Introduction
Avian influenza viruses (AIVs), have repeatedly
caused worldwide pandemics and are one of the leading infectious
diseases with high rates of morbidity and mortality (1). AIV genomes consist of a
single-stranded negative RNA with the following eight segments:
Polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acid
(PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA),
matrix (M) and non-structural polypeptides (NS). These eight
segments encode 15 known proteins, including PA, PB2, PB1, HA, NP,
NA, M1, M2, NS1 and NS2, as well as supplementary proteins
(2). According to two viral
membrane proteins, namely, HA and NA, AIVs can be divided into
subtypes (H1-18 and N1-11, respectively) (2,3).
HAs, which are a surface glycoprotein, are encoded
by the fourth RNA segment. HAs facilitate the binding to and
entrance of viruses into host cells via sialic acid (SAa) residues
that are present on host cell surfaces (4). HAs appear to be predominantly
involved in facilitating viral entry into the cell by specifically
binding to the SAa surface receptor in host cells, and HA is likely
able to alter surface antigenic properties by creating genetic
mutations or reassortments (5).
HAs are often used to design vaccines or major immunogenic
components of influenza vaccines. HAs isolated from human strains
specifically identify and bind SAa-2, 6, which is a receptor found
on cells of the upper respiratory tract in humans. HAs also bind to
SAa-2, 3, which is a receptor on lower respiratory tract cells in
humans. The binding of HA and SAa is correlated with viral
pneumonia. Moreover, mutations in HA are associated with
epidemiology and clinical symptoms and outcomes (6,7).
H7N9, which is a novel AIV subtype, is transmitted
to humans and may be fatal by causing acute respiratory distress
syndrome or severe pneumonia; however, the transmission mechanism
of AIVs from person to person remain unclear (8,9).
More than 400 individuals were infected with H7N9 from March 2013
to April 2014, in China. Patients with H7N9 infections present with
intense systemic inflammatory response syndrome that is concomitant
with an anti-inflammatory response (10). HAs from the H7N9 virus cause
pathological responses in hosts, and simultaneously hosts may
resist or regulate this pathological damage by triggering
protective homeostatic responses. However, the exact molecular
mechanisms of the pathogenesis of H7N9 infection are not fully
understood.
MicroRNAs (miRNAs), which are small non-coding RNAs,
control the expression of several target mRNAs. miRNAs control the
activity of ~30% of all protein-coding genes in mammals (11). miRNAs also regulate immune and
inflammatory responses (12,13).
In a previous study, miR-181a was involved in the homeostasis
response to inflammatory stimulus (14). Patients infected with H7N9 showed
increased expression of miR-20a, miR-106a, miR-17 and miR-376c, and
decreased expression of let-7e in the blood (15). However, the functions of the miRNAs
induced by H7N9 remain unclear. The present study aimed to assess
the inflammatory response and self-regulatory mechanisms of the
miRNAs, which are induced by HAs of H7N9 in THP-1 cells.
Materials and methods
Cell culture
THP-1, which is a human acute monocytic leukemia
cell line, grows mainly in a suspended state. The THP-1 cell line
was selected for the present study due to its shared
characteristics with native monocyte-derived macrophage cells, as
well as the fact that it is a well-developed cell model for
inflammation research. The THP-1 cells were supplied by KeyGEN
Biotech (Nanjing, China). The cells were cultured in Dulbecco's
modified Eagle's medium (Invitrogen Life Technologies, Guangzhou,
China) with 10% fetal bovine serum (Gibco, Life Technologies,
Guangzhou, China) and incubated at 5% CO2 at 37°C. The
cells were seeded in a 6-well tissue plate at a density of
4.0×105 per well. An HA protein of the influenza virus
(H7N9) A/Anhui/1/2013 (ACRO Biosystems, Beijing, China) was added
to the incubated cells after the cells had been seeded for 12 h.
Cell samples were collected at 0, 2, 4, 8, 12 and 24 h following
treatment with 100 ng/ml HA to analyze the proinflammatory factors
and miRNAs. This procedure was performed to determine the optimal
time of HA incubation, which would elicit the strongest effects on
the cells. The THP-1 cells were then treated with HA at 0, 10, 100
and 200 ng/ml for 4 h. Cell and medium samples, which were required
for the assay of the proinflammatory factors and miRNAs, were
collected. In another separate experiment, the THP-1 cells were
incubated with 1 ng/ml lipopolysaccharide (LPS; Sigma-Aldrich, St.
Louis, MO, USA) alone, 100 ng/ml HA alone, and a combination of 1
ng/ml LPS and 100 ng/ml HA for 4 h. The incubated THP-1 cells were
used to investigate the synergic effect of HA and LPS on the
expression of proinflammatory factors. This process was conducted
as H7N9 virus infections are usually associated with bacterial
infections.
