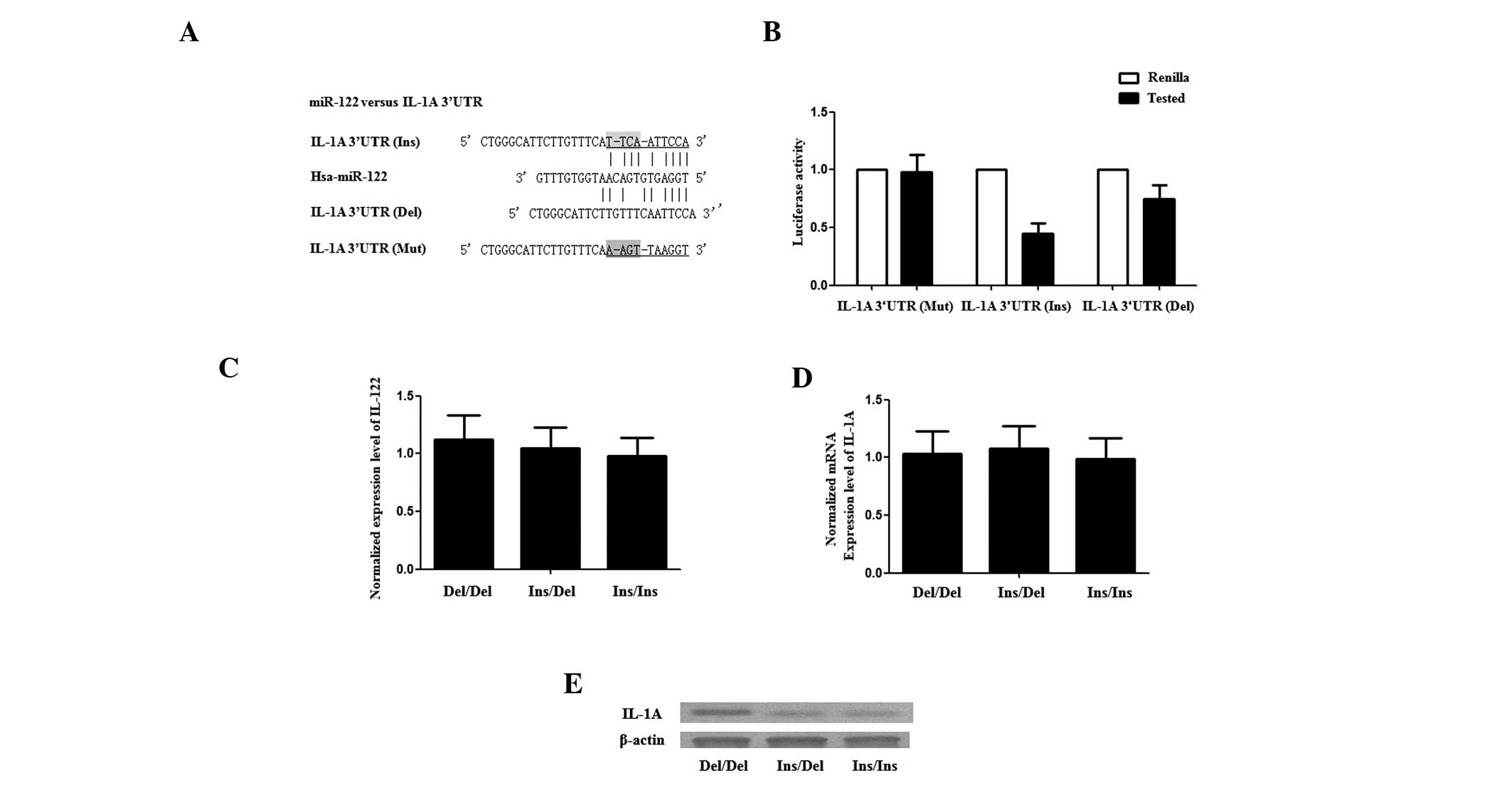
To investigate how the rs3783553 genotypes affect
IL-1A expression, the 3′-UTR of a Renilla luciferase
reporter gene was replaced with the full length IL-1A 3′-UTR
containing either allele of the rs3783553 polymorphism. In
transiently transfected human synovial cells, compared with the
constructs containing TTCA Del alleles, luciferase activity from
constructs containing the TTCA Ins allele was significantly lower
in the presence of miR-122 (Fig. 1A
and B; P<0.01). This result indicated that miR-122 binds to,
and negatively regulates the transcription of, IL-1A and that this
regulation is negatively influenced by the presence of the TTCA Ins
allele, which is likely to affect the binding of miR-122 to the
IL-1A transcript.
