Introduction
Glioblastoma multiforme (GBM; WHO grade IV glioma)
is the most malignant brain tumor in adults. Even following
treatment with surgical resection, radiotherapy and concomitant
chemotherapy, the median survival time of patients with GBM is only
14.6 months (1). A number of
molecular markers for GBM have been identified and are associated
with diagnosis, prognosis and treatment. For example, somatic
mutations in isocitrate dehydrogenase 1 (IDH1) have been
identified in GBM patients, particularly in secondary GBM, which
evolves from lower-grade gliomas (2). In 90% of IDH1 mutations in gliomas,
the arginine at position 132 is replaced by a histidine (R132H
mutation) (3). Patients with
IDH1R132H gliomas also have significantly improved
survival rates (4). However, the
mechanism responsible for this improved survival rate remains to be
elucidated.
MicroRNAs (miRNAs) are a class of non-coding,
single-stranded RNAs comprised of ~22 nucleotides (5). miRNAs are post-transcriptional
regulators that bind complementary sequences on target mRNA
transcripts. Target binding usually results in gene regulation by
translational repression or mRNA degradation, thereby modulating a
variety of biological processes (6). Several miRNAs are known to associate
with clinical outcomes in GBM (5,7,8),
including miR-128a, a crucial regulator in brain development
(9-11). Aberrant expression of miR-128a has
been detected in glioma (12) and
is involved in cancer-associated biological processes, including
cell proliferation, differentiation and apoptosis (13,14).
In order to evaluate the effect of the
IDH1R132H mutation and its association with miR-128a, a
clonal U87 cell line overexpressing the IDH1R132H mutant
protein was generated. The present study also investigated the
functional molecules upstream and downstream of miR-128a in
IDH1R132H overexpressing cells.
Materials and methods
Cell culture and reagents
U87 glioma cells and HEK 293T cells were obtained
from the American Type Culture Collection (Manassas, VA, USA) and
were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Invitrogen Life Technologies, Carlsbad, CA, USA). SYBR Green PCR
master mix and the TaqMan microRNA reverse transcription kit were
purchased from Applied Biosystems (Foster City, CA, USA). YC-1 was
obtained from Sigma-Aldrich (St. Louis, MO, USA) and dissolved in
dimethyl sulfoxide.
Generation of constructs
The full-length human IDH1 coding sequence was
amplified from HEK 293T cells (Sangon Biotech Co., Ltd., Shanghai,
China). The cDNA was fused in-frame with a FLAG tag at the
N-terminus using the following synthesized primers: Forward primer
with MluI site: TTTCGTACGATGGATTACAAGGACGACGAT GACAA
GTCCAAAAAAAT and reverse primer with BsiWI site:
TTTACGCGTGGTATGAACTTAAAGTTTGG. The amplified target was inserted
into the MluI- and BsiWI-linearized pHR-SIN vector.
The IDH1R132H mutation was generated in the vector
described above by QuickChange (Stratagene, Santa Clara, CA, USA)
with the following primer sequences: R132H, forward
5′-ACCTATCATCATAGGTCATCATGCTTA TGGG-3′ and reverse
5′-TGACCTATGATGATAGGTT TTACCCATCCAC-3′.
Stable overexpression of
IDH1WT and IDH1R132H constructs in U87
cells
HEK 293T cells were seeded in 60 mm plates
(4×106 cells/plate) in DMEM with 10% heat-inactivated
FBS 1 day prior to transfection. Cells were transfected with 5.2
μg of either pHR-SIN-IDH1WT or pHR-SIN-IDH1R132H, along with
2.36 μg of pSPAX2 and 0.8 μg of pMD2 G plasmids using
Lipofectamine 2000 in DMEM. After 6 h, the transfection media was
removed and replaced with antibiotic-free DMEM with 10%
heat-inactivated FBS. Lentiviral particles were harvested at 72 h
post-transfection. U87 glioma cells were plated 1 day prior to
transduction, at 20% confluency, in 60 mm plates with DMEM
containing 10% phosphate-buffered saline (PBS). To transduce cells,
3 ml of conditioned media containing viral particles and 6
μg/ml polybrene (Sigma-Aldrich) were added to each culture.
After 16 h, the conditioned media was removed and replaced with
DMEM containing 15% FBS.
Protein extraction and western
blotting
Cells prepared for protein extraction were
immediately placed on ice and washed with ice-cold PBS solution.