Knockdown of TLR4
The small interfering (si)RNAs were transiently
transfected into the cells at a density of 4×105
cells/well and cultured in a 6-well plate using a Lipofectamine
2000 reagent (Invitrogen Life Technologies). siTLR4
(GGAAUGAGCUAGUAAAGAA) was designed using siRNA Explorer software
(developed by the group of Professor Yaou Zhang, Shenzhen Key Lab
of Health Science and Technology, Division of Life Science and
Health, Graduate School at Shenzhen, Tsinghua University, Shenzhen.
China) and validated in preliminary trials. siRNA duplexes with
random sequence were used as a negative control
(UUGUACUACACAAAAGUACUG). These RNAs were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The medium was replaced
with a fresh medium containing 100 ng/ml HA after 12 h of
transfection, and cell samples were collected for further
biochemical assays (assaying levels of inflammatory factors and
let-7e) 4 h after treatment with HA. All samples were immediately
stored at −80°C for future biochemical assays.
mRNA reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was extracted from the THP-1 cells
using TRIzol® (Invitrogen Life Technologies) and from
the cell medium using TRIzol LS Reagent (Invitrogen Life
Technologies) according to the manufacturer's instructions. The RNA
was reverse-transcribed to cDNA using a PrimeScript™ 1st Strand
cDNA Synthesis kit (Takara Biotechnology (Dalian) Co., Ltd.,
Dalian, China) with an Alpha™ Unit Block Assembly for DNA EngineH
systems (Bio-Rad, Hercules, CA, USA). The RNA was then amplified
using SYBR Green I dye (Takara Biotechnology (Dalian) Co., Ltd.)
with an ABI PRISM 7300 Real time PCR system (Applied Biosystems,
Foster, CA, USA) according to the manufacturer's instructions. In
addition, the primers for the target genes were synthesized by
Invitrogen Life Technologies, and actin was selected as an internal
control for normalization (Table
I). The fold change was calculated using the 2−ΔΔCt
method of relative quantification.
 | Table IPrimers for tested genes in THP-1
cells. |
Table I
Primers for tested genes in THP-1
cells.
| Gene name | NCBI accession
no. | Primers (5′ to
3′) | Size (bp) |
|---|
| IL1A | NM_000575 | Forward:
ATCATGTAAGCTATGGCCCACT
Reverse: CTTCCCGTTGGTTGCTACTAC | 131 |
| IL1B | NM_000576 | Forward:
CTCGCCAGTGAAATGATGGCT
Reverse: GTCGGAGATTCGTAGCTGGAT | 144 |
| IL6 | NM_000600 | Forward:
ACTCACCTCTTCAGAACGAATTG
Reverse: CCATCTTTGGAAGGTTCAGGTTG | 149 |
| TNFA | NM_000594 | Forward:
CCTCTCTCTAATCAGCCCTCTG
Reverse: GAGGACCTGGGAGTAGATGAG | 220 |
| GAPDH | NM_001256799 | Forward:
GGAGCGAGATCCCTCCAAAAT
Reverse: GGCTGTTGTCATACTTCTCATGG | 197 |
miRNA RT-qPCR
The expression of hsa-let-7e was analyzed using an
miRNA assay kit (Shanghai GenePharma Co., Ltd.) according to the
manufacturer's instructions. miRNA RT-qPCR was performed using the
same systems and machines as mRNA RT-qPCR. The hsa-let-7e was
selected as let-7e can target interleukin (IL)-6 (http://www.targetscan.org/). A previous study
demonstrated that let-7e decreased in the sera of patients with
H7N9 (15). A U6 gene was used as
the internal reference for normalization.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Statistical significance of the data was evaluated by
conducting a one-way analysis of variance using SPSS 13.0 (SPSS,
Inc., Chicago, IL, USA). Newman-Keuls comparison was used to
determine the source of significant difference where appropriate.
P<0.05 was considered to indicate a statistically significant
difference.
