Introduction
tRNase ZL-utilizing efficacious gene silencing (TRUE
gene silencing) is an RNA-mediated gene expression control
technology with therapeutic potential (1–6).
This technology is based on a unique enzymatic property of
mammalian tRNase ZL, which is that it can cleave any target RNA at
a desired site by recognizing a pre-tRNA-like or
micro-pre-tRNA-like complex formed between the target RNA and an
artificial small guide RNA (sgRNA).
The efficacy of TRUE gene silencing has been
demonstrated by using it to introduce various artificially-designed
sgRNAs into living cells either by their expression plasmids or by
2′-O-methyl RNAs (2,7). sgRNA is divided into four groups:
5′-half-tRNA, RNA heptamer, hook RNA and ~14-nt linear RNA
(1,2,8,9).
Linear-type sgRNAs can downregulate the expression of the human
genes Bcl-2 and GSK-3β (7).
Luciferase gene expression in mouse liver can be inhibited by a
heptamer-type sgRNA (7). In
addition, the efficacy of TRUE gene silencing can be compared with
that of RNA interference technology (6–8).
sgRNA can be easily taken up by cultured cells without any
transfection reagents, and naked sgRNAs targeting Bcl-2 or WT1 mRNA
can reduce the levels of mRNA and protein, as well as inducing the
apoptosis of leukemia cells (9,10).
However, little is known regarding the mechanism of naked sgRNA
uptake into cultured cells. The 'gymnotic' mechanism (11–13)
and/or the adaptor protein AP2M1 (14) are reported to be involved in the
cellular uptake of naked oligonucleotides by cultured cells. In the
present study, the effects of various inhibitors of apoptotic
induction were investigated using naked sgRNA treatment in order to
elucidate uptake pathways and mechanisms for sgRNA in cultured
cells.
Materials and methods
RNA/DNA synthesis
The following 5′- and/or 3′-phosphorylated sgRNAs
with full 2′-O-methyl modifications were chemically synthesized by
Nippon Bioservice (Asaka, Saitama, Japan): mh1 (Bcl-2),
5′-pGGGCCAGp-3′; Luc-hep2, 5′-pGAUCGAG-3′; mh3 (EGFP),
5-pUUGCCGUp-3; Hep5 (HSP90AA1) 5′-pAGAUCCUp-3′; mh2 (FGFR3),
5′-pCCAGCACp-3′ (2,9,10).
The DNA primers for polymerase chain reaction (PCR) were obtained
from Hokkaido System Science (Sapporo, Japan).
Cell cultures
HL-60 or HEK-293 human cells were obtained from
RIKEN BioResource Center (Tsukuba, Japan) and were cultured in
RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA) or α-minima
essential medium (MEM; Sigma-Aldrich) containing 100 µg/ml
kanamycin (Meiji Seika Pharma International Ltd., Tokyo, Japan) and
10% fetal bovine serum (FBS; SAFC Biosciences, Inc. Lenexa, KS,
USA) at 37°C in culture flasks or 100-mm culture dishes (Corning,
Corning, NY, USA) in a humidified atmosphere of 5% CO2.
The cells were incubated with medium containing chlorpromazine
(CPZ), nystatin (Nys), methyl-β-cyclodextrin (MbCD), heparan
sulfate, 10 K dextran sulfate, chloroquine, brefeldin A or none
(vehicle group). The cells were cultured either with 1 µM
naked 2′-O-methyl RNA or without naked 2′-O-methyl RNA (control
cultures).
Compounds and reagents
CPZ, Nys, MbCD, heparan sulfate, 10 K dextran
sulfate, chloroquine and brefeldin A were purchased from
Sigma-Aldrich.
