Introduction
Enzootic nasal tumor (ENT) is caused by the enzootic
nasal tumor virus (ENTV) and is a chronic, progressive, sexually
transmitted disease. Excluding Australia and New Zealand, ENT has
spread throughout the majority of countries which keep goats
(1). At present, an effective
method for the early diagnosis of ENT remains to be developed, and
affected goats are only removed subsequent to the appearance of
symptoms (1). Due to the fact that
goats with ENT cannot be distinguished from those which are
healthy, disease spreads in the group, causing a greater number of
goats to become infected, which can threaten the security of the
population. An increasing number of studies have demonstrated that
abnormal DNA methylation is closely associated with the presence of
neoplasms as well as autoimmune and neurodegenerative diseases
(1–3). DNA methylation involves the addition
of a methyl group to the carbon of the cytosine pyrimidine ring in
5-position via the action of DNA methyltransferase (DNMT) (2–4).
Abnormal DNA methylation often leads to tumor formation, which is
closely associated with the occurrence of cancer (5). Investigating the promoter methylation
status and mRNA expression of goat tumor-associated genes and DNMT
genes in primary tumors in ENT can provide a foundation for
screening of tumor-specific markers for early diagnosis of ENT and
further investigations into the epigenetic mechanisms of
ENTV-induced nasal epithelial cell carcinoma.
Materials and methods
Tissue specimens
A total of 24 nasal tumor tissue (from goats with
ENT) specimens and 20 normal nasal epithelial tissue specimens from
healthy animals were obtained from Yiyou farms (Sichuan, China).
The animals ~1 year old and were sacrificed by the owner prior to
sample collection. The present study was approved by the National
Institute of Animal Health Animal Care and Use Committee at Sichuan
Agricultural University (Ya'an, China; approval no. 2010–020).
Quarantine of control groups
A pair of primers were designed and synthesized
based on the reported genomic RNA sequence of ENTV and the genetic
variation range of gag. Sequences of the polymerase chain reaction
(PCR) primers were: gag (850 bp) forward, 5′-TTCCTCGCCACTACTCTTG-3′
and reverse, 5′-AGTCGCTGTGCTTGTTTCA-3 (6,7).
According to the manufacturer's instructions, RNA was extracted,
then reverse transcription and PCR amplification were conducted.
Amplified PCR products were evaluated by 2% agarose gel [Tiangen
Biotech (Beijing) Co., Ltd., Beijing, China] electrophoresis.
mRNA expression of DNMT
Total RNA from nasal tumor tissues and normal nasal
epithelial tissues was isolated using the RNeasy Micro kit
(Sigma-Aldrich Shanghai Trading Co., Ltd., Shanghai, China) and the
quantities were determined spectrophotometrically (722n; Shanghai
Jinghua Science & Technology Instruments Co., Ltd., Shanghai,
China). First-strand cDNA was synthesized from 2 µg total RNA using
the RevertAid kit (Thermo Fisher Scientific, Waltham, MA, USA). The
PCR conditions included an initial cDNA synthesis reaction at 42°C
for 1 h using the RevertAid kit, followed by traditional PCR
amplification. The positive standard was prepared according to the
conventional methods and the recombinant plasmids were sent to a
sequencing facility (Shanghai Invitrogen Biotechnology Co., Ltd.,
Shanghai, China). Subsequent to sequencing, the results were
analzyed using the Basic Local Alignment Search Tool algorithm
(BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared
with the corresponding subtypes.
Optimal reaction conditions of reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) were
used. The RT-qPCR was performed on a CFX96 Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A total of
2 µl positive standard, 12.5 µl SYBR® Premix Ex Taq™ II
(Tli RNase H Plus; 2X) (Takara Biotechnology Co., Ltd., Dalian,
China), 1 µl each of 10 µM forward and reverse primers, and 8.5 µl
dH2O were added to each reaction well. The PCR
conditions included a denaturation step for 30 sec at 95°C and 40
cycles of 5 sec at 95°C and 30 sec at a temperature gradient from
55–64°C. Following the last cycle, melting curve analysis was
performed at 65-95°C with an increment of 0.5°C/sec. The
housekeeping gene 18S-rRNA was used as an internal control. The
sequences of the PCR primers were as follows: DNMT3a (191 bp)
forward, 5′-ACGGAGAAGCCTAAGGTCAAG-3′ and reverse,
5′-TGACGAAGGACCTTACGCG-3′; DNMT3b (212 bp) forward,
5′-CAGACAGCACCGAGTATCAGG-3′ and reverse, 5′-CAACCACGTAACCCTAAC-3′;
DNMT1 (176 bp) forward, 5′-CCCCAGGATTACAAGGAAG-3′ and reverse,
5′-CTGGATGTAACTCGACGTCTCT-3′; MGMT (101 bp) forward,
5′-CAACCCTATTCCCATCCTCATCC-3′ and reverse,
5′-GCACTTCCTCACCGACGACC-3′; 18S-rRNA (198 bp) forward,
5′-GAGAAACGGCTACCACAT C-3′ and reverse, 5′-GCTATTGGAGCTGGAATTAC-3′.
The optimal annealing temperature of each gene was determined using
the Ct values, the highest fluorescence value and melting curve
analysis.
The standard and melting curves were established
using RT-qPCR amplification of the positive standard with a
multiple proportion dilution (1×101~1×106
copies/µl). RT-qPCR amplified the positive standard with an optimal
annealing temperature. The experiments were repeated three times to
evaluate the repeatability and reproducibility. The data were
analyzed using the 2−∆∆Ct method (8) and SPSS, version 13.0 (SPSS, Inc.,
Chicago, IL, USA).
mRNA expression of goat tumor-associated
genes
For detection of tumor-associated genes, the same
methods were used as for detecting the expression of DNMT. The
sequences of the PCR primers used were as follows: P73 (219 bp)
forward, 5′-CCCTCCAACACCGACTACCC-3′ and reverse,
5′-GGTCACATGCTCCGCCTTCTTAT-3′; P53 (148 bp) forward,
5′-CCACCATCCACTACAACTTCA-3′ and reverse,
5′-CCAGGACAGGCACAAACACG-3′; KRAS (198 bp) forward,
5′-GGCTCAGGACTTAGCAAGAA-3′ and reverse, 5′-CGTCAACACCCAGATTACAT-3′;
NRAS (118 bp) forward, 5′-TCAGTGAGCCAATTAGCATC-3′ and reverse,
5′-TAACCGACTTCTTTCCTTGC′; GADD45G (325 bp) forward,
5′-ATCTCACCACCTCTTGCTCG-3′ and reverse, 5′-AGTCGTTGACGCTGCGGCTC-3′;
EGFR (254 bp) forward, 5′-TGTGACTTGCGTTGATAGAA-3′ and reverse,
5′-CGGCGTTGCTGCGTGAAT-3′; CHFR (202 bp) forward,
5′-AACAAGAGCCATCAACCG-3′ and reverse, 5′-AGGAGATGAGCACCGAAG-3′;
C-myc (136 bp) forward, 5′-ACATCCTGTCGGTCCAAGCA-3′ and reverse,
5′-CCTCCCTCCAATAGGTCAAT-3′; THBS1 (354 bp) forward,
5′-CTGTGCGGGCGGAGAAAGGT-3′ and reverse,
5′-TGCTGAGTCTGGCGATGCTG-3′.