Results
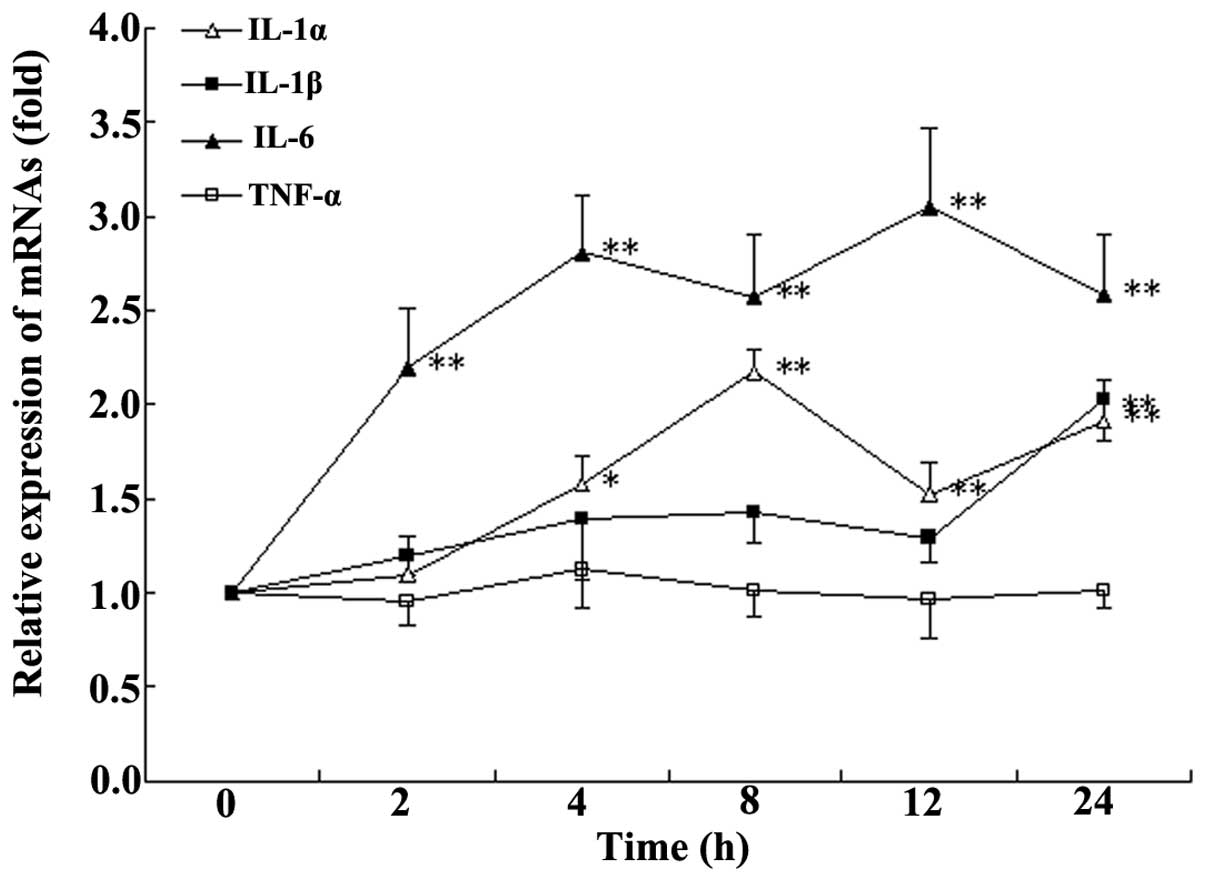
Incubation time with HA
After 4 h of incubation with HA, at a final
concentration of 100 ng/ml, a stable increase in the expression of
IL-1α and IL-6 (Fig. 1) was
observed compared with that at 0 h. Therefore, 4 h incubation was
selected for following experiments.
Effect of HA concentration on cytokine
expression
HA at 10, 100, and 200 ng/ml significantly promoted
the IL-1α and IL-6 expression in THP-1 cells (Fig. 2A). HA also showed a marked increase
in the expression of IL-1β; however, this was not identified to be
significant. HA did not exhibit a significant effect on the
expression of TNF-α. In the following experiment, a concentration
of 100 ng/ml HA was selected for further investigation.
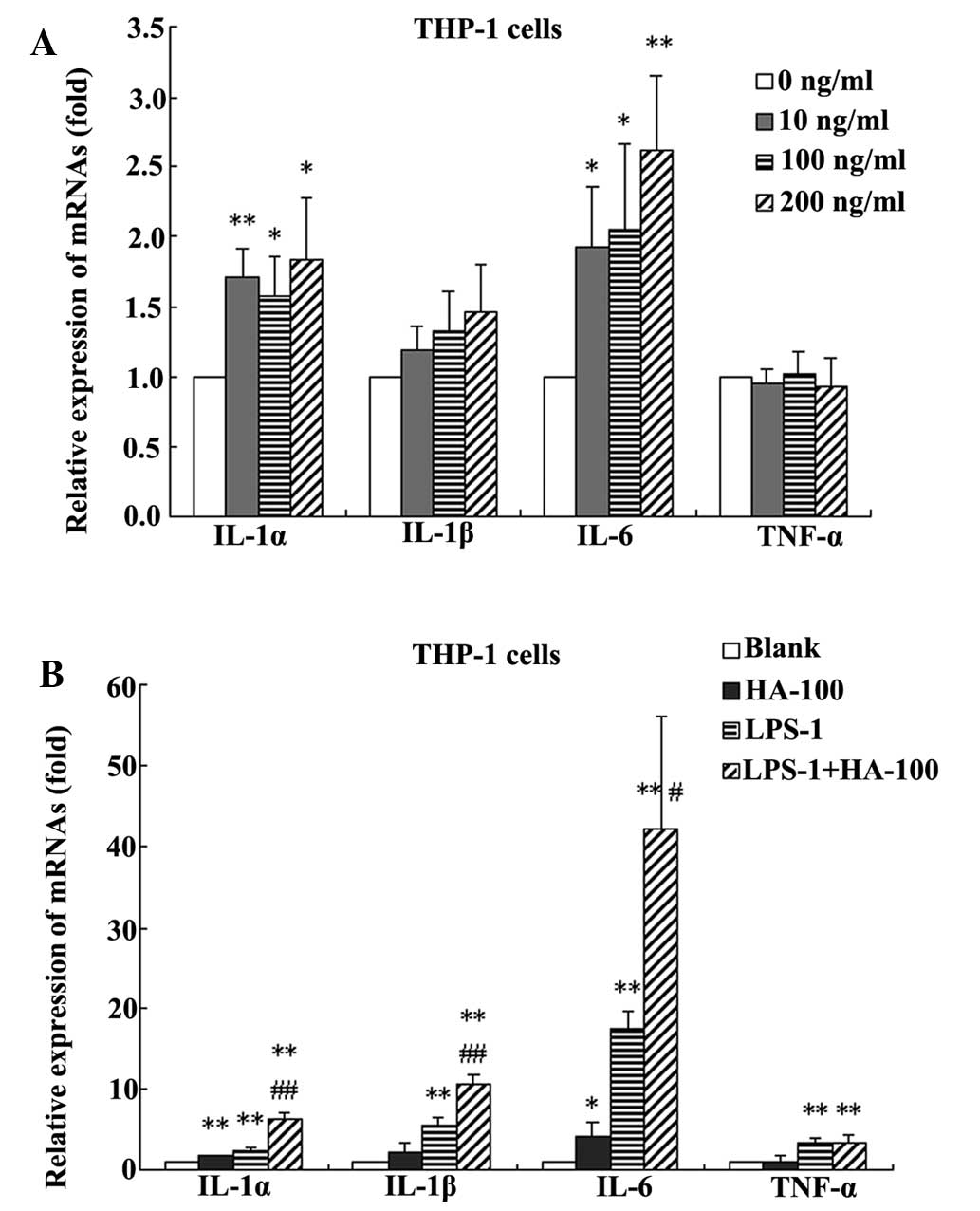 | Figure 2Changes in IL-1α, IL-1β, IL-6 and
TNF-α in (A) THP-1 cells treated with HA at different
concentrations (0, 10, 100 and 200 ng/ml). *P<0.05
and **P<0.01, vs. the 0 ng/ml-treated group. (B)
THP-1 cells treated without HA (blank), with 100 ng/ml HA (HA-100),
with 1 ng/ml LPS (LPS-1) or a combination of the two
(HA-100+LPS-1). *P<0.05, **P<0.01, vs.
the blank group; #P<0.05, ##P<0.01, vs.
the LPS-1 group. Data are expressed as the mean ± standard
deviation (n=4). IL, interleukin; TNF, tumor necrosis factor; HA,
hemagglutinin; LPS, lipopolysaccharide. |
Effect of HA in combination with LPS on
cytokine levels
The synergic effects of HA and LPS on the
proinflammatory factors in THP-1 cells were also observed. Compared