Quantitation of living cell numbers
HL-60 cells were seeded at a density of
1×103 cells/well in a 96-well plate in RPMI-1640 medium
and were incubated for 30 min with medium containing CPZ, Nys,
MbCD, heparan sulfate, 10 K dextran sulfate, chloroquine or
brefeldin A, as appropriate, at the concentrations shown. The cells
were cultured with 1 µM of naked 2′-O-methyl RNA;
appropriate vehicle buffer was added to control cultures. After 3
days, to quantitate the number of living cells by the
tetrazolium-based colorimetric MTT assay, 20 µl TetraColor
ONE (Seikagaku Biobusiness Corp., Tokyo, Japan) was added to each
well. After 2–4 h of incubation at 37°C, the optical density was
measured at a wavelength of 450 nm using a microplate reader
(iMark; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse transcription-PCR (RT-PCR)
PCR was used to analyze the transcript levels of
hsp90AA1. Total RNA was extracted from the cells at the indicated
time points using Isogen (Nippongene, Toyama, Japan) and treated
with RNase-free DNase (Qiagen, Hilden, Germany) to remove any
contaminating genomic DNA. RT-PCR was performed as previously
described (15). The primer
sequences for each gene were as follows: hsp90AA1,
5′-TGCACCTTGGCTCTGTCTGAA-3′ (forward), 5′-CACCTGTTAACTGGTACCAAG-3′
(reverse); glyceraldehyde-3-phosphate dehydrogenase (GAPDH),
5′-TCCACCACCCTGTTGCTGTA-3′ (forward), 5′-ACCACAGTCCATGCCATCAC-3′
(reverse). To account for any difference in the quantity of RNA,
GAPDH was selected as the endogenous control and amplified using
the primers described above. The amplification products were
electrophoresed on 2% agarose gels.
Quantitation of gene expression using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
RT-qPCR was performed using assay-on-demand TaqMan
probes (Applied Biosystems, Foster City, CA, USA) and the
StepOne® real time PCR system as previously described
(16). The relative level of gene
expression was quantified using the comparative Ct method with
GAPDH as the endogenous control.
Statistical analysis
All experiments were repeated at least three times
and representative results are shown. In the RT-qPCR analysis and
the cell viability assay, differences between control and
experimental groups are reported as the mean ± standard deviation,
and were analyzed by Student's t-test, in which values of P<0.05
were considered to indicate a statistically significant
difference.
Results and Discussion
To investigate the properties of heptamer-type
sgRNA, our group designed a heptamer, mh1 (Bcl-2), targeting the
human Bcl-2 mRNA (9). mh1 (Bcl-2)
has the potential to bind Bcl-2 mRNA at four sites, each of which
has an immediate upstream sequence that can form a hairpin
structure resembling a T-arm of tRNA. As a result, a
micro-pre-tRNA-like complex can be formed at each site, which acts
as a substrate for tRNase ZL (2).
It was identified that mh1 (Bcl-2) downregulated Bcl-2 mRNA
compared with mock or mh3 (EGFP) sgRNA which have no potential
target sites on Bcl-2 mRNA. This suggested that naked mh1 (Bcl-2)
directs Bcl-2 mRNA cleavage by cellular tRNase ZL (9). First, the effects of fully
synthesized 2′-O-methylated mh1 (Bcl-2) containing 5′ and 3′
phosphates were tested on living cell numbers by adding it to HL-60
cell culture medium without any transfection reagents. When 1
µM of naked mh1 (Bcl-2) was added to the culture medium, the
living cell number reduced in 3 days compared with cultures without
sgRNA (9). By contrast, mh3
(EGFP), that has no potential target site on Bcl-2 mRNA, had no
effect on the living cell number (9).