Exogenous expression of miR-122
suppresses the expression of IL-1A with Ins/Ins and Ins/Del
genotypes, but not the Del/Del genotype
The synovial tissues of each rs3783553 genotype were
obtained from the patients who received a synovectomy, and the
cells were digested, passaged and cultured prior to the functional
analysis. The endogenous expression levels of miR-122 and IL-1A
were determined in each cell group using RT-qPCR and western blot
analysis, and the expression of miR-122 and IL-1A mRNA was found to
be comparable between each group (Fig.
1C and E); however, the IL-1A protein expression level in the
Del/Del group was markedly higher than in the Ins/Ins and Ins/Del
groups (Fig. 1D). Additionally,
hsa-miR-122 mimics were transfected into the synovial cells of each
genotype, and the mRNA and protein expression levels of IL-1A in
the synovial cells were examined again. This demonstrated that the
protein level of IL-1A in the Del/Del group was not changed;
however in the Ins/Ins and Ins/Del groups, the protein level was
substantially lowered (Fig. 2A)
and the mRNA expression level remained unchanged (Fig. 2B).
Demographic characteristics
A total of 931 OA patients and 952 healthy control
subjects were recruited; the demographic characteristics and OA
radiographic severity are presented in Table I. No statistically significant
differences were observed with respect to age (P=0.2599), gender
(P=0.1698) and smoking status (P=0.0704). Higher BMI values were
detected among the OA patients (P=0.0001).
 | Table IDistribution of selected variables
between the OA cases and control subjects. |
Table I
Distribution of selected variables
between the OA cases and control subjects.
| Variable | OA, n=931 | Control, n=952 | P-value |
|---|
| Gender, n (%) | | | 0.1698 |
| Male | 614 (66) | 599 (63) | |
| Female | 317 (34) | 353 (37) | |
| Age, years | 63.2±12.9 | 62.6±11.2 | 0.2599 |
| Body mass index,
kg/m2 | 27.2±3.9 | 24.6±3.7 | <0.001 |
| Smoking status, n
(%) | | | 0.0704 |
| Yes | 633 (68) | 609 (64) | |
| Never | 298 (32) | 343 (36) | |
| Kellgren-Lawrence
score, n (%) | | | |
| Grade 1 and 2 | 670 (72) | | |
| Grade 3 and 4 | 261 (28) | | |
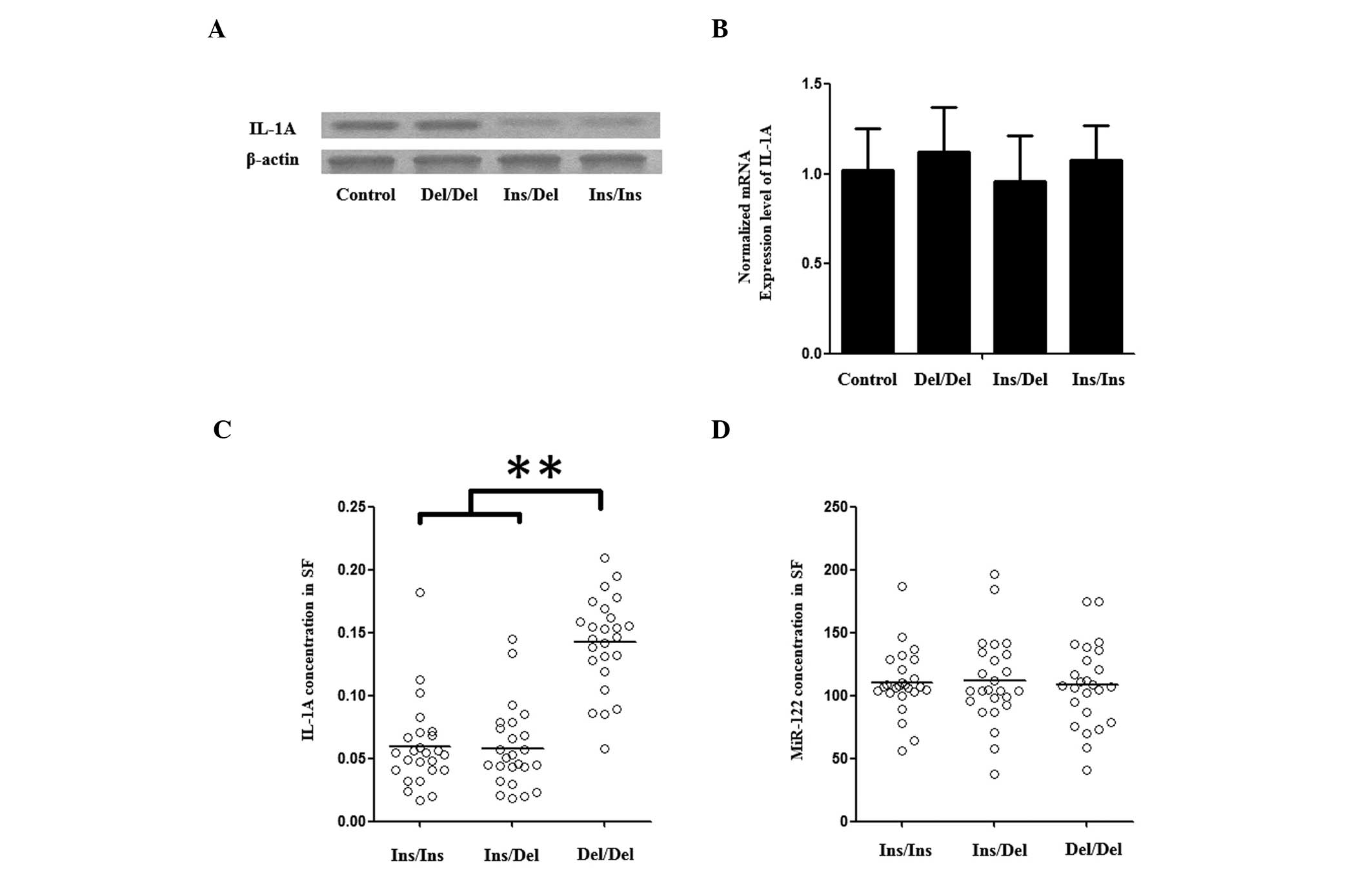