with the untreated control, treatment with 100 ng/ml HA
significantly increased the mRNA expression levels of IL-1α, IL-1β
and IL-6, by 1.6, 2.0 and 4.2 fold, respectively. Treatment with
LPS at 1 ng/ml significantly increased the mRNA expression levels
of IL-1α, IL-1β, IL-6 and TNF-α by 2.3, 5.4, 17.4 and 3.2 fold,
respectively. However, treatment with HA at 100 ng/ml in
combination with LPS at 1 ng/ml significantly increased the mRNA
expression levels of IIL-1α, IL-1β, IL-6 and TNF-α, by 6.3, 10.5,
42.1 and 3.4 fold, respectively. Furthermore, compared with HA
alone, treatment with HA in combination with LPS significantly
increased the mRNA expression levels of IL-1α, IL-1β and IL-6, by
4.0, 5.2 and 10.0 fold, respectively. Compared with LPS alone,
treatment with HA in combination with LPS significantly increased
the mRNA expression levels of IL-1α, IL-1β and IL-6 by 2.8, 2.0 and
2.4 fold, respectively (Fig.
2B).
Effect of HA on TLR4
The mechanisms of upregulation of these
proinflammatory factors by HA remain unclear. TLRs are important
pattern recognition receptors that receptors mediate inflammatory
stress. In the following experiment, it was determined whether HA
resulted in inflammatory stress mediated by the toll-like receptor
(TLR) pathway. siTLR4 was used, and the inhibition efficiency of
the TLR4 mRNA expression was ~37% (Fig. 3). Following transfection with
siTLR4, levels of TLR4, IL-1α, IL-6, and TNF-α significantly
decreased (Fig. 3) compared with
the control.