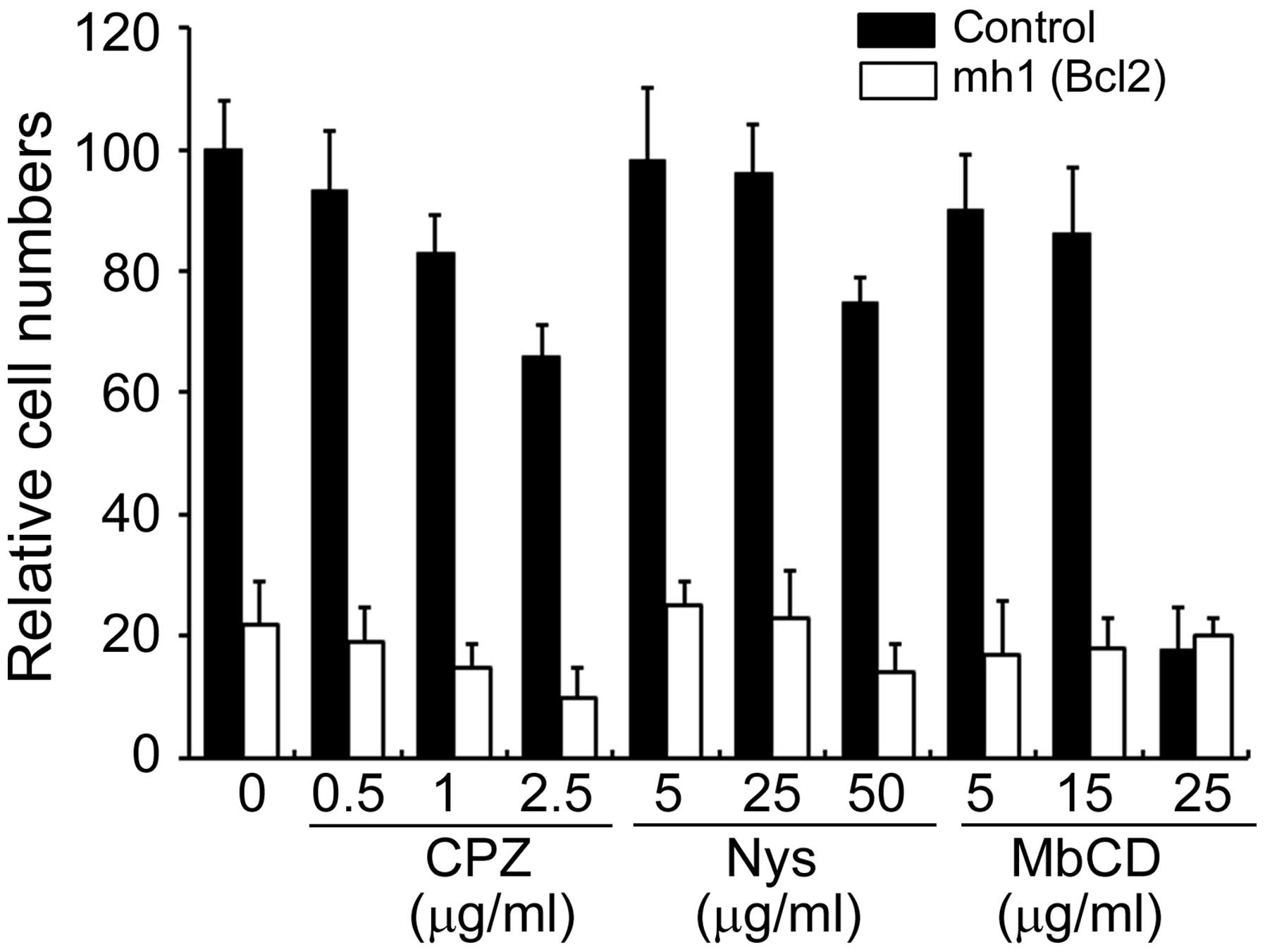
To elucidate the mechanism of naked sgRNA uptake
using cultured cells, the effects of various endocytosis inhibitors
on sgRNA-induced apoptosis were examined. CPZ disrupts
clathrin-dependent endocytosis, while Nys selectively disrupts
caveolae- and lipid raft-dependent endocytosis but has no effect on
clathrin-dependent endocytosis. MbCD disrupts both lipid raft- and
clathrin-dependent endocytosis (17). HL-60 cells were incubated for 30
min without (control) or with the different inhibitors, and then
incubated with 1 µM of mh1 (Bcl-2) for 3 days. Addition of
any concentration of the compounds in combination with naked
effective sgRNA was unable to diminish its effect on apoptosis in
HL-60 cells (Fig. 1), indicating
that sgRNA-induced apoptosis does not depend upon caveolae-, lipid
raft- or clathrin-dependent endocytic mechanisms in these
cells.