Association of rs3783553 genotypes with
the concentrations of IL-1A and miR-122 in SF
Three different genotypic synovial fluid samples
were used to further investigate the impacts of rs3783553 on the
transcription of IL-1A: 25 Homozygous for the TTCA Del allele, 25
heterozygous and 25 homozygous for the TTCA Ins allele. Using an
ELISA detection kit, the IL-1A level observed in the TTCA Ins
homozygous group was identified to be comparable to the
heterozygous group, and the two were found to be notably lower than
that of the TTCA Del homozygous group (Fig. 2C). To eliminate possible effects
from miR-122, the concentration of miR-122 in SFs of the same 75
patients was measured, and no difference was identified between the
IL-1A rs3783553 genotype groups (Fig.
2D).
Association of rs3783553 genotypes with
the risk for OA and its radiographic severity
To analyze the association between the IL-1A
rs3783553 polymorphism and the risk for OA in a Chinese Han
population, 931 OA patients and 952 healthy individuals were
determined for the allelic and genotypic frequencies of the IL-1A
rs3783553 polymorphism. The rs3783553 polymorphism was in HWE and
was assessed for correlation with the risk for OA and its
radiographic severity. A significant association was noted between
the IL-1A rs3783553 polymorphism rare allele (Del) and an increased
risk for OA [following adjustment for multiple potential risk
factors (Table II)]. The
frequency of the IL-1A rs3783553 polymorphism, Del allele was
identified to be notably higher in the OA patients than in the
control subjects (Table II),
which was demonstrated by an allelic frequency analysis, following
adjustment for known risk factors. Additionally, a significant
association between rs3783553 homozygote (Del/Del) and an increased
OA severity was noted in the recessive genetic model, compared with
its heterozygote and homozygote (Table II; OR=0.71, 95% CI=0.55–0.92, and
P=0.010). Due to the relatively large sample size, a stratified
analysis by patient age, gender, BMI and smoking status was
performed. It was noted that the association was even more marked
in young subjects (<62 years) (Table III; OR=0.53; P<0.001).