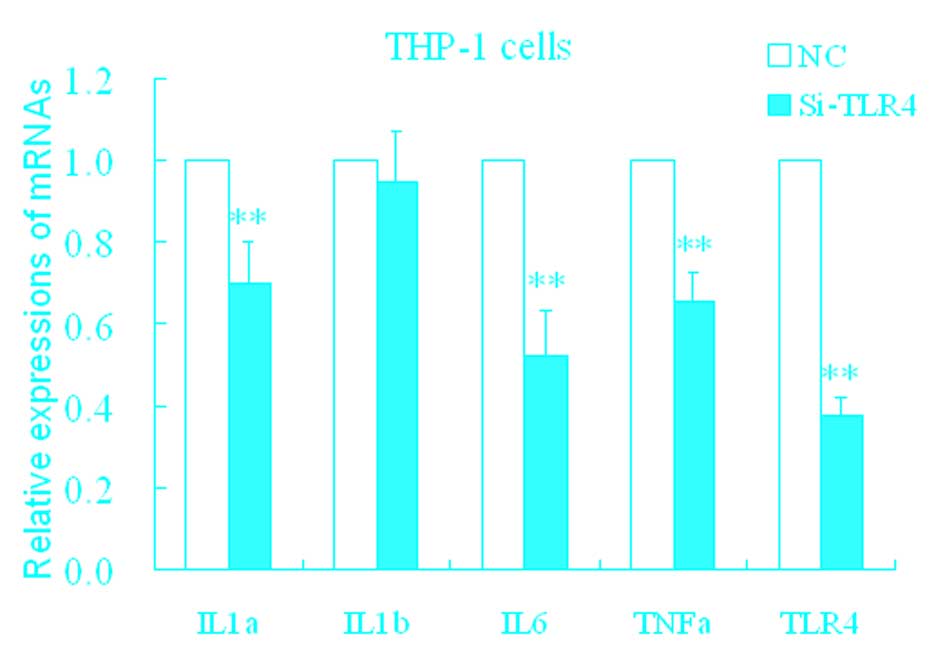 | Figure 3Changes in IL-1α, IL-1β, IL-6, TNF-α
and TLR4 in THP-1 cells treated with siTLR4. *P<0.05
and **P<0.01 vs. NC. Data are expressed as the mean ±
standard deviation (n=4). NC, HA-induced THP-1 cell treated with
negative control; siTLR4, HA-induced THP-1 cell treated with
siTLR4. IL, interleukin; TNF, tumor necrosis factor; TLR4,
toll-like receptor 4; HA, hemagglutinin; si, small interfering. |
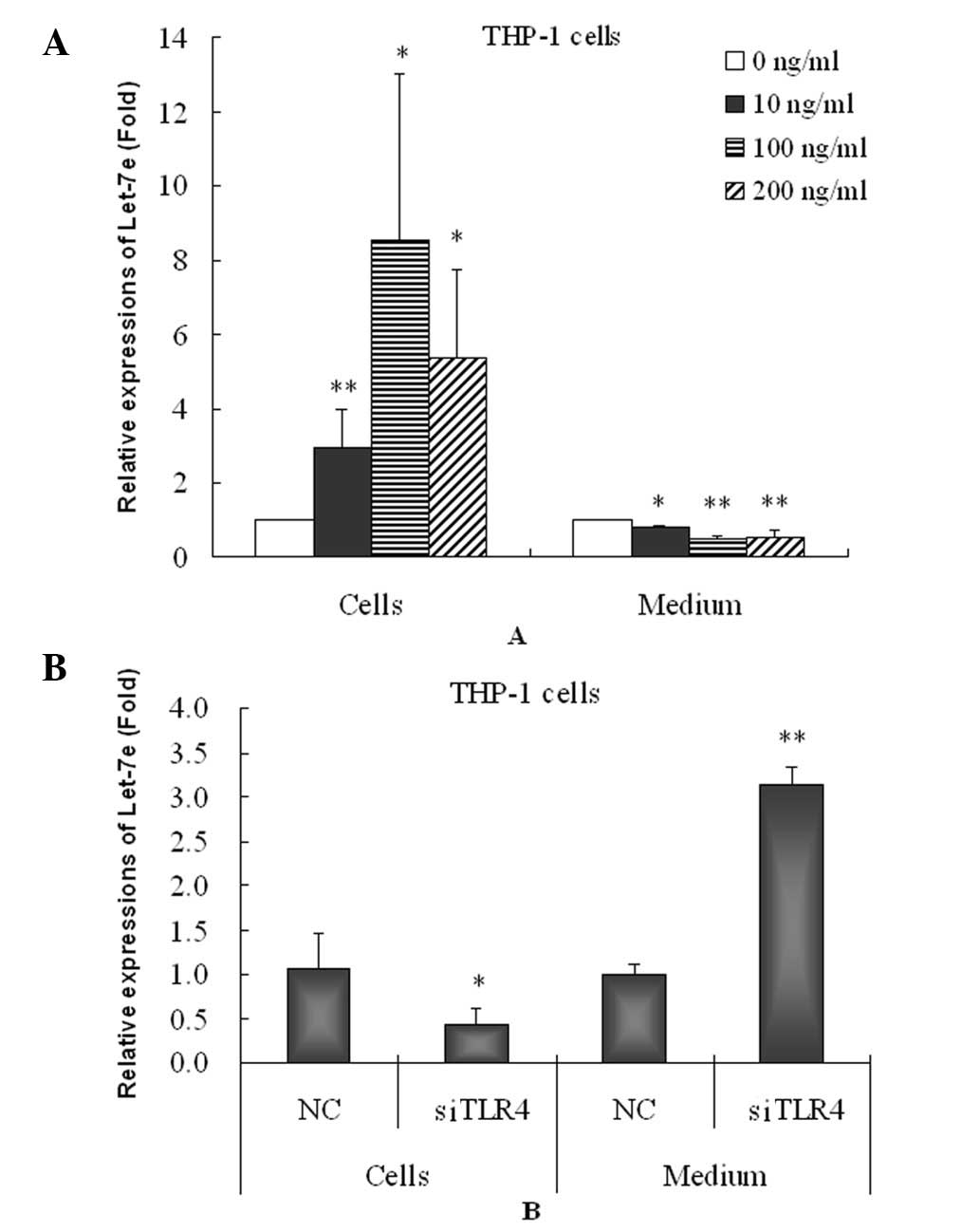
Homeostatic response to inflammatory
stress
Upregulation of proinflammatory factors usually
activates the expression of anti-inflammatory factors and causes
homeostasis responses to inflammatory stress. mRNAs also mediate
the homeostasis response. Let-7 is a type of miRNA that binds to
the 3′-untranslated region of IL-6 mRNA and inhibits IL-6 mRNA and
protein expression. In this study, let-7e was significantly
increased in the HA-induced THP-1 cells (Fig. 4A). However, HA significantly
decreased the let-7e level in THP-1 cell culture medium and thus
may inhibit the secretion of let-7e from the THP-1 cells enabling
the maintenance of high intracellular let-7e levels.
Effect of TLR4 on let-7e
Furthermore, the knockdown of TLR4 caused
significant downregulation of let-7e in the THP-1 cells but
increased the level of let-7e in the cell culture medium (Fig. 4B) compared with negative controls.
These results suggested that knockdown of TLR4 reverses the effects
of HA on let-7. It is likely that the increase in let-7e in the
THP-1 cells and decrease in let-7e in cell culture medium by HA may
be mediated by activating the TLR4 pathway.
Discussion
Molecular mechanisms of the pathogenesis of H7N9
infection remain unclear. By employing high throughput
RNA-sequencing analysis of samples with and without H7N9 viral
infection, Mei et al (16)
found that differentially expressed genes are involved in several
biological pathways associated with immunity and inflammation. In a
study by Wu et al, the majority of H7N9-infected patients
presented with systemic inflammatory response syndrome (10). The Th2-type inflammation in
H7N9-infected patients with pre-existing chronic diseases likely
contributes to the pathogenesis of H7N9 infection; this