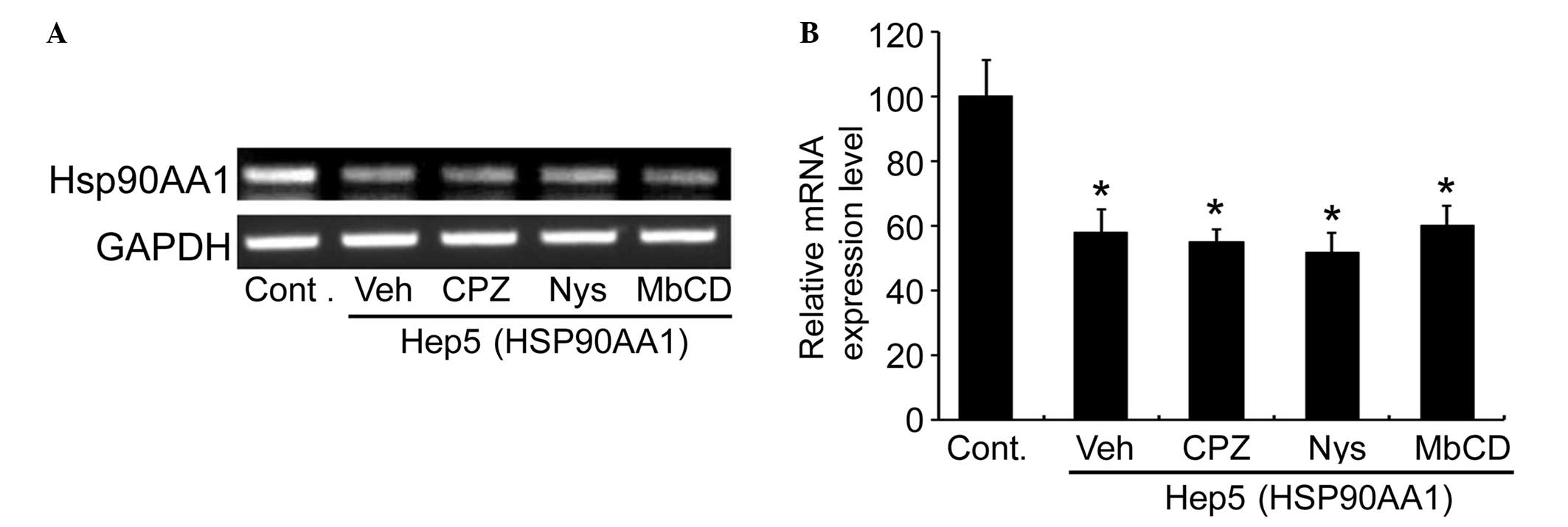
The effects of various endocytosis inhibitors on
sgRNA-directed mRNA reduction in HEK-293 cells were examined.
Treatment with Hep5 (HSP90AA1) resulted in a reduction of hsp90AA1
mRNA levels in HEK-293 cells (Fig.
2). None of the endocytosis inhibitors CPZ, NYS or MbCD had any
inhibitory effect on the action of sgRNA in reducing hsp90AA1 mRNA
as compared with the vehicle (Fig.
2). These observations also suggested that functional uptake of
sgRNA by cells is clathrin-, caveolae- and lipid
raft-independent.