Assessment of the promoter methylation
status of goat tumor-associated genes
Total DNA from nasal tumor tissues, normal nasal
epithelial tissues and peripheral-blood cells was isolated using
the Mammalian Genomic DNA Extraction kit (Takara Biotechnology Co.,
Ltd.) and the quantities were determined spectrophotometrically.
The CpG island of peripheral-blood cell DNA was modified using CpG
methyltransferase (Zymo Research, Irvine, CA, USA) and used as the
positive control template of methylation following modification.
The DNA of nasal tumor tissues, normal nasal epithelial tissues,
peripheral blood cells and the DNA modified by CpG
methyltransferase was modified using the EZ DNA Methylation-Gold™
Kit (Zymo Research). The DNA of peripheral blood cells was used as
unmethylated positive-control template following modification.
A total of 2 µl DNA sample, 10 µl EmeraldAmp PCR
Master Mix (2xPremix) (Takara Biotechnology Co., Ltd.), 0.5 µl each
of 20 µM forward and reverse primers and 7 µl dH2O was
added to each reaction well. Amplifications were performed using
the following methylation-specific PCR (MSP) conditions: 95°C for 5
min, 35 cycles of 95°C for 30 sec, 57°C (methylation, M) or 55°C
(unmethylation, U) for 45 sec, and 30 sec at 72°C. Following the
last cycle, a final extension was performed at 72°C for 10 min. The
sequences of the MSP primers were as follows: GADD45G (M) (134 bp)
forward, 5′-CGGGGTTTTTTTATTTTATTTAGC-3′ and reverse,
5′-GACTCGTTTACATTTCTATACCGAA-3′; GADD45G (U) (134 bp) forward,
5′-TGG,GGTTTTTTTATTTTATTTAGTG-3′ and reverse,
5′-AACTCATTTACATTTCTATACCAAA-3′; P73 (M) (274 bp) forward,
5′-TAGTGAATTTTATTTTTTAGTTCGT-3′ and reverse,
5′-AACCTAAAACTCTAAATTACTCCG C-3′; P73 (U) (273 bp) forward,
5′-TAGTGAATTTTATTTTTTAGTTTGT-3′ and reverse,
5′-ACCTAAAACTCTAAATTACTCCACC-3′; CEBPA (M) (165 bp) forward,
5′-ATGGGTTTGTTTTTTGTTTTTTTC-3′ and reverse,
5′-ACGTAATTACTACCTCTCCCAACG-3′; CEBPA (U) (166 bp) forward,
5′-ATGGGTTTGTTTTTTGTTTTTTTT-3′ and reverse,
5′-CACATAATTACTACCTCTCCCAACAA-3′; KRAS (M) (184 bp) forward,
5′-ATTTTTGTTAATTGTTTTGGAGAAC-3′ and reverse,
5′-TACTCTATATCCTATAAACCCACC G-3′; KRAS (U) (180 bp) forward,
5′-TTTTGTTAATTGTTTTGGAGAATGA-3′ and reverse,
5′-CTCTATATCCTATAAACCCACCACA-3′; NRAS (M) (149 bp) forward,
5′-TTTTTTTTATTTTTTCGTATTCGA-3′ and reverse,
5′-TCTACTCCTCTTAAAACATTACGCT-3′; NRAS (U) (150 bp) forward,
5′-TTTTTTTATTTTTTTGTATTTGA-3′ and reverse,
5′-TTCTACTCCTCTTAAAACATTACACT-3′; P53 (M) (198 bp) forward,
5′-TGTATAGTATATAATGAGATATTGTATTC-3′ and reverse,
5′-AAAATTTATACCTTTTTACTACCTTCGTC-3′; P53 (U) (197 bp) forward,
5′-GTATAGTATATAATGAGATATTGTATTTGT-3′ and reverse,
5′-AAAATTTATACCTTTTTACTACCTTCATC-3′; EGFR (M) (172 bp) forward,
5′-TAATTTTAGGTTTGAAGGGGTAGC-3′ and reverse,
5′-CAACACAAAACACAAAATAATAAACG; EGFR (U) (169 bp) forward,
5′-TAATTTTAGGTTTGAAGGGGTAGTG-3′ and reverse,
5′-CACAAAACACAAAATAATAAACACC-3′; CHFR (M) (213 bp) forward,
5′-TTGCGATGTTTGTTGATAGTAGC-3′ and reverse,
5′-GTCCGATAAAACCCTAACCG-3′; CHFR (U) (219 bp) forward,
5′-GATTTGTGATGTTTGTTGATAGTAGTG-3′ and reverse,
5′-AACATCCAATAAAACCCTAACCAC-3′; THBS1 (M) (185 bp) forward,
5′-TTTAGTCGTAGTTTTCGAATTTAC G-3′ and reverse,
5′-ATAACTAACCAAAACAACACTCGTC-3′; THBS1 (U) (183 bp) forward,
5′-TTAGTTGTAGTTTTTGAATTTATGG-3′ and reverse,
5′-TAACTAACCAAAACAACACTCATC-3′; C-myc,(M) (151 bp) forward,
5′-TTGAAATTTTAGGTTTTTGAGATCG-3′ and reverse,
5′-CGTAATAAAAAAACTCATCCACGTA-3′; C-myc (U) (151 bp) forward,
5′-TGAAATTTTAGGTTTTTGAGATTGG-3′ and reverse,
5′-CATAATAAAAAAACTCATCCACATA-3′. Amplified PCR products were
evaluated by 2% agarose gel electrophoresis.
Results
Quarantine of control groups
The results of the PCR amplification of the gag gene
were all negative (Fig. 1). This
illustrates the fact that the samples of the control group were
uninfected.