Total protein extraction was achieved using RIPA lysis buffer
containing protease inhibitors (Roche Diagnostics, Mannheim,
Germany) and PMSF. Proteins were resolved on 8-12%
SDS-polyacrylamide gels (Sangon Biotech Co. Ltd., Shanghai, China)
and transferred onto nitrocellulose membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The membranes were blocked
in Tris-buffered saline and Tween 20 (TBST) with 5% non-fat dry
milk for 1 h and incubated overnight at 4°C with the following
respective primary antibodies in TBST with 5% bovine serum albumin:
Rabbit monoclonal anti-human IDH1 (1:1,000; cat. no. 8137; Cell
Signaling Technology, Inc., Danvers, MA, USA), mouse monoclonal
anti-human IDH1R132H (1:200; cat. no. SAB4200548; Sigma-Aldrich),
mouse monoclonal anti-FLAG M2 (1:500; cat. no. F3165;
Sigma-Aldrich) or mouse monoclonal anti-human HIF-1α (1:1,000; cat.
no. AH339; Beyotime Institute of Biotechnology, Shanghai, China).
Immunospecific bands were detected using an ECL substrate
(Sigma-Aldrich). β-actin immunoblotting was performed as a loading
control.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen Life Technologies). Total RNA (1 μg) was
used as a template for a Moloney murine leukemia virus-RT reverse
transcriptase reaction, which was performed according to the
manufacturer's instructions (Fermentas, Vilnius, Lithuania). qPCR
was performed using SYBR Green Master mix (Applied Biosystems).
β-actin amplification was used as an internal control. To
quantitate the expression level of miR-128a, 1 μg of total
RNA was reverse transcribed in 50 μl reaction using TaqMan
Reverse Transcription Reagents (Applied Biosystems) with the stem
loop primer. Reverse transcribed cDNA (2 μl) was subjected
to SYBR Green mix and the results were normalized to RNU48.
The following primers were used: RNU48, forward
5′-TGATGATGACCCCAGGTAACTC-3′ and reverse 5′-GAG CGCTGCGGTGATG-3′;
Bmi-1, forward 5′-CTGCAGCTC GCTTCAAGATG-3′ and reverse
5′-CACACACATCAGGT GGGGAT-3′; β-actin, forward 5′-ATGACTTAGTTGCGTTAC
ACC-3′ and reverse 5′-TGCTGTCACCTTCACCGTTC-3′; miR-128a stem loop
primer: 5′-GTCGTATCCAGTGCAGGG TC CGAGGTATTCGCACTGGATACGACAAAGAG-3′;
miR-128a, forward 5′-TCGCGTTCACAGTGAA-3′ and reverse
5′-GTGCAGGGTCCGAGG-3′.
Chromatin immunoprecipitation (ChIP)
assays
ChIP assays were performed using a ChIP kit (cat.
no. 17-371; EMD Millipore, Billerica, MA, USA) according to the
manufacturer's instructions. Cells were crosslinked with 1%
formaldehyde for 20 min at 37°C and quenched in 0.125 M glycine.
DNA was immunoprecipitated from sonicated cell lysates and
quantified using SYBR Green Real-time PCR analysis. The following
primer sequences were used: Forward: 5′-GTAATAGAATTTTCATATTG-3′ and
reverse: 5′-ATTTTGCCATGTTGAAGAAC-3′. Fold enrichment was calculated
based on Ct as 2−Δ(ΔCt), where ΔCt = CtIP –
CtInput and Δ(ΔCt) = ΔCtantibody −
ΔCtIgG.
Cell proliferation
Briefly, 5,000 cells per well were plated in 96-well
plates and cultured for 96 h. Following incubation, CCK-8 (10
μl) was added to each well and allowed to incubate at 37°C
for 2 h. Cell proliferation was then quantified by measuring the
optical density at a wavelength of 450 nm (SpectraMax M2 Multimode
Microplate Reader; Molecular Devices, LLC, Sunnyvale, CA, USA).
Each experiment was performed in triplicate.
Statistical analysis
The data are presented as the mean ± standard
deviation. The samples were analyzed using a two-tailed unpaired
Student's t-test, unless otherwise noted. P<0.05 was considered
to indicate a statistically significant difference.