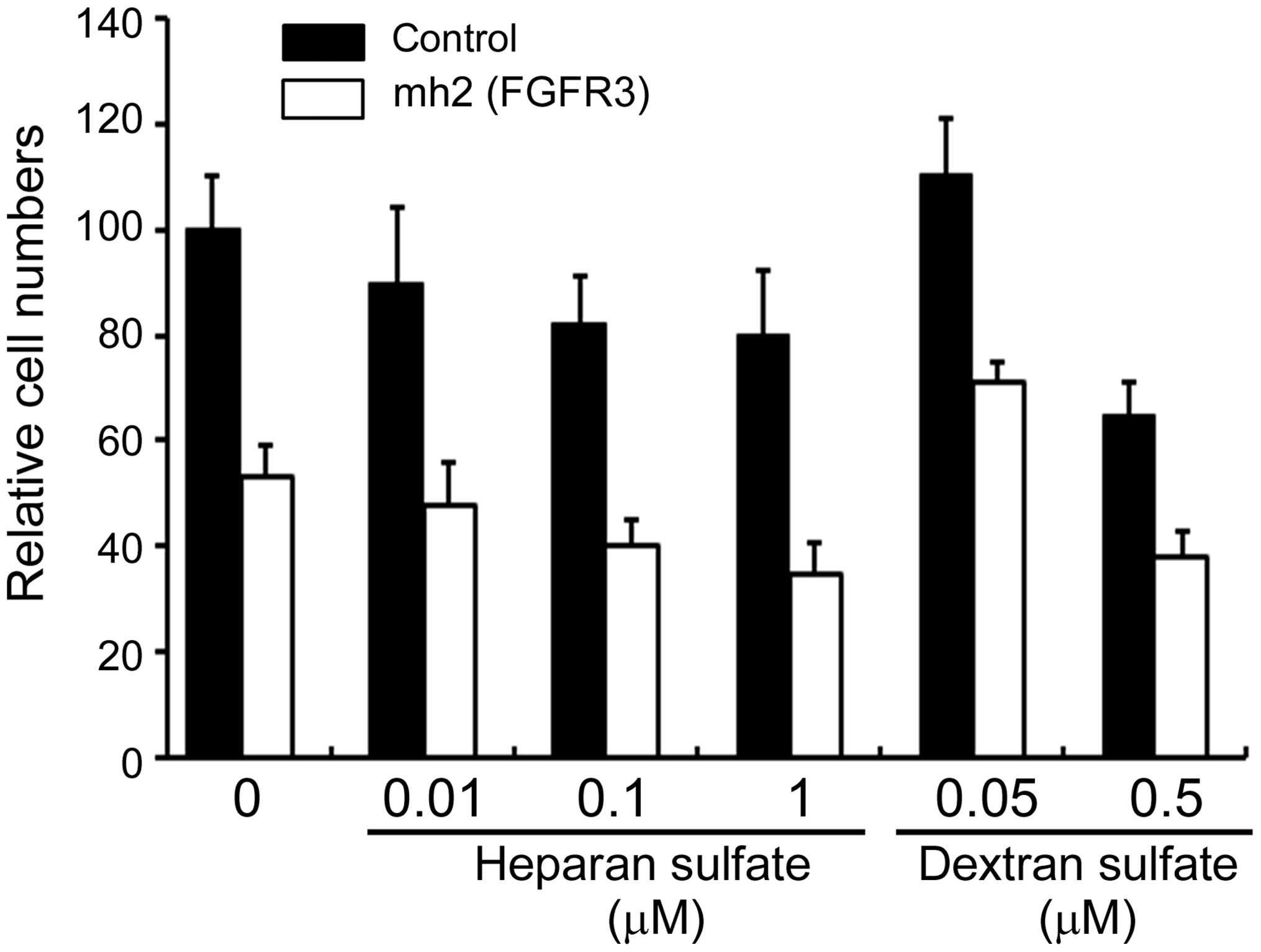
To determine the ability of polyanions to compete
for functional cellular sgRNA uptake and sgRNA-induced apoptosis,
heparan sulfate and dextran sulfate were used. Neither heparan
sulfate nor dextran sulfate were able to attenuate the effect of
mh2 (FGFR3) on apoptosis in HL-60 cells (Fig. 3). These results demonstrate that
sgRNA activities do not only occur via interactions of polyanions
with membrane proteins, but appear to be also due to recognition of
both base and sugar.