PCR amplification of DNMTs and MGMT
The results of the agarose gel electrophoresis were
as expected (Fig. 2), and the
results of DNA sequencing were identical to those of the reference
sequence (National Center for Biotechnology Information, Bethesda,
MA, USA). The absorbance (A)260/A280 nm value of purified plasmid
DNA was between 1.8 and 2.0, which conformed to the requirement of
follow-on experiments.
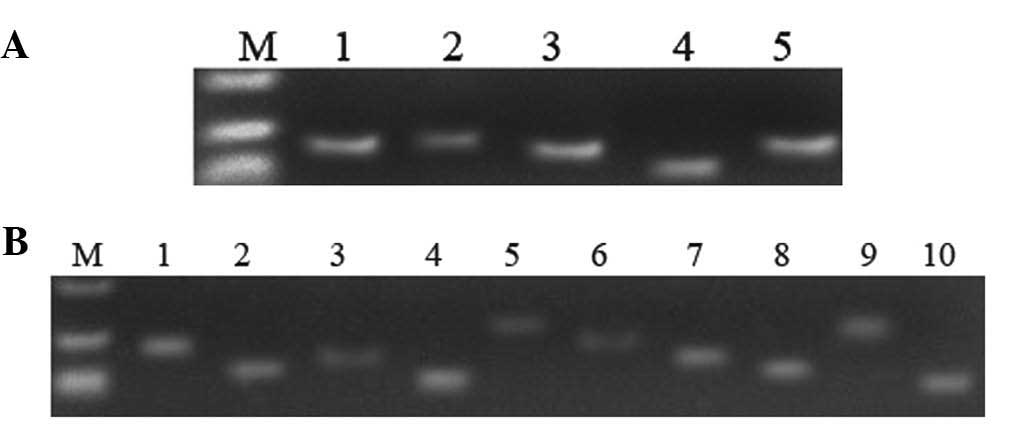 | Figure 2Electrophoretic analysis of
recombinant plasmids amplified by conventional polymerase chain
reaction in cancerous tissue samples. (A) Lane 1, DNMT3a; 2,
DNMT3b; 3, DNMT1; 4, MGMT; 5, 18S-Rrna; 6, negative control; M,
DL2000 marker. (B) Lane 1, P73; 2, P53; 3, KRAS; 4, NRAS; 5,
GADD45G; 6, EGFR; 7, CHFR; 8, C-myc; 9, THBS1; 10, CEBPA; M, DL2000
marker. |
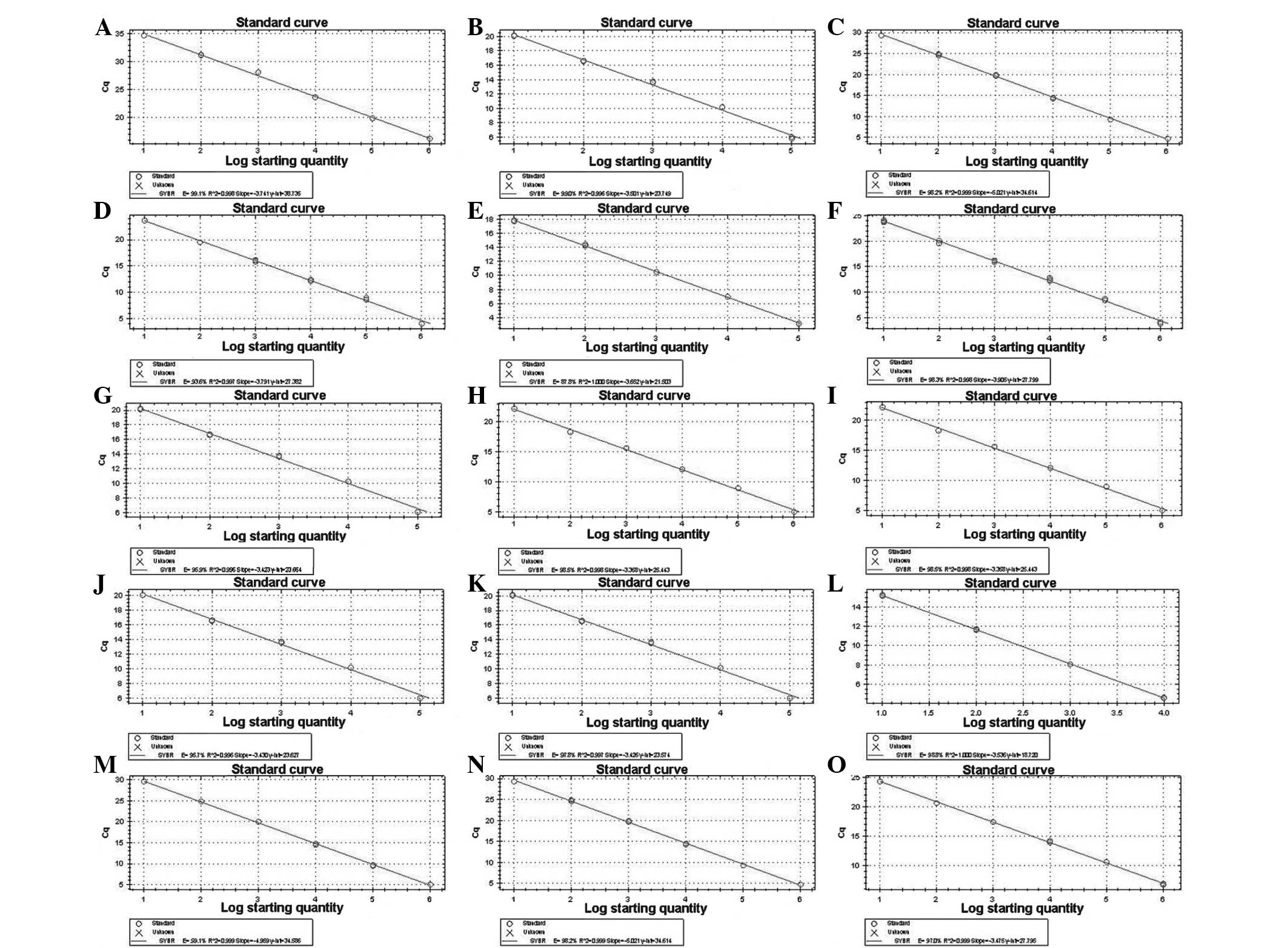
Establishment of standard curves
The standard and melting curves were established by
RT-qPCR amplification of the positive standard with a multiple
proportion dilution (1×101–1×106 copies/µl).
According to the Ct values of each gene with different dilution
degrees, standard curves (Fig. 3)
and amplification standard curves (Fig. 4) were produced. As demonstrated in
Fig. 4, the correlation
coefficient was >0.999 and the amplification efficiency was ~1,
which conformed to the requirement of follow-on experiments.