 | Table IIGenotype and allele frequencies of
the rs3783553 polymorphism among cases and control subjects, and
the associations with OA risk. |
Table II
Genotype and allele frequencies of
the rs3783553 polymorphism among cases and control subjects, and
the associations with OA risk.
| rs3783553
polymorphism | Control, n=952
(%) | OA, n=931 (%) | OR (95% CI) | P-value |
|---|
| Del/Del | 114 (12) | 158 (17) | 1.00 | |
| Ins/Del | 447 (47) | 418 (45) | 0.67
(0.55–0.88) | 0.0051 |
| Ins/Ins | 391 (41) | 355 (38) | 0.65
(0.49–0.86) | 0.0031 |
| Ins/Del +
Ins/Ins | 838 (88) | 773 (83) | 0.66
(0.51–0.86) | 0.0021 |
 | Table IIIStratification analysis between the
different genotypes of the rs3783553 polymorphism and the OA
risk. |
Table III
Stratification analysis between the
different genotypes of the rs3783553 polymorphism and the OA
risk.
| Variable | Genotype, n
| Ins/Del + Ins/Ins
vs. Del/Del [OR (95% CI)] | P-value |
|---|
| Del/Del
(Case/Control) | Ins/Del + Ins/Ins
(Case/Control) |
|---|
| Age, years |
| ≤62 | 78/54 | 366/460 | 0.53
(0.36–0.77) | 0.0010 |
| >62 | 80/60 | 407/378 | 0.78
(0.54–1.13) | 0.018 |
| Gender |
| Male | 104/72 | 510/528 | 0.66
(0.48–0.92) | 0.015 |
| Female | 54/42 | 263/310 | 0.66
(0.42–1.02) | 0.061 |
| Smoker |
| Never | 107/72 | 510/528 | 0.69
(0.50–0.96) | 0.028 |
| Yes | 51/42 | 263/310 | 0.70
(0.45–1.09) | 0.110 |
| BMI |
| ≤25 | 55/59 | 271/435 | 0.66
(0.44–1.00) | 0.047 |
| >25 | 103/55 | 502/403 | 0.67
(0.47–0.95) | 0.023 |
| Total | 158/114 | 773/838 | 0.66
(0.51–0.86) | 0.0021 |
Discussion
An important step toward the clinical application of
this novel subclass of genetic variations in health management will
be to improve understanding of distinct polymorphisms in
miRNA-associated gene and protein expression regulation in a
variety of diseases at the population level. In the present study,
performance of a luciferase assay demonstrated that rs3783553
genotypes affect IL-1A expression by regulating the miR-122 binding
in human synovial cells. Furthermore, the rs3783553 polymorphism
was shown to be significantly associated with OA risk in a Chinese
Han population (OR=0.71; P=0.010), and an even more marked
association was noted in younger subjects (OR=0.45; P<0.001).
Additionally, it was found that the genotype was associated with
the radiographic severity in the same population (OR=0.72;
P=0.023).
Modulation of the immune system depends on the
complex interaction within the cytokine network, where an optimal
physiological environment must be maintained by the delicate
modulation of the products of cytokine genes and their respective
receptor genes. The IL-1 system is an effective example of the
highly regulated mechanism, which is necessary for this to be
achieved. An important signaling pathway in the inflammatory
process, innate immunity and the immune response is IL-1 signaling
via the IL-1 receptor type I (IL-1R) (32). It has various effects, including
the stimulation of fibroblasts, keratinocyte proliferation,
cartilage breakdown, angiogenesis and incremental production of
acute phase response proteins by the liver (32). The activation of c-Jun N-terminal
kinases and p38 mitogen-activated protein kinases and subsequent
upregulation of the expression of genes via the transcription
factors, nuclear factor-κ-light-chain-enhancer of activated B
cells, activator protein-1 and CCAAT-enhancer-binding proteins,
mediate these physiological effects at the molecular level
(33). It is hypothesized that the
products of the IL-1 gene cluster are involved in various diseases,
one of which is OA. IL-1 may stimulate chondrocytes to produce
cartilage matrix degrading enzymes, thereby contributing to this
loss. In support of this hypothesis, the specific cleavage of the
aggrecan core protein at the Glu373-Ala374 bond was found to be
strongly correlated with the release of aggrecan catabolites in
response to IL-1 (34). The SFs of
OA patients are known to possess fragments bearing this epitope
(35). Furthermore, compared with
normal synovium, cells from OA synovium secrete more IL-1b
(36,37), and similarly, chondrocytes from OA
cartilage produce IL-1A and IL-1b that is quantitatively different
from normal chondrocytes (38,39).