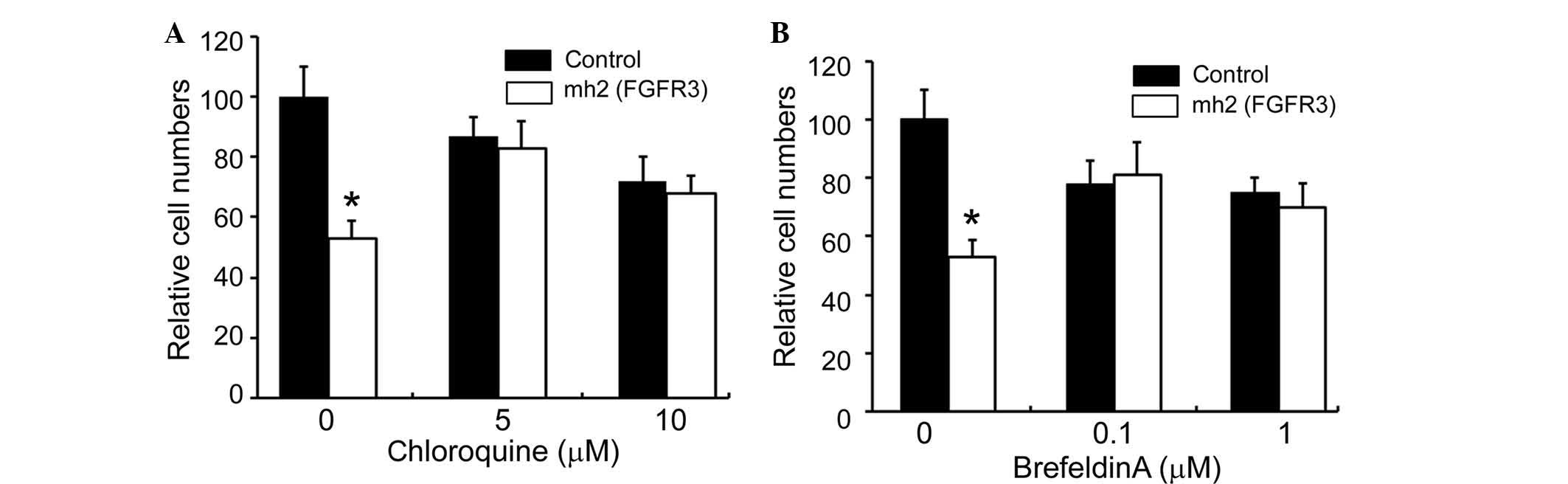
Next, two pharmacological inhibitors of endocytic
processes were used to further define the pathways through which
sgRNA could induce apoptosis in the functional compartment of
cells. Chloroquine, an inhibitor of lysosome acidification, and
brefeldin A (BFA), a lactone antibiotic that blocks retrograde
protein transport from the Golgi apparatus to the endoplasmic
reticulum (18) were administered.
In the presence of these compounds, the induction of apoptosis by
naked effective mh2 (FGFR3) was reduced (Fig. 4). These results suggest that the
uptake of functional sgRNA into cells involves a vesicular
transport process.
Chemically modified single-stranded
oligonucleotides, delivered systemically, rapidly disperse out of
the plasma and are taken up by cells within tissues (19–21).
Although transfection agents are required to produce effects of
single-stranded oligonucleotides in the majority of cultured cells,
several investigations have indicated that single-stranded
oligonucleotides can be taken up and exert their effects without
any transfection agent (11). It
was also demonstrated that a heptamer, mh1 (Bcl-2), which targets
human Bcl-2 mRNA, can be taken up by cells without any transfection
reagents and that it can induce apoptosis of leukemia cells
(9). However, it is not fully
understood how polyanionic oligonucleotides gain access to the
cytoplasmic and/or nuclear compartments of the cell where they can
bind to the target RNA.
Endocytosis describes the de novo production
of internal membranes from the plasma membrane lipid bilayer
(22). In clathrin-mediated
endocytosis, proteins involved in this process recruit cargo into
developing clathrin-coated pits, and subsequently form
clathrin-coated vesicles. By contrast, clathrin-independent
endocytosis is hypothesized to occur by seemingly distinct
pathways, based on the reliance of these pathways on certain
proteins and lipids. In the present study, none of the endocytosis
inhibitors CPZ, Nys or MbCD were able to diminish the effects of
apoptosis induction or target mRNA reduction by sgRNA. These
results indicate that functional uptake by cells in culture does
not appear to be mediated by clathrin-, caveolae- or lipid
raft-dependent pathways. Similarly to our studies, Koller et
al (14) reported that
functional uptake of a single-stranded phosphorothioate modified
antisense oligonucleotide is not mediated by clathrin or
caveolin.