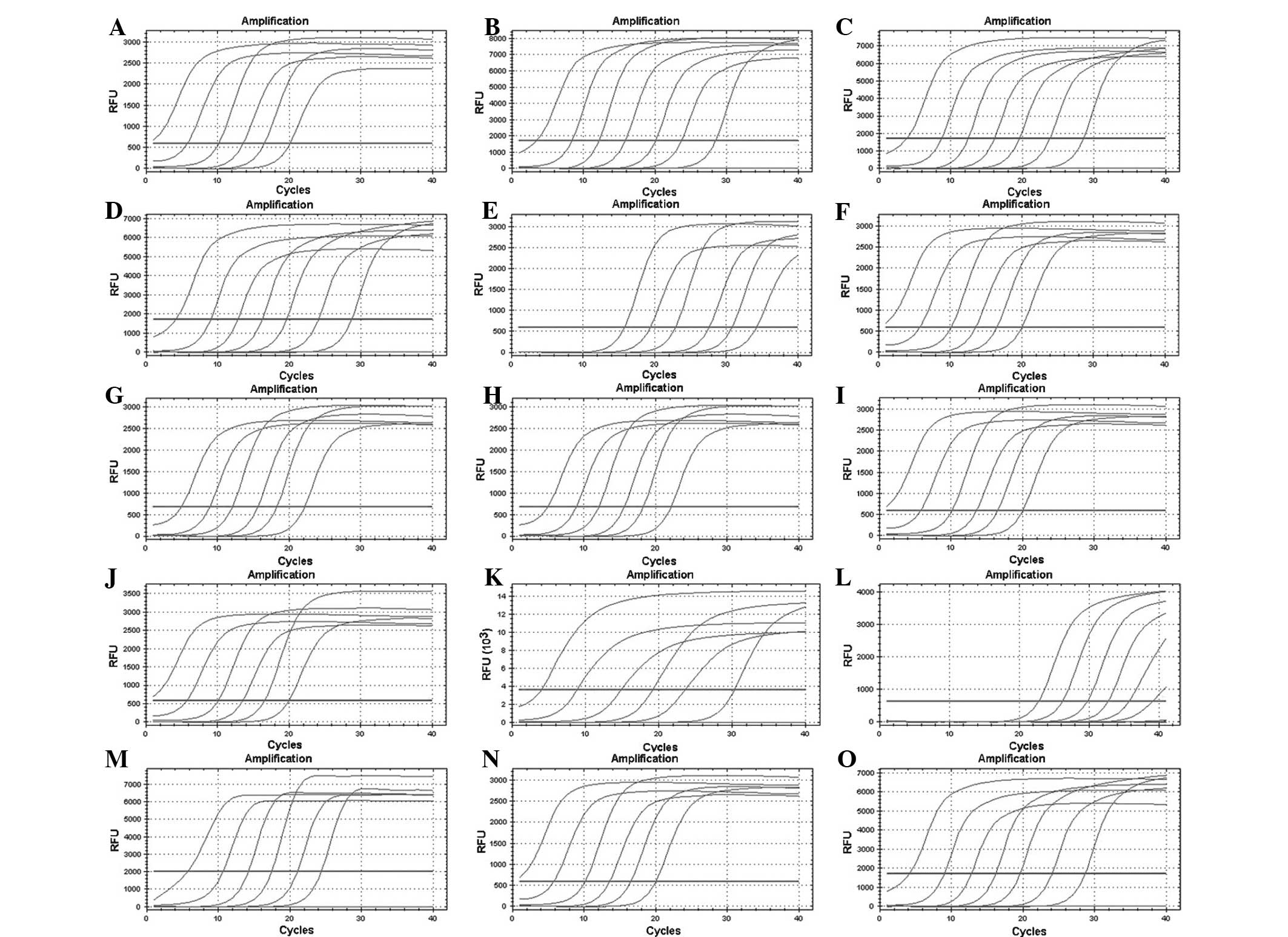
Sensitivity, specificity and
repeatability of RT-qPCR
Based on the electrophoretic analysis of PCR
products (Fig. 2) and the PCR
melting curve (Fig. 5), it was
concluded that the method had a good specificity as no primer
dimers and no additional peaks in the melting curves were observed.
The minimum detection concentration was 100 copies/µl when detected
with a standard sample of a 10X gradient dilution (Fig. 3). The intra-assay variation
coefficient ranged between 0.13 and 1.05% and the inter-assay
variation coefficient was between 0.67 and 3.29%, which suggested
that the experiments had improved repeatability (Tables I and II).
 | Table IIntra-assay reproducibility of DNMT
and tumor-associated genes. |
Table I
Intra-assay reproducibility of DNMT
and tumor-associated genes.
| Gene | Plasmid concentration
(ng) | Intra-assay data
|
|---|
| Repeat 1 | Repeat 2 | Repeat 3 | Mean ± SD | Variation coefficient
(%) |
|---|
| 18S-rRNA | 59.33 | 14.52 | 14.37 | 14.25 | 14.38±0.11 | 0.76 |
| DNMT1 | 36.01 | 22.02 | 21.85 | 21.99 | 21.95±0.074 | 0.38 |
| DNMT3a | 17.67 | 17.58 | 17.32 | 17.53 | 17.48±0.11 | 0.63 |
| DNMT3b | 12.67 | 34.56 | 34.30 | 34.27 | 34.38±0.13 | 0.38 |
| MGMT | 28.33 | 20.85 | 21.29 | 20.86 | 21.00±0.21 | 0.01 |
| P73 | 19.01 | 15.37 | 15.25 | 15.52 | 15.38±0.11 | 0.72 |
| P53 | 13.06 | 21.57 | 21.33 | 21.53 | 21.48±0.11 | 0.51 |
| KRAS | 27.33 | 24.04 | 23.87 | 23.98 | 23.96±0.075 | 0.31 |
| NRAS | 13.67 | 18.35 | 18.23 | 18.51 | 18.36±0.10 | 0.54 |
| GADD45G | 33.01 | 19.47 | 19.84 | 19.94 | 19.75±0.18 | 0.91 |
| EGFR | 32.09 | 23.94 | 23.83 | 23.92 | 29.90±0.18 | 0.60 |
| CHFR | 14.09 | 21.85 | 21.29 | 21.86 | 21.67±0.12 | 0.55 |
| C-myc | 17.81 | 17.08 | 16.96 | 17.19 | 17.08±0.094 | 0.55 |
| THBS1 | 18.09 | 25.39 | 25.44 | 25.36 | 25.40±0.033 | 0.13 |
| CEBPA | 17.08 | 21.29 | 20.85 | 20.86 | 20.00±0.21 | 1.05 |
 | Table IIInter-assay reproducibility of DNMT
and tumor-associated genes. |
Table II
Inter-assay reproducibility of DNMT
and tumor-associated genes.