inflammation is correlated with poor clinical outcomes (17). In the LA-4 mouse lung cell line,
CK1 (an H7N9 virus isolated from chickens) also induced high levels
of IL-6 and cyclooxygenase-2 mRNA (18). In the present study, it was
demonstrated that HA from H7N9 can induce an inflammatory response
in THP-1 cells; HA is likely an important pathological protein
component of H7N9. In addition, HA and LPS exhibited a synergic
effect on the promotion of expression of the inflammatory factors.
The results suggested that combined H7N9 viral and bacterial
infections may elicit enhanced inflammatory responses, compared
with virus or bacterial infections alone.
Despite this, inflammatory stress can cause
anti-inflammatory homeostasis response. Patients with avian H7N9
virus infection display intense systemic inflammatory response
syndrome that is concomitant with an anti-inflammatory response
(10). Anti-inflammatory
mechanisms include increased anti-inflammatory factor (IL-4 and
IL-10) levels (19). miRNAs also
regulate inflammatory responses and mediate anti-inflammatory
processes (20,21); for example, Let-7 can target IL-6
and inhibit IL-6 expression (22).
In this study, the inflammatory response was associated with
increased expression of let-7e. Increased let-7e may be associated
with a homeostatic response to the inflammatory stimulus induced by
HA since let-7e was predicted to target IL-6 and inhibit its
production.
TLRs are important pathogen-recognition receptors in
epithelial immune responses to microbial infection (23). TLRs are involved in the early
innate viral inhibition of naturally occurring influenza (24). TLRs can activate inflammatory
responses (25). Interaction of HA
of the influenza A viruses (IAV) with the cell surface is a key
factor for virus entry and productive infection of the cell
(26). HA proteins from H5N1 and
2009 H1N1 viruses were able to induce dendritic cell (DC)
activation of murine DCs in a MyD88/TLR4 dependent manner (27). Nucleic acid-recognizing receptors
(mainly RIG-I and TLRs) are the most extensively investigated
pattern recognition receptors for IAVs (28). In this study, HA proteins from H7N9
appeared to not only activate inflammatory responses but also cause
miRNA homeostasis to this inflammatory stress, dependent on the
TLR4 pathway.