Following cellular uptake, oligonucleotides are
sequestered in intracellular compartments of unknown nature, from
which they are hypothesized to escape by an undetermined and
presumably inefficient mechanism. Based on relief by monensin of
fluorescence quenching, fluoresceinated phosphorothioate
oligonucleotides were reported to be identified in acidic
compartments, compatible with lysosomes (23). The present study suggested that
functional uptake of sgRNA is an intracellular vesicular uptake
process, since it is blocked by choroquine and brefeldin A.
Recently, it has been reported that inhibition of the adaptor
protein AP2M1 with small interfering RNA attenuated the antisense
effects in cultured hepatocytes (14,24).
Additional studies are required to determine whether AP2M1 mediates
the effects of naked sgRNAs and to fully identify methods of sgRNA
uptake. A greater understanding of how naked sgRNAs enter cells and
how they reach their target RNA may aid in the design of more
specifically-targeted and more potent sgRNA drugs.
Acknowledgments
This study was supported by the Adaptable and
Seamless Technology Transfer Program through Target-driven R&D,
Japan Science and Technology Agency (to Professor Masayuki
Nashimoto).
Abbreviations:
|
TRUE gene silencing
|
tRNase ZL-utilizing efficacious gene
silencing
|
|
sgRNA
|
small guide RNA
|
|
CPZ
|
chlorpromazine
|
|
Nys
|
nystatin
|
|
MbCD
|
methyl-β-cyclodextrin
|
References
|
1
|
Nashimoto M: Specific cleavage of target
RNAs from HIV-1 with 5′ half tRNA by mammalian tRNA 3′ processing
endoribo-nuclease. RNA. 2:523–524. 1996.PubMed/NCBI
|
|
2
|
Tamura M, Nashimoto C, Miyake N, Daikuhara
Y, Ochi K and Nashimoto M: Intracellular mRNA cleavage by 3′ tRNase
under the direction of 2′-O-methyl RNA heptamers. Nucleic Acids
Res. 31:4354–4360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nashimoto M: Anomalous RNA substrates for
mammalian tRNA 3′ processing endoribonuclease. FEBS Lett.
472:179–186. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takaku H, Minagawa A, Takagi M and
Nashimoto M: A novel 4-base-recognizing RNA cutter that can remove
the single 3′ terminal nucleotides from RNA molecules. Nucleic
Acids Res. 32:e912004. View Article : Google Scholar
|
|
5
|
Shibata HS, Takaku H, Takagi M and
Nashimoto M: The T loop structure is dispensable for substrate
recognition by tRNase ZL. J Biol Chem. 280:22326–22334. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Elbarbary R, Takaku H, Tamura M and
Nashimoto M: Inhibition of vascular endothelial growth factor
expression by TRUE gene silencing. Biochem Biophys Res Commun.
379:924–927. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakashima A, Takaku H, Shibata HS, Negishi
Y, Takagi M, Tamura M and Nashimoto M: Gene silencing by the tRNA
maturase tRNase Z(L) under the direction of small-guide RNA. Gene
Ther. 14:78–85. 2007. View Article : Google Scholar
|
|
8
|
Sano T, Takahashi M, Nozaki T, Takahashi
Y, Tamura M and Nashimoto M: Expanding the utility of heptamer-type
sgRNA for TRUE gene silencing. Biochem Biophys Res Commun.
416:427–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi M, Elbarbary RA, Nakashima A,
Abe M, Watanabe N, Narita M, Takahashi M, Tamura M, Yoshida T and
Nashimoto M: A naked RNA heptamer targeting the human Bcl-2 mRNA
induces apoptosis of HL60 leukemia cells. Cancer Letters.