Compared with similar explants from non-arthritic cartilage
(40), OA cartilage explants tend
to be more susceptible to the effects of IL-1, and the expression
of IL-1R and -2R on chondrocytes is relevant to this susceptibility
(41).
The risk allele is likely to be more prevalent
during early tumor development, which creates an improved binding
site for miR-122, and inhibits IL-1A expression and immune
surveillance. Gao et al performed an in-silicon analysis,
and predicted that miR-122 and miR-378 were binding tightly to
IL-1A mRNA transcripts, which contain the TTCA Del allele and
modulate IL-1A expression in a negative manner (26). By contrast, binding with mRNA
transcripts, which contain the TTCA Ins allele would be
interrupted, causing upregulation of IL-1A expression. The
luciferase assay, which compared IL-1A 3′-UTR with different
rs3783553 genotypes was in support of this hypothesis, revealing
the following relative transcription activities: Del/Del, 1.0;
Del/Ins, 3.8; and Ins/Ins, 5.5 (17). Consistently, the highest IL-1A mRNA
level was detected in the TTCA Ins homozygous group, followed by
the heterozygous and TTCA Del groups, as demonstrated by a
TaqMan® Gene Expression analysis of the tumor tissues of
different rs3783553 genotypes from hepatocellular carcinoma
patients. When compared with the TTCA Del homozygous group, the
average IL-1A concentrations were 3.76- and 5.57-fold higher in the
heterozygous and TTCA Ins homozygous groups, respectively, and the
average IL-1A expression levels were 3.76- and 5.57-fold higher in
the heterozygous and TTCA Ins homozygous groups, respectively. In
the present study, a luciferase reporter system was used to
establish IL-1A as an effective target gene of miR-122 in synovial
cells that were obtained from patients who had received a
synovectomy. This finding was further validated by the observation
that exogenous over-expression of miR-122 in the synovial cells
significantly downregulated the expression of IL-1A in the cells
with Ins/Ins and Ins/Del genotypes, although this was not observed
in the Del/Del group. Furthermore, the expression pattern of
miR-122 and IL-1A in the synovial cells and SF of different
genotypes was examined, and the miR-122 concentration was
identified to be comparable among each group, although the IL-1A
expression levels in the Ins/Ins and Ins/Del genotype groups were
markedly lower than those of the Del/Del group in the synovial
cells and the SF.
In conclusion, the risk caused by a polymorphism in
the IL-1A gene and a potential biological mechanism for an
increased risk of OA have been identified in the present study via
genetic association. Due to the high expression of miR-122 in human
synovium, further studies are required to determine whether miR-122
and IL-1A may be applied to the diagnosis and treatment of OA.
References
|
1
|
Dinarello CA: The interleukin-1 family: 10
years of discovery. FASEB J. 8:1314–1325. 1994.PubMed/NCBI
|
|
2
|
Lotz M, Blanco FJ, von Kempis J, Dudler J,
Maier R, Villiger PM and Geng Y: Cytokine regulation of chondrocyte
functions. J Rheumatol Suppl. 43:104–108. 1995.PubMed/NCBI
|
|
3
|
van den Berg WB, Joosten LA and van de Loo
FA: TNF alpha and IL-1 beta are separate targets in chronic
arthritis. Clin Exp Rheumatol. 17(6 Suppl 18): S105–S114.