Results
Overexpression of IDH1R132H
decreases U87 cell proliferation
To determine the effects of glioma-associated
IDH1R132H, N-terminal FLAG-tagged IDH1WT,
IDH1R132H and control expression constructs were
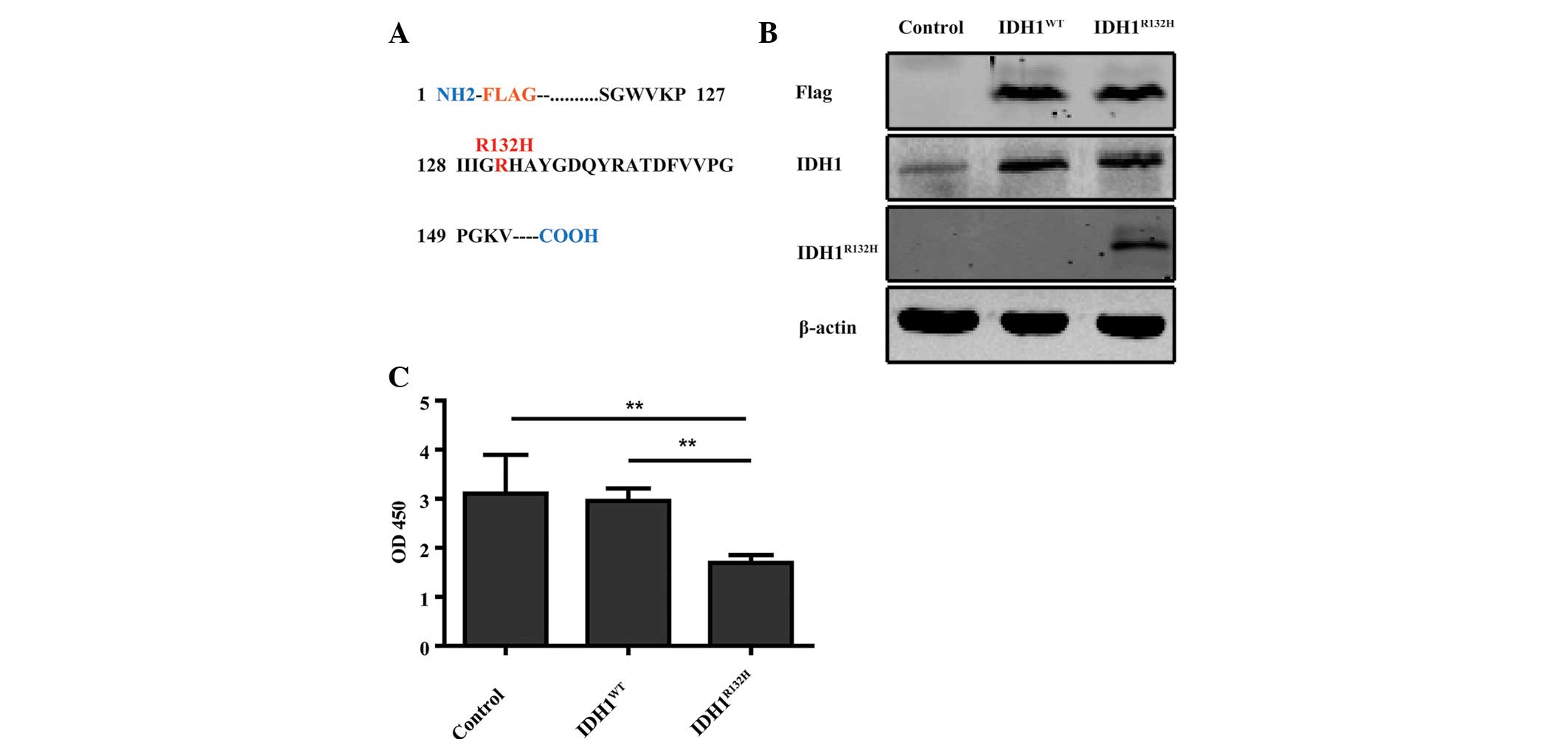
generated for stable expression in U87 cells (Fig. 1A). Western blot analysis with FLAG,
IDH1 and IDH1R132H specific antibodies confirmed the
expression of the appropriate IDH1 protein products in U87 cell
lines (Fig. 1B). Previous clinical
studies demonstrated that patients with
IDH1R132H-positive tumors have a significant survival
advantage (4,15). However, the role of
IDH1R132H in glioma cell proliferation is not fully
understood. To assess the role of IDH1R132H in U87 cell
proliferation, CCK-8 proliferation assays were performed. The
proliferation of U87 cells overexpressing IDH1R132H was
significantly decreased compared with the control and
IDH1WT-expressing cells over a 4-day incubation period
(Fig. 1C; P<0.01).