328:362–368. 2013. View Article : Google Scholar
|
|
10
|
Watanabe N, Narita M, Yamahira A,
Taniguchi T, Furukawa T, Yoshida T, Miyazawa T, Nashimoto M and
Takahashi M: Induction of apoptosis of leukemic cells by TRUE gene
silencing using small guide RNAs targeting the WT1 mRNA. Leuk Res.
37:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein CA, Hansen JB, Lai J, Wu S,
Voskresenskiy A, Høg A, Worm J, Hedtjärn M, Souleimanian N, Miller
P, et al: Efficient gene silencing by delivery of locked nucleic
acid antisense oligonucleotides, unassisted by transfection
reagents. Nucleic Acids Res. 38:e32010. View Article : Google Scholar :
|
|
12
|
Soifer HS, Souleimanian N, Wu S,
Voskresenskiy AM, Collak FK, Cinar B and Stein CA: Direct
regulation of androgen receptor activity by potent CYP17 inhibitors
in prostate cancer cells. J Biol Chem. 287:3777–3787. 2012.
View Article : Google Scholar :
|
|
13
|
Souleimanian N, Deleavey GF, Soifer H,
Wang S, Tiemann K, Damha MJ and Stein CA: Antisense 2′-deoxy,
2′-fluroarabino nucleic acids (2′F-ANAs) oligonucleotides: In vitro
gymnotic silencers of gene expression whose potency is enhanced by
fatty acids. Mol Ther Nucleic Acids. 1:e432012. View Article : Google Scholar
|
|
14
|
Koller E, Vincent TM, Chappell A, De S,
Manoharan M and Bennett CF: Mechanisms of single-stranded
phosphorothioate modified antisense oligonucleotide accumulation in
hepatocytes. Nucleic Acids Res. 39:4795–4807. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sato MM, Nakashima A, Nashimoto M, Yawaka
Y and Tamura M: Bone morphogenetic protein-2 enhances
Wnt/beta-catenin signaling-induced osteoprotegerin expression.
Genes Cells. 14:141–153. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uyama M, Sato MM, Kawanami M and Tamura M:
Regulation of osteoblastic differentiation by the proteasome
inhibitor bortezomib. Genes Cells. 17:548–558. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dutta D and Donaldson JG: Search for
inhibitors of endocytosis: Intended specificity and unintended
consequences. Cell Logist. 2:203–208. 2012. View Article : Google Scholar
|
|
18
|
Donaldson JG, Finazzi D and Klausner RD:
Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine
nucleotide onto ARF protein. Nature. 360:350–352. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kastelein JJ, Wedel MK, Baker BF, Su J,
Bradley JD, Yu RZ, Chuang E, Graham MJ and Crooke RM: Potent
reduction of apolipoprotein B and low-density lipoprotein
cholesterol by short-term administration of an antisense inhibitor
of apolipo-protein B. Circulation. 114:1729–1735. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sazani P, Gemignani F, Kang SH, Maier MA,
Manoharan M, Persmark M, Bortner D and Kole R: Systemically
delivered antisense oligomers upregulate gene expression in mouse
tissues. Nat Biotechnol. 20:1228–1233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vickers TA, Zhang H, Graham MJ, Lemonidis
KM, Zhao C and Dean NM: Modification of MyD88 mRNA splicing and
inhibition of IL-1beta signaling in cell culture and in mice with a
2′-O-methoxyethyl-modified oligonucleotide. J Immunol.
176:3652–3661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Doherty GJ and McMahon HT: Mechanisms of
endocytosis. Annu Rev Biochem. 78:857–902. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tonkinson JL and Stein CA: Patterns of
intracellular compartmentalization, trafficking and acidification
of 5′-fluorescein labeled phosphodiester and phosphorothioate
oligodeoxynucleotides in HL60 cells. Nucleic Acids Res.
22:4268–4275. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bennett CF and Swayze EE: RNA targeting
therapeutics: Molecular mechanisms of antisense oligonucleotides as
a therapeutic platform. Annu Rev Pharmacol Toxicol. 50:259–293.
2010. View Article : Google Scholar : PubMed/NCBI
|