| Gene | Plasmid
concentration (ng) | Intra-assay data
|
|---|
| Repeat 1 | Repeat 2 | Repeat 3 | Mean ± SD | Variation
coefficient (%) |
|---|
| 18S-rRNA | 59.33 | 13.48 | 14.85 | 14.92 | 14.42±0.19 | 1.32 |
| DNMT1 | 36.01 | 21.95 | 22.83 | 23.40 | 22.73±0.59 | 2.60 |
| DNMT3a | 17.67 | 17.51 | 17.79 | 17.10 | 17.47±0.28 | 1.60 |
| DNMT3b | 12.67 | 34.38 | 34.03 | 34.94 | 34.45±0.64 | 1.86 |
| MGMT | 28.33 | 20.40 | 21.85 | 21.80 | 21.35±0.67 | 3.14 |
| P73 | 19.01 | 21.94 | 20.38 | 21.03 | 21.11±0.64 | 3.03 |
| P53 | 13.06 | 22.83 | 21.95 | 23.40 | 22.73±0.59 | 2.60 |
| KRAS | 27.33 | 22.83 | 24.38 | 23.90 | 23.70±0.65 | 2.74 |
| NRAS | 13.67 | 18.85 | 17.88 | 18.00 | 18.58±0.41 | 2.21 |
| GADD45G | 33.01 | 19.92 | 19.48 | 19.85 | 19.75±0.19 | 0.96 |
| EGFR | 32.09 | 22.83 | 22.92 | 23.24 | 22.00±0.17 | 0.77 |
| CHFR | 14.09 | 23.85 | 22.40 | 23.80 | 23.35±0.67 | 2.87 |
| C-myc | 17.81 | 17.10 | 17.51 | 17.79 | 17.46±0.28 | 1.60 |
| THBSI | 18.09 | 25.26 | 25.08 | 25.85 | 25.40±0.33 | 1.30 |
| CEBPA | 17.08 | 19.41 | 20.79 | 20.85 | 20.35±0.67 | 3.29 |
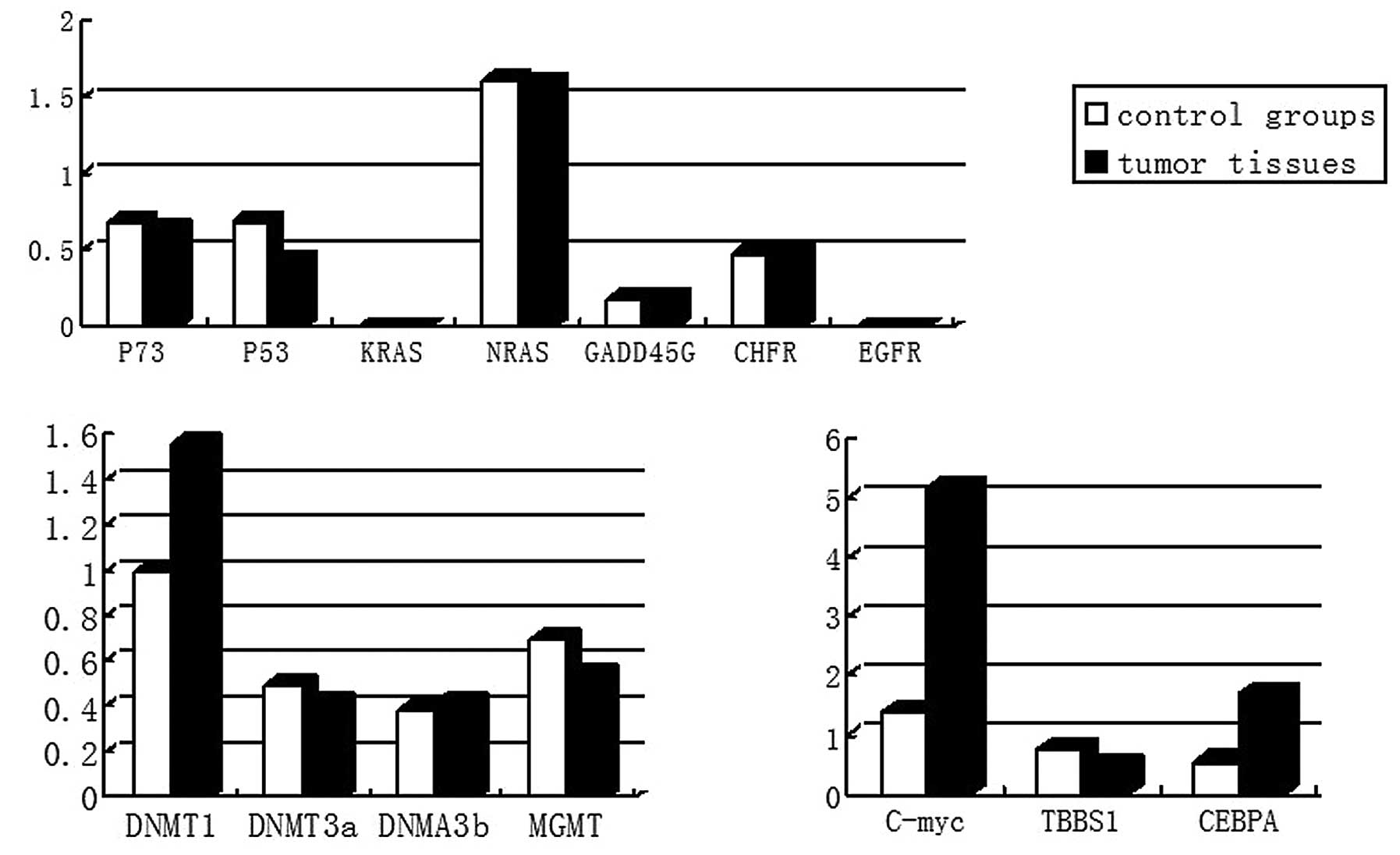
mRNA expression of DNMT
The expression levels of DNMT1 were observed to
increase by 56% compared with those in normal nasal epithelial
tissues, while MGMT, DNMT3a and DNMT3b had similar expression
levels in the two tissue types (Table III and Fig. 6).
 | Table IIIRelative quantification of DNMT. |
Table III
Relative quantification of DNMT.