Overexpression of IDH1R132H in
U87 cells induces the expression of miR-128a
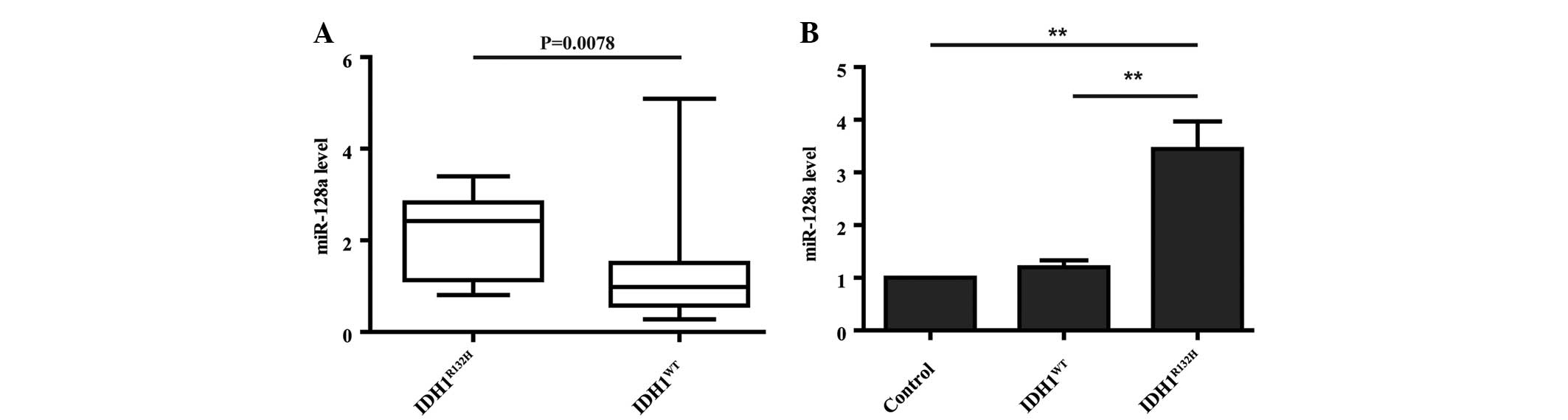
Several miRNAs associated with IDH1 mutations
have been revealed via miRNA expression profiling and the miRNA
expression signature has been identified as a prognostic biomarker
for GBM patients (7,11). However, whether
IDH1R132H induces a specific miRNA that controls cell
proliferation remains to be elucidated. According to the
IDH1 mutation-specific 23-miRNA signature analyzed by Wang
et al (11), miR-128a was
significantly overexpressed in IDH1 mutant patient samples
compared with wild-type samples. miRNA expression data was analyzed
using The Cancer Genome Atlas (TCGA) portal (https://tcga-data.nci.nih.gov/tcga/), which contains
data from seven IDH1R132H and 133 wild-type IDH1
patients. In this dataset, miR-128a was overexpressed in
IDH1R132H compared with wild-type IDH1 patients
(Fig. 2A; P=0.0078). To confirm
this expression pattern, qPCR was performed to detect miR-128a
levels in IDH1-expressing U87 cells. In agreement with the clinical
samples, U87 cells overexpressing IDH1R132H had
significantly higher levels of miR-128a compared with control and
IDH1WT cells (Fig. 2B;
P<0.01).
HIF-1α is required for miR-128a
expression induced by IDH1R132H
The ectopic expression of mutant IDH1 in cultured
cells also increases the levels of HIF-1α (16). It was hypothesized that increased
HIF-1α expression may therefore control
IDH1R132H-induced miR-128a expression. In agreement with
this hypothesis, HIF-1α protein levels were found to be
significantly higher in U87 cells overexpressing
IDH1R132H compared with the control and
IDH1WT cells (Fig. 3A).
In order to assess whether HIF-1α is required for miR-128a
overexpression in IDH1R132H cells, cells were treated
with YC-1, an inhibitor of HIF-1α activity (17). YC-1 significantly decreased the
miR-128a level in IDH1R132H cells, however, similar
effects were not observed in the control and IDH1WT
cells (Fig. 3B; P<0.01).
Notably, there is a conserved HIF-1α binding sequence GCGTG
(hypoxia response element; HRE) ~2.5 kilobases upstream of the gene
encoding pri-miR-128a (Fig. 3C).
Therefore, HIF-1α could potentially regulate miR-128a expression by
binding to this HRE. To investigate whether HIF-1α binds this
element in the promoter of the miR-128a gene, ChIP assays were
performed. In IDH1R132H overexpressing U87 cells, HIF-1α
was in fact enriched at this HRE suggesting that it regulates
miR-128a expression (Fig. 3D;
P<0.01). Taken together, the data demonstrated that HIF-1α binds
the HRE in the pri-miR-128a promoter and is required for
IDH1R132-induced miR-128a expression.