In addition, miRNAs can be transported from donor
cells to recipient cells; these RNAs (i.e., let-7b) function in a
novel manner as ligands of TLRs (29). The structure of let-7e is similar
to let-7b. In this study, the HA of H7N9 inhibited the secretion of
let-7e. This effect may be useful in preventing an excessive
inflammatory response. Zhu et al demonstrated that patients
infected with H7N9 showed decreased let-7e in the sera (15), indicating that cells of these
patients attempted to trigger a protective response to avoid severe
inflammatory damage in specific organs or tissues. A number of the
H7N9-infected patients did not exhibit obvious systemic
inflammation compared with the healthy controls (17), although the majority of the
H7N9-infected patients (~74%) presented with systemic inflammatory
response syndrome (10). However,
the mechanisms that mediated the inhibitory effect on let-7
secretion by HA remain unclear although its secretion was also
dependedent on the TLR4 pathway.
In conclusion, this study showed that HA
significantly promoted the expression of proinflammatory factors,
such as IL-1α and IL-6. HA also produced a homeostatic response to
inflammatory stress by regulating let-7e production and secretion.
The inflammatory stress and miRNA homeostatic responses were
mediated by TLR4 mechanisms. The present study provided evidence
that HA, one of the most important proteins of the H7N9 virus,
directly causes the inflammatory response of immune cells in
humans, but simultaneously triggers compensatory protective
responses via miRNA mechanisms. Furthermore, the present study also
demonstrated that the interaction between miRNAs and mRNAs may be
regulated by immune cell signaling pathways. Therefore, host cells
have a delicate biological network associating miRNAs and mRNAs,
which protects against the invasion of H7N9 viruses. The results of
the present study may prove useful in understanding the potential
pathological mechanisms underlying H7N9 infection, or to identify
novel biomarkers for H7N9 virus infection. To further elucidate the
interaction between miRNAs and mRNAs against the invasion of H7N9
viruses, future studies are required to identify more functional
proteins of the H7N9 virus, or miRNAs in host cells.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81373460
and 81072680), the Shenzhen Science and Technology R&D
Foundation (grant nos. SGLH20121008144756945 and ZYC201105170341A),
the Natural Science Foundation of Guangdong Province (grant no.
2014A030313744), the China Scholarship Council (grant no.
201308440130), the ITF Grant (grant no. GHX/002/12SZ) and the Key
Lab Promotion Grant of Shenzhen (grant no.
ZDSY20120619141025668).
Abbreviations:
|
AIVs
|
avian influenza viruses
|
|
HA
|
hemagglutinin
|
|
IL-1α
|
interleukin 1 α
|
|
IL-1β
|
interleukin 1 β
|
|
IL-6
|
interleukin 6
|
|
TLRs
|
toll-like receptors
|
|
TNF-α
|
tumor necrosis factor-α
|
References
|
1
|
Hayward AC, Fragaszy EB, Bermingham A,
Wang L, Copas A, Edmunds WJ, Ferguson N, Goonetilleke N, Harvey G,
Kovar J, et al: Comparative community burden and severity of
seasonal and pandemic influenza: Results of the Flu watch cohort
study. Lancet Respir Med. 2:445–454. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Le TH and Nguyen NT: Evolutionary dynamics
of highly pathogenic avian influenza A/H5N1 HA clades and vaccine
implementation in Vietnam. Clin Exp Vaccine Res. 3:117–127. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alamares-Sapuay JG, Martinez-Gil L, Stertz
S, Miller MS, Shaw ML and Palese P: Serum- and
glucocorticoid-regulated kinase 1 is required for nuclear export of
the ribonucleoprotein of influenza A virus. J Virol. 87:6020–6026.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Casalegno JS, Ferraris O, Escuret V,
Bouscambert M, Bergeron C, Linès L, Excoffier T, Valette M, Frobert
E, Pillet S, et al: Functional balance between the hemagglutinin
and neuraminidase of influenza a (H1N1) pdm09 HA D222 variants.
PLoS One. 9:e1040092014. View Article : Google Scholar
|
|
5
|
Miotto O, Heiny AT, Albrecht R,
García-Sastre A, Tan TW, August JT and Brusic V: Complete-proteome
mapping of human influenza a adaptive mutations: Implications for
human transmissibility of zoonotic strains. PLoS One. 5:e90252010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Houng HS, Garner J, Zhou Y, Lyons A,
Kuschner R, Deye G, St Clair K, Douce RW, Chicaiza W, Blair PJ, et
al: Emergent 2009 influenza A (H1N1) viruses containing HA D222N
mutation associated with severe clinical outcomes in the Americas.
J Clin Virol. 53:12–15. 2012. View Article : Google Scholar
|
|
7
|
Wang B, Dwyer DE, Soedjono M, Shi H,
Matlho K, Ratnamohan M, Blyth C, McPhie K, Cunningham AL and
Saksena NK: Evidence of the circulation of pandemic influenza
(H1N1) 2009 with D222D/G/N/S hemagglutinin polymorphisms during the
first wave of the 2009 influenza pandemic. J Clin Virol.