1999.PubMed/NCBI
|
|
4
|
Luo L, Cruz T and McCulloch C: Interleukin
1-induced calcium signalling in chondrocytes requires focal
adhesions. Biochem J. 324:653–658. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pritchard S and Guilak F: Effects of
interleukin-1 on calcium signaling and the increase of filamentous
actin in isolated and in situ articular chondrocytes. Arthritis
Rheum. 54:2164–2174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hubbard JR, Steinberg JJ, Bednar MS and
Sledge CB: Effect of purified human interleukin-1 on cartilage
degradation. J Orthop Res. 6:180–187. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kahle P, Saal JG, Schaudt K, Zacher J,
Fritz P and Pawelec G: Determination of cytokines in synovial
fluids: correlation with diagnosis and histomorphological
characteristics of synovial tissue. Ann Rheum Dis. 51:731–734.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Apte RN and Voronov E: Is interleukin-1 a
good or bad 'guy' in tumor immunobiology and immunotherapy? Immunol
Rev. 222:222–241. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McNulty AL, Rothfusz NE, Leddy HA and
Guilak F: Synovial fluid concentrations and relative potency of
interleukin-1 alpha and beta in cartilage and meniscus degradation.
J Orthop Res. 31:1039–1045. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henderson B and Pettipher ER:
Arthritogenic actions of recombinant IL-1 and tumor necrosis factor
alpha in the rabbit: evidence for synergistic interactions between
cytokines in vivo. Clin Exp Immunol. 75:306–310. 1989.PubMed/NCBI
|
|
11
|
Smith AJ, Keen LJ, Billingham MJ, Perry
MJ, Elson CJ, Kirwan JR, Sims JE, Doherty M, Spector TD and Bidwell
JL: Extended haplotypes and linkage disequilibrium in the
IL1R1-IL1A-IL1B-IL1RN gene cluster: association with knee
osteoarthritis. Genes Immun. 5:451–460. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valdes AM and Spector TD: The contribution
of genes to osteoarthritis. Rheum Dis Clin North Am. 34:581–603.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valdes AM and Spector TD: Genetic
epidemiology of hip and knee osteoarthritis. Nat Rev Rheumatol.
7:23–32. 2011. View Article : Google Scholar
|
|
14
|
Pillai RS: MicroRNA function: multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiemer EA: The role of microRNAs in
cancer: no small matter. Eur J Cancer. 43:1529–1544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen K, Song F, Calin GA, Wei Q, Hao X and
Zhang W: Polymorphisms in microRNA targets: a gold mine for
molecular epidemiology. Carcinogenesis. 29:1306–1311. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin
Y, Wang E, Wu M and Shen SH: Aberrant allele frequencies of the
SNPs located in microRNA target sites are potentially associated
with human cancers. Nucleic Acids Res. 35:4535–4541. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saunders MA, Liang H and Li WH: Human
polymorphism at microRNAs and microRNA target sites. Proc Natl Acad
Sci USA. 104:3300–3305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chin LJ, Ratner E, Leng S, Zhai R, Nallur
S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al: A SNP in a
let-7 microRNA complementary site in the KRAS 3′untranslated region
increases non-small cell lung cancer risk. Cancer Res.
68:8535–8540. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Landi D, Gemignani F, Naccarati A, Pardini
B, Vodicka P, Vodickova L, Novotny J, Försti A, Hemminki K, Canzian
F and Landi S: Polymorphisms within micro-RNA-binding sites and
risk of sporadic colorectal cancer. Carcinogenesis. 29:579–584.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brendle A, Lei H, Brandt A, Johansson R,
Enquist K, Henriksson R, Hemminki K, Lenner P and Försti A:
Polymorphisms in predicted microRNA-binding sites in integrin genes
and breast cancer: ITGB4 as prognostic marker. Carcinogenesis.
29:1394–1399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang G, van der Walt JM, Mayhew G, Li YJ,
Züchner S, Scott WK, Martin ER and Vance JM: Variation in the
miRNA-433 binding site of FGF20 confers risk for Parkinson disease
by overex-pression of alpha-synuclein. Am J Hum Genet. 82:283–289.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sethupathy P, Borel C, Gagnebin M, Grant
GR, Deutsch S, Elton TS, Hatzigeorgiou AG and Antonarakis SE: Human
microRNA-155 on chromosome 21 differentially interacts with its
polymorphic target in the AGTR1 3′ untranslated region: a mechanism
for functional single-nucleotide polymorphisms related to
phenotypes. Am J Hum Genet. 81:405–413. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Clop A, Marcq F, Takeda H, Pirottin D,
Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al:
A mutation creating a potential illegitimate microRNA target site
in the myostatin gene affects muscularity in sheep. Nat Genet.