 | Figure 3HIF-1α is required for miR-128a
expression induced by IDH1R132H. (A) Western blot
analysis of HIF-1α expression in IDH1WT,
IDH1R132H and control overexpressing U87 cells. (B) U87
cells were treated with 5 μM YC-1, a HIF-1α inhibitor, and
the level of miR-128a was determined by quantitative polymerase
chain reaction. (C) Diagram of the putative HRE site in the
promoter of miR-128a. (D) Chromatin immunoprecipitation was
performed in IDH1R132H-overexpressing U87 cells with an
HIF-1α-specific antibody. **P<0.01. IDH1, isocitrate
dehydrogenase 1; HIF-1α, hypoxia inducible factor-1α; HRE, hypoxia
response element; miR-128a, microRNA-128a; DMSO, dimethyl
sulfoxide; IgG, immunoglobulin G; NC, negative control. |
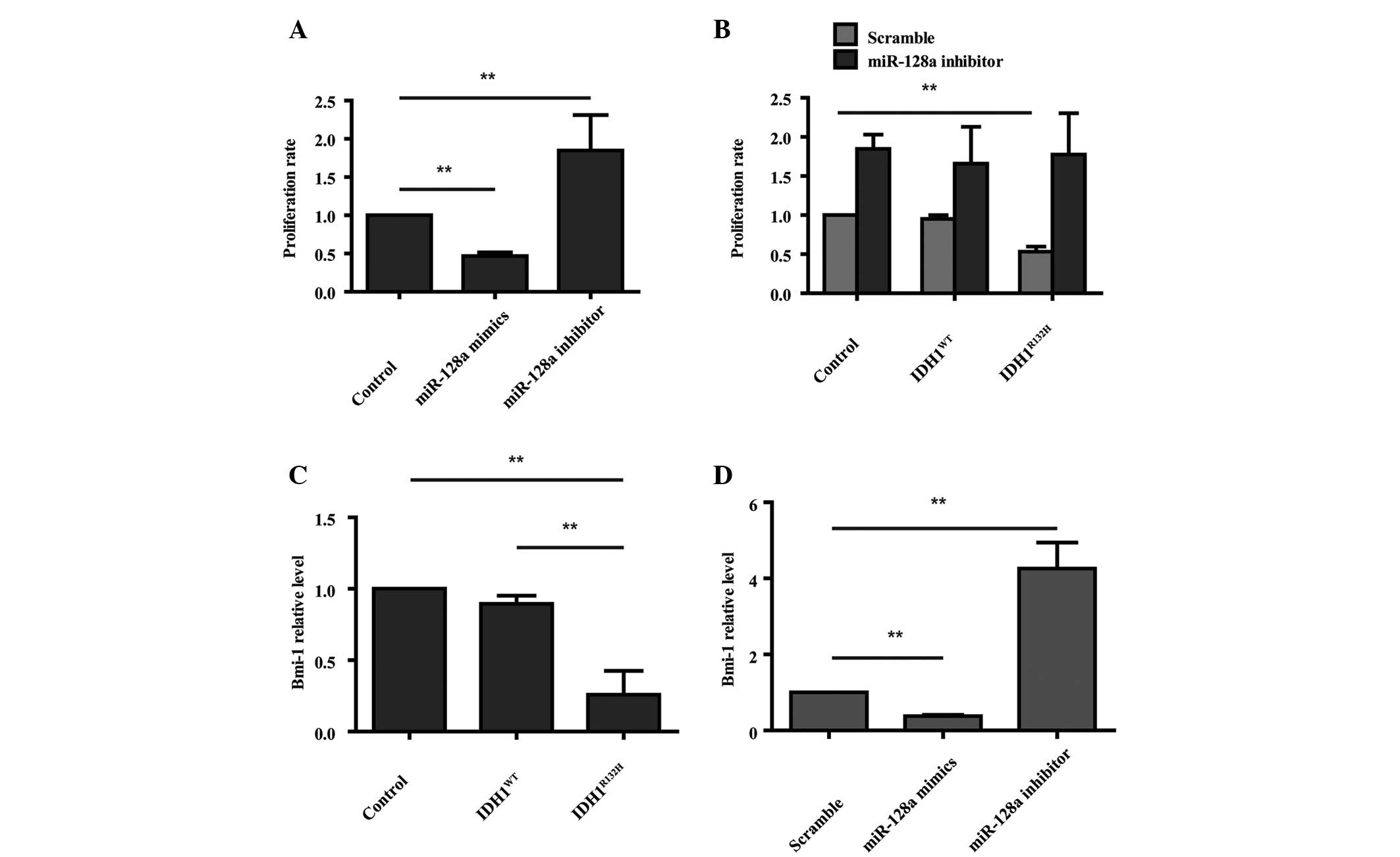
miR-128a regulates the decreased
proliferation rate induced by IDH1R132H
Previous studies have reported that miR-128a is
downregulated in human glioblastoma (18) and its expression inhibits cell
growth in vitro and in vivo (19). To examine the biological function
of miR-128a expression, U87 cells were transfected with miR-128a
mimics or the miR-128a inhibitor and CCK-8 proliferation assays
were performed. Overexpression of miR-128a decreased the cell
proliferation rate, whereas, knockdown of miR-128a with the
miR-128a inhibitor enhanced cell proliferation (Fig. 4A; P<0.01). IDH1R132H
overexpressing cells were also treated with the miR-128a inhibitor,
which rescued the decreased proliferation rate induced by
IDH1R132H (Fig. 4B;
P<0.01). miR-128a causes a marked decrease in the expression of
the oncogene Bmi-1 in U87 cells by direct regulation of the Bmi-1
mRNA 3′-untranslated region (19).