| Gene | Expression of the
DNMT mRNA
|
|---|
| Cases (n) | ΔΔCt (mean ±
SD) |
2−ΔΔCt |
|---|
| DNMT1 (control
groups) | 20 | 0.00±0.53 | 1.00
(0.693–1.444) |
| DNMT1 (nasal tumor
tissues) | 24 | –0.64±0.56 | 1.56
(0.678–1.474) |
| DNMT3a (control
groups) | 20 | 1.01±0.74 | 0.497
(0.599–1.670) |
| DNMT3a (nasal tumor
tissues) | 24 | 1.25±0.73 | 0.420
(0.603–1.659) |
| DNMT3b (control
groups) | 20 | 1.39±0.88 | 0.382
(0.503–1.840) |
| DNMT3b (nasal tumor
tissues) | 24 | 1.25±0.63 | 0.420
(0.646–1.548) |
| MGMT (control
groups) | 20 | 0.53±1.40 | 0.693
(0.379–2.639) |
| MGMT (nasal tumor
tissues) | 24 | 0.87±0.75 | 0.547
(0.595–1.682) |
mRNA expression of tumor-associated
genes
The expression levels of P53 were reduced by 36.8%
and those of THBS1 by 43%, C-myc expression increased 2.9-fold and
CEBPA increased by 2-fold compared with that in normal nasal
epithelial tissues. GADD45, P7, CHFR and NRAS had similar
expression levels in the two tissues. No EGFR or KRAS expression
was observed in either of the two tissue types (Table IV and Fig. 6).
 | Table IVRelative quantification of goat
tumor-associated genes. |
Table IV
Relative quantification of goat
tumor-associated genes.
| Gene | Expression of the
tumor-associated gene mRNA
|
|---|
| Cases (n) | ΔΔCt (mean ±
SD) |
2−ΔΔCt |
|---|
| P73 (control
groups) | 20 | 0.54±0.94 | 0.688
(0.521–1.919) |
| P73 (nasal tumor
tissues) | 24 | 0.65±0.70 | 0.637
(0.616–1.625) |
| P53 (control
groups) | 20 | 0.54±0.35 | 0.688
(0.785–1.275) |
| P53 (nasal tumor
tissues) | 24 | 1.20±0.63 | 0.435
(0.646–1.548) |
| KRAS (control
groups) | 20 | – | – |
| KRAS (nasal tumor
tissues) | 24 | – | – |
| NRAS (control
groups) | 20 | –0.69±0.42 | 1.613
(0.747–1.338) |
| NRAS (nasal tumor
tissues) | 24 | –0.68±0.53 | 1.602
(0.693–1.444) |
| GADD45G (control
groups) | 20 | 2.48±0.82 | 0.179
(0.566–1.765) |
| GADD45G (nasal
tumor tissues) | 24 | 2.47±0.72 | 0.180
(0.607–1.647) |
| EGFR (control
groups) | 20 | – | – |
| EGFR (nasal tumor
tissues) | 24 | – | – |
| CHFR (control
groups) | 20 | 1.02±0.99 | 0.493
(0.503–1.986) |
| CHFR (nasal tumor
tissues) | 24 | 1.07±1.02 | 0.476
(0.493–2.028) |
| C-myc (control
groups) | 20 | −0.42±1.90 | 1.338
(0.268–3.732) |
| C-myc (nasal tumor
tissues) | 24 | −2.37±0.58 | 5.169
(0.669–1.495) |
| CEBPA (control
groups) | 20 | −0.85±0.24 | 0.555
(0.847–1.818) |
| CEBPA (nasal tumor
tissues) | 24 | −0.76±0.26 | 1.693
(0.835–1.197) |
| THBS1 (control
groups) | 20 | 0.38±0.25 | 0.768
(0.801–1.189) |
| THBS1 (nasal tumor
tissues) | 24 | 1.05±0.32 | 0.483
(0.801–1.248) |
Promoter methylation status
The amplified products of the positive control were
consistent with the anticipated results. The only tumor suppressor
genes observed to be methylated were CHFR, GADD45G and THBS1. The
methylation expression rate of the CHFR gene was ~60% in the two
tissues. The methylation rate of THBS1 was 100% in nasal tumor
tissues and 20% in normal nasal epithelial tissues. The exhaustive
methylation expression rate of GADD45G was 62.5% and the partial
methylation expression rate was 37.5% in nasal tumor tissue, while
no methylation was observed in the normal nasal epithelial tissues.
C-myc was the only proto-oncogene observed to be methylated. The
methylation expression rate of C-myc was 87.5 and 15% in the nasal
tumor tissues and normal nasal epithelial tissues, respectively.
The methylation expression rate of CEBPA was 100% in the nasal
tumor tissues and 40% in normal nasal epithelial tissues. EGFR had
similar expression levels in the two tissues. The methylation
expression rate of the EGFR gene was ~80% in the two tissues
(Table V, Figs. 7 and 8).
 | Table VMethylation status of the
tumor-associated genes. |
Table V
Methylation status of the
tumor-associated genes.
| Gene | Methylation status
of the tumor-associated genes (%)
|
|---|
| Cases (n) | Exhaustive
methylation | Partial
methylation | Unmethylated |
|---|
| P73 (control
groups) | 20 | 0 (0/20) | 0 (0/20) | 100 (20/20) |
| P73 (nasal tumor
tissues) | 24 | 0 (0/24) | 0 (0/24) | 100 (24/24) |
| P53 (control
groups) | 20 | 0 (0/20) | 0 (0/20) | 100 (20/20) |
| P53 (nasal tumor
tissues) | 24 | 0 (0/24) | 0 (0/24) | 100 (24/24) |
| KRAS (control
groups) | 20 | 0 (0/20) | 0 (0/20) | 100 (20/20) |
| KRAS (nasal tumor
tissues) | 24 | 0 (0/24) | 0 (0/24) | 100 (24/24) |
| NRAS (control
groups) | 20 | 0 (0/20) | 0 (0/20) | 100 (20/20) |
| NRAS (nasal tumor
tissues) | 24 | 0 (0/24) | 0 (0/24) | 100 (24/24) |
| GADD45G (control
groups) | 20 | 15 (3/20) | 0 (0/20) | 85 (17/20) |
| GADD45G (nasal
tumor tissues) | 24 | 62.5 (15/24) | 37.5 (9/24) | 0 (0/24) |
| EGFR (control
groups) | 20 | 75 (15/20) | 0 (0/20) | 25 (5/20) |
| EGFR (nasal tumor
tissues) | 24 | 87.5 (21/24) | 0 (0/24) | 12.5 (3/24) |
| CHFR (control
groups) | 20 | 60 (12/20) | 0 (0/20) | 40 (8/20) |
| CHFR (nasal tumor
tissues) | 24 | 58.3 (14/24) | 0 (0/24) | 41.7 (10/24) |
| C-myc (control
groups) | 20 | 15 (3/20) | 0 (0/20) | 85 (17/20) |
| C-myc (nasal tumor
tissues) | 24 | 87.5 (21/24) | 0 (0/24) | 12.5 (3/24) |
| CEBPA (control
groups) | 20 | 40 (8/20) | 0 (0/20) | 60 (12/20) |
| CEBPA (nasal tumor
tissues) | 24 | 100 (24/24) | 0 (0/24) | 0 (0/24) |
| THBS1 (control
groups) | 20 | 20 (4/20) | 0 (0/20) | 80 (16/20) |
| THBS1 (nasal tumor
tissues) | 24 | 100 (24/24) | 0 (0/24) | 0 (0/24) |
Discussion
The present study established a method to screen
animals for ENT using RT-qPCR analysis. In the PCR analyses
performed, pure single products were obtained and agarose gels
appeared according to the expected outcome. No primer dimers and no
unexpected peaks in the melting curves were observed, and DNA
sequencing results were similar to those of the reference sequence,
thus suggesting that the experiments were specific, repeatable and
accurate. The correlation coefficients of each gene were >0.999,
which indicated the Ct value and concentration of cDNA had a strong
linear association (8,9). The amplification efficiency was ~1,
which indicated that RT-qPCR products were of high quality. In
terms of repeatability of the experiments, the intra-assay and
inter-assay variation coefficient remained <3.29%, which
demonstrated that the method was repeatable and may be used to
detect gene transcription (8).