38:813–818. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martin MM, Buckenberger JA, Jiang J,
Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD and Elton
TS: The human angiotensin II type 1 receptor +1166 A/C polymorphism
attenuates microRNA-155 binding. J Biol Chem. 282:24262–24269.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao Y, He Y, Ding J, Wu K, Hu B, Liu Y, Wu
Y, Guo B, Shen Y, Landi D, et al: An insertion/deletion
polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′
untranslated region confers risk for hepatocellular carcinoma.
Carcinogenesis. 30:2064–2069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fermor B, Jeffcoat D, Hennerbichler A,
Pisetsky DS, Weinberg JB and Guilak F: The effects of cyclic
mechanical strain and tumor necrosis factor alpha on the response
of cells of the meniscus. Osteoarthritis Cartilage. 12:956–962.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuettner KE, Pauli BU, Gall G, Memoli VA
and Schenk RK: Synthesis of cartilage matrix by mammalian
chondrocytes in vitro. 1 Isolation, culture characteristics and
morphology. J Cell Biol. 93:743–750. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maroudas A and Evans H: A study of ionic
equilibria in cartilage. Connect Tissue Res. 1:69–77. 1972.
View Article : Google Scholar
|
|
30
|
Dayer JM, Krane SM, Russell RG and
Robinson DR: Production of collagenase and prostaglandins by
isolated adherent rheumatoid synovial cells. Proc Natl Acad Sci
USA. 73:945–949. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Attur M, Wang HY, Kraus VB, Bukowski JF,
Aziz N, Krasnokutsky S, Samuels J, Greenberg J, McDaniel G,
Abramson SB and Kornman KS: Radiographic severity of knee
osteoarthritis is conditional on interleukin 1 receptor antagonist
gene variations. Ann Rheum Dis. 69:856–861. 2010. View Article : Google Scholar :
|
|
32
|
Dinarello CA: Biologic basis for
interleukin-1 in disease. Blood. 87:2095–2147. 1996.PubMed/NCBI
|
|
33
|
O'Neill LA and Greene C: Signal
transduction pathways activated by the IL-1 receptor family:
ancient signaling machinery in mammals, insects, and plants. J
Leukoc Biol. 63:650–657. 1998.PubMed/NCBI
|
|
34
|
Arner EC, Hughes CE, Decicco CP, Caterson
B and Tortorella MD: Cytokine-induced cartilage proteoglycan
degradation is mediated by aggrecanase. Osteoarthritis Cartilage.
6:214–228. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fosang AJ, Last K, Knäuper V, Murphy G and
Neame PJ: Degradation of cartilage aggrecan by collagenase-3
(MMP-13). FEBS Lett. 380:17–20. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Westacott CI, Taylor G, Atkins R and Elson
C: Interleukin 1 alpha and beta production by cells isolated from
membranes around aseptically loose total joint replacements. Ann
Rheum Dis. 51:638–642. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Elson CJ, Mortuza FY, Perry MJ, Warnock
MG, Webb GR and Westacott CI: Cytokines and focal loss of cartilage
in osteoarthritis. Br J Rheumatol. 37:106–107. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Towle CA, Hung HH, Bonassar LJ, Treadwell
BV and Mangham DC: Detection of interleukin-1 in the cartilage of
patients with osteoarthritis: a possible autocrine/paracrine role
in pathogenesis. Osteoarthritis Cartilage. 5:293–300. 1997.
View Article : Google Scholar
|
|
39
|
Moos V, Rudwaleit M, Herzog V, Höhlig K,
Sieper J and Müller B: Association of genotypes affecting the
expression of interleukin-1beta or interleukin-1 receptor
antagonist with osteoarthritis. Arthritis Rheum. 43:2417–2422.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ismaiel S, Atkins RM, Pearse MF, Dieppe PA
and Elson CJ: Susceptibility of normal and arthritic human
articular cartilage to degradative stimuli. Br J Rheumatol.
31:369–373. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Martel-Pelletier J, McCollum R, DiBattista
J, Faure MP, Chin JA, Fournier S, Sarfati M and Pelletier JP: The
interleukin-1 receptor in normal and osteoarthritic human articular
chondrocytes. Identification as the type I receptor and analysis of
binding kinetics and biologic function. Arthritis Rheum.
35:530–540. 1992. View Article : Google Scholar : PubMed/NCBI
|