Therefore, it was hypothesized that Bmi-1 may be involved in the
decreased proliferation of IDH1R132H cells. qPCR was
performed to detect the Bmi-1 level in IDH1R132H
overexpressing cells. The results demonstrated that it was
decreased compared with the control and IDH1WT cells
(Fig. 4C; P<0.01). Furthermore,
overexpression of miR-128a decreased the mRNA expression of Bmi-1,
whereas inhibition of miR-128a enhanced Bmi-1 expression (Fig. 4D; P<0.01). Taken together, it
can be concluded that IDH1R132H restricts cell
proliferation by inducing the expression of miR-128a, which
downregulates Bmi-1 expression.
Discussion
Clinical data demonstrate that patients with the
IDH1R132H mutation have an improved outcome compared
with those with the wild-type sequence. However, the mechanistic
role by which IDH1R321H regulates tumor development is
not fully understood. In the present study, the functional impact
of the tumor-associated IDH1R132H mutation was examined
in U87 glioma cells in vitro. Following establishing stable
expression of IDH1R132H in U87 cells, it was found that
it reduced cell proliferation, which is consistent with the
findings of Bralten et al (20). This lends support to the theory
that IDH1 mutations protect against tumor proliferation and
may explain why IDH1R132H is an independent favorable
prognostic marker in glioma patients (15,21).
Wang et al reported that the IDH1
mutation-specific microRNA signature predicted a favorable
prognosis in glioblastoma patients with the IDH1 wild type
(11). However, it remained to be
determined whether the IDH1R132H mutation regulated cell
growth through microRNAs. miR-128a expression is markedly reduced
in human glioblastoma specimens compared with adjacent brain
samples without tumors (18,19,22).
A previous study suggested that loss of miR-128 expression is an
early event in gliomagenesis (23). By analyzing the TCGA data, miR-128a
was found to be overexpressed in human IDH1R132H GBM
patients compared with wild-type IDH1 patients. In addition,
miR-128a expression was increased in IDH1R132H
overexpressing U87 glioma cells. The result demonstrating that
miR-128a is upregulated in IDH1R132H gliomas is
noteworthy given that miR-128a is downregulated in
IDH1WT glioblastomas (11). This demonstrates that increased
miR-128a expression is an important characteristic of mutant
IDH1R132H gliomas.
miR-128a is downregulated in GBM, suggesting that it
may function as a tumor suppressor in brain tumor progression
(24) and may be associated with
the survival of GBM patients (11). In U87 cells, overexpression of
miR-128a suppressed cell proliferation. By contrast, knockdown of
miR-128a increased cell proliferation. Our findings are consistent
with a study on miR-128a in medulloblastoma cells (25). Furthermore, the present study
illustrated that inhibition of miR-128a in IDH1R132H
overexpressing U87 cells reversed the cell proliferation induced by
IDH1R132H, suggesting that miR-128a is responsible for
IDH1R132H-induced cell proliferation.
HIF-1α, an important transcription factor of the
cellular response to hypoxia, is known to facilitate tumor growth
when oxygen is low (26). The
stability of HIF-1α is regulated by α-ketoglutaric acid, which is
an important enzyme product of IDH1 (27). The results demonstrated that
overexpression of the IDH1R132H mutant increased HIF-1α
protein levels in U87 glioma cells, which is consistent with a
former study by Zhao et al (16). In the present study, miR-128a
expression was significantly decreased when the activity of HIF-1α
was inhibited, which indicated that HIF-1α may regulate miR-128a in
IDH1R132H cells. In addition, an HRE located in the
promoter upstream of the gene encoding pri-miR-128a was identified
and HIF-1α was found to specifically bind the site in
IDH1R132H over-expressing U87 cells. This suggests that
the transcription factor HIF-1α regulates miR-128a expression.
Currently, to the best of our knowledge, the present study is the
first to demonstrate that HIF-1α acts as a regulator of
IDH1R132-dependent miR-128a expression by directly
binding an HRE within its promoter.
Bmi-1, a direct target of miR-128a, is upregulated
in several types of cancer, including gliomas and is a positive
regulator of neural stem cell renewal (19,28–31).
The present study demonstrated that Bmi-1 was downregulated by
miR-128a in IDH1R132H overexpressing cells. Bmi-1 is an
oncogene involved in regulating cell proliferation as a
transcriptional repressor (19),
and studies in transgenic mice revealed a critical role for Bmi-1
in driving glioma growth (32).