52:304–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dai J, Zhou X, Dong D, Liu Y, Gu Q, Zhu B,
Wu C and Cai H: Human infection with a novel avian-origin influenza
A (H7N9) virus: Serial chest radiographic and CT findings. Chin Med
J (Engl). 127:2206–2211. 2014.
|
|
9
|
Ren L, Yu X, Zhao B, Wu F, Jin Q, Zhang X
and Wang J: Infection with possible precursor of avian influenza A
(H7N9) virus in a child, China, 2013. Emerg Infect Dis.
20:1362–1365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu W, Shi Y, Gao H, Liang W, Sheng J and
Li L: Immune derangement occurs in patients with H7N9 avian
influenza. Crit Care. 18:R432014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Marin-Muller C, Bharadwaj U, Chow
KH, Yao Q and Chen C: MicroRNAs: Control and loss of control in
human physiology and disease. World J Surg. 33:667–684. 2009.
View Article : Google Scholar
|
|
12
|
Zhu S, Pan W and Qian Y: MicroRNA in
immunity and autoimmunity. J Mol Med (Berl). 91:1039–1050. 2013.
View Article : Google Scholar
|
|
13
|
Rebane A and Akdis CA: MicroRNAs:
Essential players in the regulation of inflammation. J Allergy Clin
Immunol. 132:15–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie W, Li Z, Li M, Xu N and Zhang Y:
miR-181a and inflammation: miRNA homeostasis response to
inflammatory stimuli in vivo. Biochem Biophys Res Commun.
430:647–652. 2013. View Article : Google Scholar
|
|
15
|
Zhu Z, Qi Y, Ge A, Zhu Y, Xu K, Ji H, Shi
Z, Cui L and Zhou M: Comprehensive characterization of serum
microRNA profile in response to the emerging avian influenza A
(H7N9) virus infection in humans. Viruses. 6:1525–1539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mei B, Ding X, Xu HZ and Wang MT: Global
gene expression changes in human peripheral blood after H7N9
infection. Gene. 551:255–260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han J, Zhang N, Zhang P, Yang C, Jin M,
Yang J, Wu X, Liu G, Ji L, Zhang C, et al: Th2-type inflammation
under conditions of pre-existing chronic disease is associated with
liver damage in patients with avian influenza H7N9 virus. Microbes
Infect. 16:672–677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Li C, Zhang AJ, To KK, Lee AC, Zhu
H, Wu HW, Chan JF, Chen H, Hung IF, et al: Avian influenza a H7N9
virus induces severe pneumonia in mice without prior adaptation and
responds to a combination of zanamivir and COX-2 inhibitor. PLoS
One. 9:e1079662014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asseman C and von Herrath M: Regulation of
viral and autoimmune responses. Novartis Found Symp. 252:239–253;
discussion 253–267. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie W, Li M, Xu N, Lv Q, Huang N, He J and
Zhang Y: MiR-181a regulates inflammation responses in monocytes and
macrophages. PLoS One. 8:e586392013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh RP, Massachi I, Manickavel S, Singh
S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH and
Rehimi H: The role of miRNA in inflammation and autoimmunity.
Autoimmun Rev. 12:1160–1165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iliopoulos D, Hirsch HA and Struhl K: An
epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA and
IL6 links inflammation to cell transformation. Cell. 139:693–706.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen XM, Splinter PL, O'Hara SP and
LaRusso NF: A cellular micro-RNA, let-7i, regulates toll-like
receptor 4 expression and contributes to cholangiocyte immune
responses against Cryptosporidium parvum infection. J Biol Chem.
282:28929–28938. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee N, Wong CK, Hui DS, Lee SK, Wong RY,
Ngai KL, Chan MC, Chu YJ, Ho AW and Lui GC: Role of human Toll-like
receptors in naturally occurring influenza A infections. Influenza
Other Respir Viruses. 7:666–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gribar SC, Anand RJ, Sodhi CP and Hackam
DJ: The role of epithelial toll-like receptor signaling in the
pathogenesis of intestinal inflammation. J Leukoc Biol. 83:493–498.
2008. View Article : Google Scholar
|
|
26
|
Ramos I and Fernandez-Sesma A: Cell
receptors for influenza a viruses and the innate immune response.
Front Microbiol. 3:1172012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu WC, Lin SC, Yu YL, Chu CL and Wu SC:
Dendritic cell activation by recombinant hemagglutinin proteins of
H1N1 and H5N1 influenza A viruses. J Virol. 84:12011–12017. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
García-Sastre A: Induction and evasion of
type I interferon responses by influenza viruses. Virus Res.
162:12–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: microRNAs are ligands of Toll-like receptors. RNA. 19:737–739.
2013. View Article : Google Scholar : PubMed/NCBI
|