DNMTl, DNMT3a and DNMT3b are the encoding genes of
methyltransferase (3,10). DNA methylation involves the
addition of a methyl group to the carbon in 5-position of the
cytosine pyrimidine ring via the action of DNMTl, DNMT3a and DNMT3b
(3,4,10).
The clustering of the CpG island of upstream genes often leads to
the downregulation of the expression of the downstream gene.
Abnormal methylation has been previously suggested to be one of the
mechanisms of tumorigenesis (11).
A previous study demonstrated that the DNMT1 gene was closely
associated with DNA methylation and that DNMT1 is important in the
maintenance of methylation (12).
The MGMT gene is a tumor suppressor gene, and a previous study
identified that abnormal expression of the MGMT gene resulted in
the activation of the oncogene or inactivation of tumor suppressor
genes, thus leading to tumor formation (13). Previous studies have identified an
association between the MGMT gene and occurrence, development,
biological behavior and prognosis of malignant tumors (13–15).
In the present study, MGMT, DNMT3a and DNMT3b were observed to have
similar expression levels in the two tissues, while the expression
of DNMT1 was upregulated in nasal tumor tissues. A total of six out
of ten tumor-associated genes were observed to exhibit methylation
in the present study. The presence of methylation was suggested to
be associated with abnormal expression levels of DNMT1 (12).
A previous study demonstrated that the inactivation
of tumor suppressor genes is important in the development of
nasopharyngeal carcinoma (16).
The inactivation of anti-oncogenes by methylation is more common in
nasopharyngeal carcinoma (16).
Nasopharyngeal cancer-associated gene methylation involves the
entire process of cell cycle regulation, DNA repair, cell apoptosis
and tumor metastasis (16). It has
been demonstrated that DNA methylation is one of three mechanisms
of tumor suppressor gene inactivation, while in certain cases, DNA
methylation is the only mechanism of tumor suppressor gene
inactivation (17). P53 has been
reported to be absent or mutated in ~50% of human tumors (18). Once induced, P53 protein can
combine with regulatory sequences and a series of trans-activating
genes (p2 J, GADD45) and act as a transcriptional regulatory factor
(18,19). Gopisetty et al (20) observed that abnormal methylation of
P53 and p14ARF may lead to activation and expression of various
apoptosis-promoting genes and induce cell apoptosis. The
hypermethylation of tumor suppressor genes, tumor metastasis
suppressor genes, hormone receptor genes, DNA repair genes and
angiogenesis-inhibiting gene promoters can result in the
downregulation of expression or loss of the corresponding gene
(19). THBS1 was first observed in
platelet particles and can be expressed by tumor cells,
macrophages, mononuclear cells and endothelial cells (19). THBS1 is one of the strongest
negative regulators of angiogenesis (21), which can additionally regulate cell
adhesion, migration, proliferation and differentiation and induce
platelet aggregation (22).
Previous studies have demonstrated that THBS1 participates in the
development and prognosis of neoplasms and the expression levels
are negatively correlated with tumor progression (23). In the present study, methylation
was identified to be absent in the P73 and P53 genes. The
methylation expression rate of THBS1 was 100% in nasal tumor
tissues and 20% in normal nasal epithelial tissues. The exhaustive
methylation expression rate of GADD45G was 62.5% and the partial
methylation expression rate was 37.5% in nasal tumor tissue, while
no methylation was observed in normal nasal epithelial tissues. The
expression levels of P53 were reduced by 36.8% and those of THBS1
were reduced by 43%.
Proto-oncogene activation is an important mechanism
in the development of various malignant tumor types (24). Point mutations (25) and low methylation levels, for
example, can lead to abnormal activation of proto-oncogenes, which
increases the proliferative and survival abilities, thus promoting
tumor progression (26). C-myc is
a multi-functional cancer gene, and as it has transcription factor
activity, it can activate cell proliferation, inhibit cell
differentiation, regulate the cell cycle and regulate cell
apoptosis (26). Abnormal
methylation can result in abnormally high expression levels of
C-myc and thus lead to the occurrence and development of tumors
(26). In the present study, the
methylation expression rate of C-myc was 87.5% in nasal tumor
tissues and 15% in normal nasal epithelial tissues. The expression
levels of C-myc increased 2.9-fold compared with those in normal
nasal epithelial tissues.
The CEBPA gene and transcription factor C/EBP α
exhibit important effects in the process of regulation of myeloid
differentiation and proliferation (27). Abnormal expression levels of the
CEBPA gene at a transcriptional and translational level have been
reported to influence the occurrence of cancer (27). In the present study, the
methylation expression rate of CEBPA was observed to be 100% in
nasal tumor tissues and 40% in normal nasal epithelial tissues. The
expression levels of CEBPA increased 2-fold compared with normal
nasal epithelial tissues.
EGFR is composed of an extracellular section,
transmembrane region and an intracellular kinase activity area
(28). Overexpression of EGFR is
associated with tumor cell metastasis and invasion (28). In the present study, the
methylation expression rate of EGFR gene was ~80% in the two tissue
types, whereas no expression of EGFR was identified.