The present data indicated that inhibition of Bmi-1 may be a
mechanism of miR-128a-mediated growth restriction in
IDH1R132H over-expressing cells.
In conclusion, the present study described the
function and mechanism of action of miR-128a in
IDH1R132H glioma cells. The results of the present study
demonstrated that the IDH1R132H mutation leads to an
increase in the levels of HIF-1α protein, which binds to an HRE in
the miR-128a promoter to induce its expression. The enhanced
expression of miR-128a subsequently inhibits Bmi-1 expression,
thereby decreasing cell proliferation in IDH1R132H
glioma cells. The present study provides novel insight into the
pathophysiology of miR-128a in IDH1 mutant glioma. Finally,
our characterization of the
IDH1R132H-HIF-1α-miR-128a-Bmi-1 pathway may have
therapeutic implications for the treatment of glioma.
Acknowledgments
This study was supported by the Shanghai Science and
Technology Committee (grant no. 13XD1402600), Shanghai Health and
Family planning Commission (grant no. 2013SY024) and the State Key
Laboratory of Oncogenes and Related Genes (grant no. 90-14-01),
Shanghai Cancer Institute, Ren Ji Hospital, School of Medicine,
Shanghai Jiao Tong University.
References
|
1
|
Johannessen TC, Prestegarden L, Grudic A,
Hegi ME, Tysnes BB and Bjerkvig R: The DNA repair protein ALKBH2
mediates temozolomide resistance in human glioblastoma cells. Neuro
Oncol. 15:269–278. 2013. View Article : Google Scholar :
|
|
2
|
Hill VK, Shinawi T, Ricketts CJ, Krex D,
Schackert G, Bauer J, Wei W, Cruickshank G, Maher ER and Latif F:
Stability of the CpG island methylator phenotype during glioma
progression and identification of methylated loci in secondary
glioblastomas. BMC Cancer. 14:5062014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agarwal S, Sharma MC and Jha P, Pathak P,
Suri V, Sarkar C, Chosdol K, Suri A, Kale SS, Mahapatra AK and Jha
P: Comparative study of IDH1 mutations in gliomas by
immunohistochemistry and DNA sequencing. Neuro Oncol. 15:718–726.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Henriksen M, Johnsen KB, Andersen HH,
Pilgaard L and Duroux M: MicroRNA expression signatures determine
prognosis and survival in glioblastoma multiforme - a systematic
overview. Mol Neurobiol. 50:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Blenkiron C and Miska EA: MiRNAs in
cancer: Approaches, aetiology, diagnostics and therapy. Hum Mol
Genet. 16(Spec No 1): R106–R113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Zhang J, Yan W, You G, Bao Z, Li
S, Kang C, Jiang C, You Y, Zhang Y, et al: Whole-genome microRNA
expression profiling identifies a 5-microRNA signature as a
prognostic biomarker in Chinese patients with primary glioblastoma
multiforme. Cancer. 119:814–824. 2013. View Article : Google Scholar
|
|
8
|
Srinivasan S, Patric IR and Somasundaram
K: A ten-microRNA expression signature predicts survival in
glioblastoma. PLoS One. 6:e174382011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Novakova J, Slaby O, Vyzula R and Michalek
J: MicroRNA involvement in glioblastoma pathogenesis. Biochem
Biophys Res Commun. 386:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sempere LF, Freemantle S, Pitha-Rowe I,
Moss E, Dmitrovsky E and Ambros V: Expression profiling of
mammalian microRNAs uncovers a subset of brain-expressed microRNAs
with possible roles in murine and human neuronal differentiation.
Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Bao Z, Yan W, You G, Wang Y, Li X
and Zhang W: Isocitrate dehydrogenase 1 (IDH1) mutation-specific
microRNA signature predicts favorable prognosis in glioblastoma
patients with IDH1 wild type. J Exp Clin Cancer Res. 32:592013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma X, Yoshimoto K, Guan Y, Hata N,
Mizoguchi M, Sagata N, Murata H, Kuga D, Amano T, Nakamizo A and
Sasaki T: Associations between microRNA expression and mesenchymal
marker gene expression in glioblastoma. Neuro Oncol. 14:1153–1162.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Papagiannakopoulos T, Friedmann-Morvinski
D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch
DH and Barres BA: Pro-neural miR-128 is a glioma tumor suppressor
that targets mitogenic kinases. Oncogene. 31:1884–1895. 2012.
View Article : Google Scholar
|
|
14
|
Cui JG, Zhao Y, Sethi P, Li YY, Mahta A,
Culicchia F and Lukiw WJ: Micro-RNA-128 (miRNA-128) down-regulation
in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key
regulators of brain cell proliferation. J Neurooncol. 98:297–304.