In conclusion, the expression of DNMT1, C-myc and
CEBPA was upregulated, and the expression of P53 and THBSI was
downregulated in nasal tumor tissues. Abnormal methylation of the
C-myc, CEBPA, GADD45G and THBS1 genes was observed in nasal tumor
tissues. The occurrence of ENT may be associated with the abnormal
expression and methylation of DNMT1, C-myc, CEBPA, P53, GADD45G and
THBS1. Therefore, it is suggested that these six genes may be used
as diagnostic marker candidate genes for ENT. The results of the
present study can provide a foundation for screening of
tumor-specific markers for early diagnosis of ENT and further
research into the epigenetic mechanisms of ENTV-induced nasal
epithelial cell carcinoma.
References
|
1
|
Kawasako K, Okamoto M, Kurosawa T, et al:
Enzootic intranasal tumour virus infection in apparently healthy
sheep in Japan. Vet Rec. 157:1182005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pan SY: DNA methylation as a biomarker and
its clinical application on molecular diagnosis and treatment. J
Mol Diagn Ther. 1:145–147. 2009.
|
|
3
|
He M: DNA methylation and tumor. J Med Mol
Biol. 8:641–645. 2011.
|
|
4
|
Kappler JW: The kinetics of DNA
methylation in cultures of a mouse adrenal cell line. J Cell
Physiol. 75:21–31. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wassenegger M: RNA-directed DNA
methylation. Plant Gene Silencing. Springer. 83–100. 2000.
|
|
6
|
Feng YW, Yan QG, Guo WZ, Wang XY and Shu
L: Construction and bioinformatics analysis of cDNA library of goat
enzootic nasal tumor virus SC strain. Chin Vet Sci. 41:126–130.
2011.
|
|
7
|
Wang XY, Feng YW, Yan QG, et al: Cloning
and sequence analysis of gag variable region of enzootic nasal
tumor virus and endogenous retrovirus of goats. Prog Vet Med.
32:29–32. 2011.
|
|
8
|
Zhao YP, Bai DY, LI B, Huang JL, Zhang YH
and Mang L: establishment of a real-time RT-PCR assay based on SYBR
Green I for detection of the expression of horse toll-like receptor
genes. Acta Vet Zootech Sin. 44:220–227. 2013.
|
|
9
|
Xiu JS, Chen XQ, Wang B and Li T:
Establishment of a real-time RT-PCR method based on SYBR Green I
for diagnosis of porcine kobuvirus. Chin J Zoonoses. 28:922–926.
2012.
|
|
10
|
Jeltsch A: Beyond Watson and Crick: DNA
methylation and molecular enzymology of DNA methyltransferases.
Chembiochem. 3:274–293. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang ZK and Wang YF: DNA methvlation and
tumor. J Med Postgra. 6:641–645. 2012.
|
|
12
|
Liu B, Gu LP, Xing CP, et al: Clinical
significance and association of caveolin-1 and Dnmt1 gene with
carcinogenesis, development of gastric carcinomas. World Chin J
Digestol. 17:1561–1566. 2009.
|
|
13
|
Sun YH, Zhang YZ, Wang ZC, Sun MZ and Zhao
DH: Relationship between the expression of O6-methylguanine-DNA
methyltransferase in glioma and the survival time of patients. Ai
Zheng. 23:1052–1055. 2004.In Chinese. PubMed/NCBI
|
|
14
|
Esteller M, Toyota M, Sanchez-Cespedes M,
et al: Inactivation of the DNA repair gene O6-methylguanine-DNA
methyhransferase by promoter hypermethylation is associated with G
to A mutations in K-ras in colorectal tumorigenesis. Cancer Res.
60:2368–2371. 2000.PubMed/NCBI
|
|
15
|
Preuss I, Eberhagen I, Haas S, et al:
O6-methylguanine-DNA methyhransferase activity in breast and brain
tumors. Int J Cancer. 61:321–326. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni HF, Huang GW, Zhang Z, Jiang P and Li
Y: Promter methyhlation and the mRNA expression of p73 gene in
human nasopharyngeal carcinoma. J Med Res. 40:24–28. 2011.
|
|
17
|
Jones PA and Laird PW: Cancer-epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M and Wang X: The biological effects of
p53 methylayion profile change. Int Genet ISTIC. 35:243–247.
2012.
|
|
20
|
Gopisetty G, Ramachandran K and Singal R:
DNA methylation and apoptosis. Mol Immunol. 43:1729–1740. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang YC, Chen SP, Li J, Wang XD, Deng CS,
Zhu YQ and Gong L: CpG is lsland methyIation and expression of
thrombospondin 1gene in coIorectal adenocarcinoma. World Chin J
Digestol. 13:189–193. 2005.
|
|
22
|
Tsuchida T, Kijima H, Tokunaga T, et al:
Expression of the thrombospondin 1 receptor CD36 is correlated with
decreased stromal vascularisation in colon cancer. Int J Oncol.
14:47–98. 1999.
|
|
23
|
Maeda K, Nishiguchi Y, Kang SM, et al:
Expression of thrombospondin-1 inversely correlated with tumor
vascularity and hematogenous metastasis in colon cancer. Oncol Rep.
8:763–766. 2001.PubMed/NCBI
|
|
24
|
Jiang BY: Clinical relevance and
underlying mechanism of altered expression of proto-oncogene BCL l
lA in non-small cell lung cance. Southern Medical University;
2010
|
|
25
|
Konopka JB, Watanabe SM, Singer JW, et al:
Cell lines and clinical isolates derived from Ph1-positive chronic
myelogenous leukemia patients express c-abl proteins with a common
structural alteration. Proc Natl Acad Sci USA. 82:1810–1814. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Li JS, Guo MZ, Meng BS, Chen XY
and Zhang JP: Effects of S-ademetionine on proliferation of gastric
cancer cell lines and methylation of c-myc and Upa genes. Chin J
Dig. 30:322–326. 2010.
|
|
27
|
Wang LMM, Xiao HW and Huang H: Roles of
CEBPA mutation and expression abnormality in acute myeloid
leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 20:1256–1260.
2012.Aritcle in Chinese. PubMed/NCBI
|
|
28
|
Wang XF, Du ZW, Wu M, Zhang YC, Jiang Y
and Zhang GZ: DNA extraction from formalin-fixed and
paraffin-embedded tissues by triton X-100 for effective
amplification of EGFR gene by polymerase chain reaction. Chem Res
Chin Univ. 25:501–505. 2011.
|