2010. View Article : Google Scholar
|
|
15
|
Sanson M, Marie Y, Paris S, Idbaih A,
Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K,
Hoang-Xuan K and Delattre JY: Isocitrate dehydrogenase 1 codon 132
mutation is an important prognostic biomarker in gliomas. J Clin
Oncol. 27:4150–4154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang
P, Yu W, Li Z, Gong L, Peng Y, et al: Glioma-derived mutations in
IDH1 dominantly inhibit IDH1 catalytic activity and induce
HIF-1alpha. Science. 324:261–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu YL, DeLay M, Jahangiri A, Molinaro AM,
Rose SD, Carbonell WS and Aghi MK: Hypoxia-induced autophagy
promotes tumor cell survival and adaptation to antiangiogenic
treatment in glioblastoma. Cancer Res. 72:1773–1783. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lakomy R, Sana J, Hankeova S, Fadrus P,
Kren L, Lzicarova E, Svoboda M, Dolezelova H, Smrcka M, Vyzula R,
et al: MiR-195, miR-196b, miR-181c, miR-21 expression levels and
O-6-methylguanine-DNA methyltransferase methylation status are
associated with clinical outcome in glioblastoma patients. Cancer
Sci. 102:2186–2190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bralten LB, Kloosterhof NK, Balvers R,
Sacchetti A, Lapre L, Lamfers M, Leenstra S, de Jonge H, Kros JM,
Jansen EE, et al: IDH1 R132H decreases proliferation of glioma cell
lines in vitro and in vivo. Ann Neurol. 69:455–463. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu J, Zuo J, Xu Q, Wang X, Wang Z and
Zhou D: Isocitrate dehydrogenase mutations may be a protective
mechanism in glioma patients. Med Hypotheses. 76:602–603. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary
glio-blastoma. Biochem Biophys Res Commun. 334:1351–1358. 2005.
View Article : Google Scholar
|
|
23
|
Peruzzi P, Bronisz A, Nowicki MO, Wang Y,
Ogawa D, Price R, Nakano I, Kwon CH, Hayes J, Lawler SE, et al:
MicroRNA-128 coordinately targets polycomb repressor complexes in
glioma stem cells. Neuro Oncol. 15:1212–1224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian
X, Yin Y, Zhao P, Wang YY, Wang XF, et al: MiR-128 inhibits tumor
growth and angiogenesis by targeting p70S6K1. PLoS One.
7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venkataraman S, Alimova I, Fan R, Harris
P, Foreman N and Vibhakar R: MicroRNA 128a increases intracellular
ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer
cell growth by promoting senescence. PLoS One. 5:e107482010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kamiyama H, Takano S, Tsuboi K and
Matsumura A: Anti-angiogenic effects of SN38 (active metabolite of
irinotecan): Inhibition of hypoxia-inducible factor 1 alpha
(HIF-1alpha)/vascular endothelial growth factor (VEGF) expression
of glioma and growth of endothelial cells. J Cancer Res Clin Oncol.
131:205–213. 2005. View Article : Google Scholar
|
|
27
|
Ichimura K: Molecular pathogenesis of IDH
mutations in gliomas. Brain Tumor Pathol. 29:131–139. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nanta R, Kumar D, Meeker D, et al:
NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal
transition and human prostate cancer stem cell growth in NOD/SCID
IL2Rγ null mice by regulating Bmi-1 and microRNA-128. Oncogenesis.
2:e422013. View Article : Google Scholar
|
|
29
|
Li J, Gong LY, Song LB, Jiang LL, Liu LP,
Wu J, Yuan J, Cai JC, He M, Wang L, et al: Oncoprotein Bmi-1
renders apoptotic resistance to glioma cells through activation of
the IKK-nuclear factor-kappaB Pathway. Am J Pathol. 176:699–709.
2010. View Article : Google Scholar :
|
|
30
|
Farivar S, Zati Keikha R, Shiari R and
Jadali F: Expression of bmi-1 in pediatric brain tumors as a new
independent prognostic marker of patient survival. Biomed Res Int.
2013:1925482013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siddique HR, Parray A, Zhong W, Karnes RJ,
Bergstralh EJ, Koochekpour S, Rhim JS, Konety BR and Saleem M:
BMI1, stem cell factor acting as novel serum-biomarker for
Caucasian and African-American prostate cancer. PLoS One.
8:e529932013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bruggeman SW, Hulsman D, Tanger E, Buckle
T, Blom M, Zevenhoven J, van Tellingen O and van Lohuizen M: Bmi1
controls tumor development in an Ink4a/Arf-independent manner in a
mouse model for glioma. Cancer Cell. 12:328–341. 2007. View Article : Google Scholar : PubMed/NCBI
|