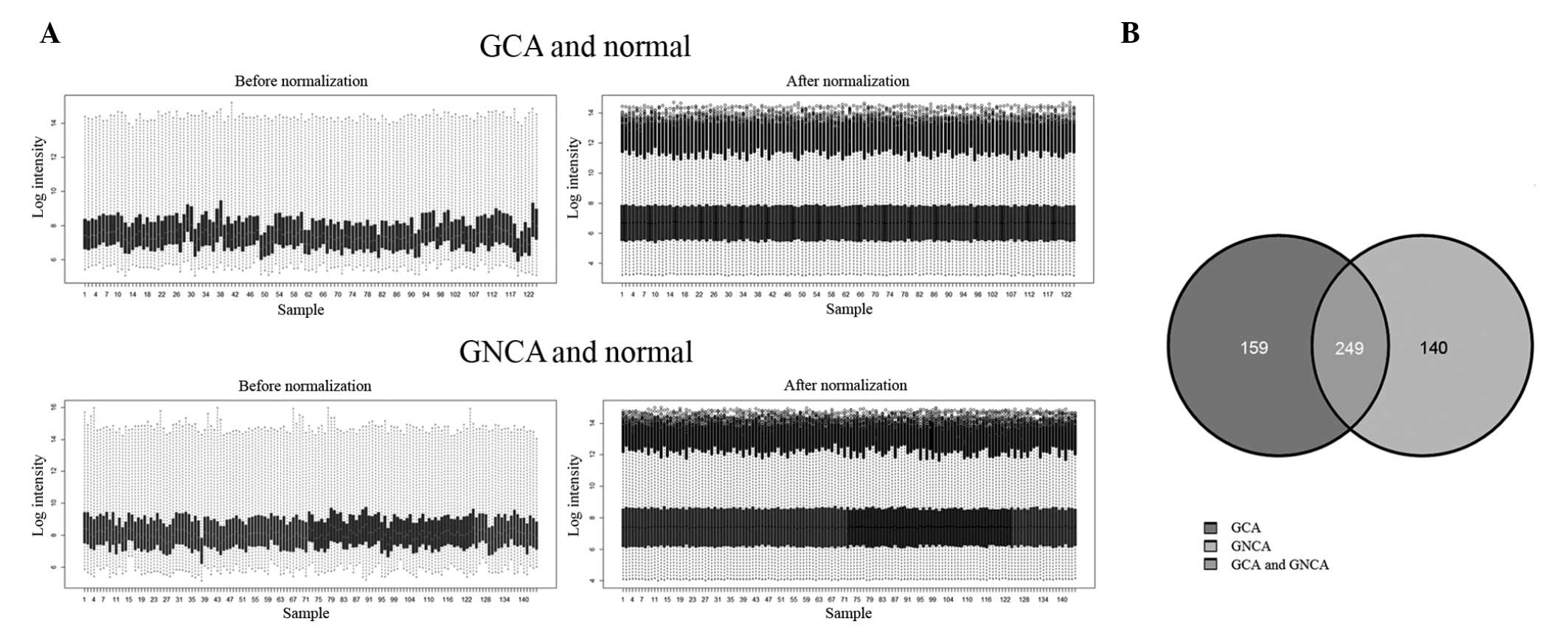
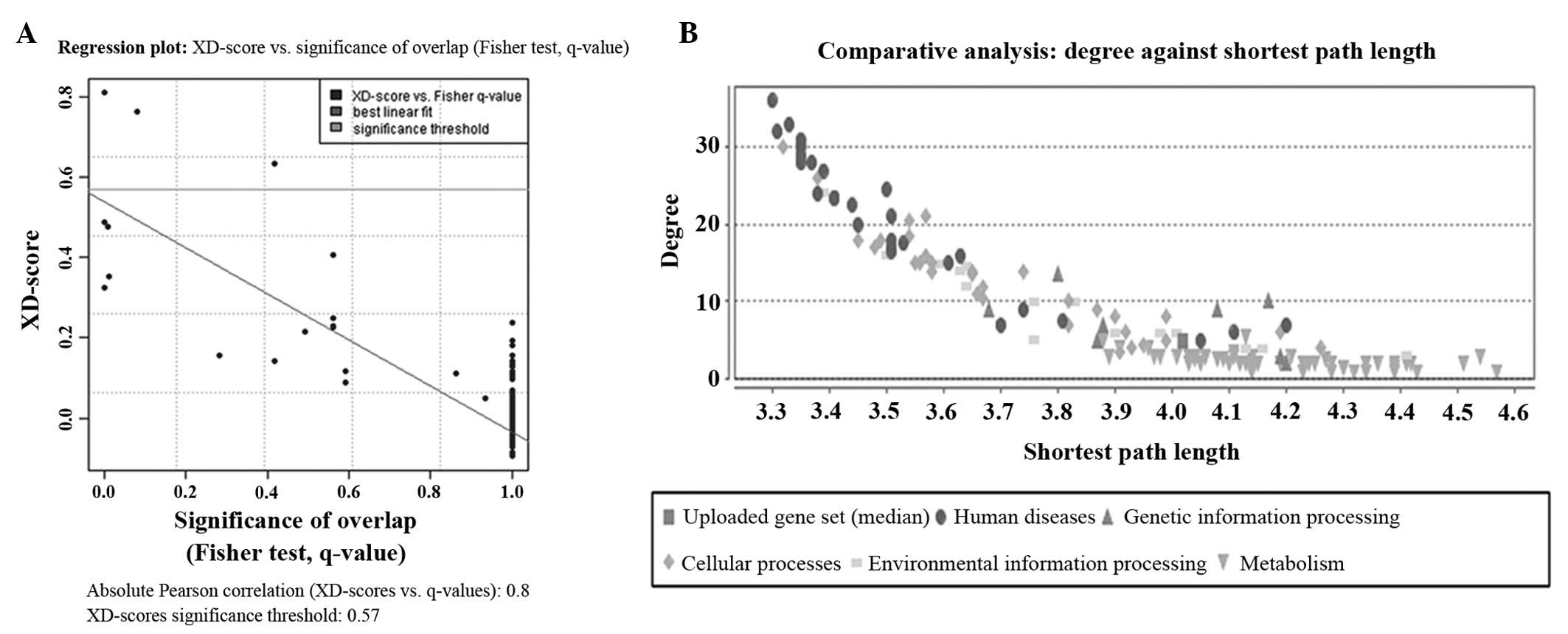
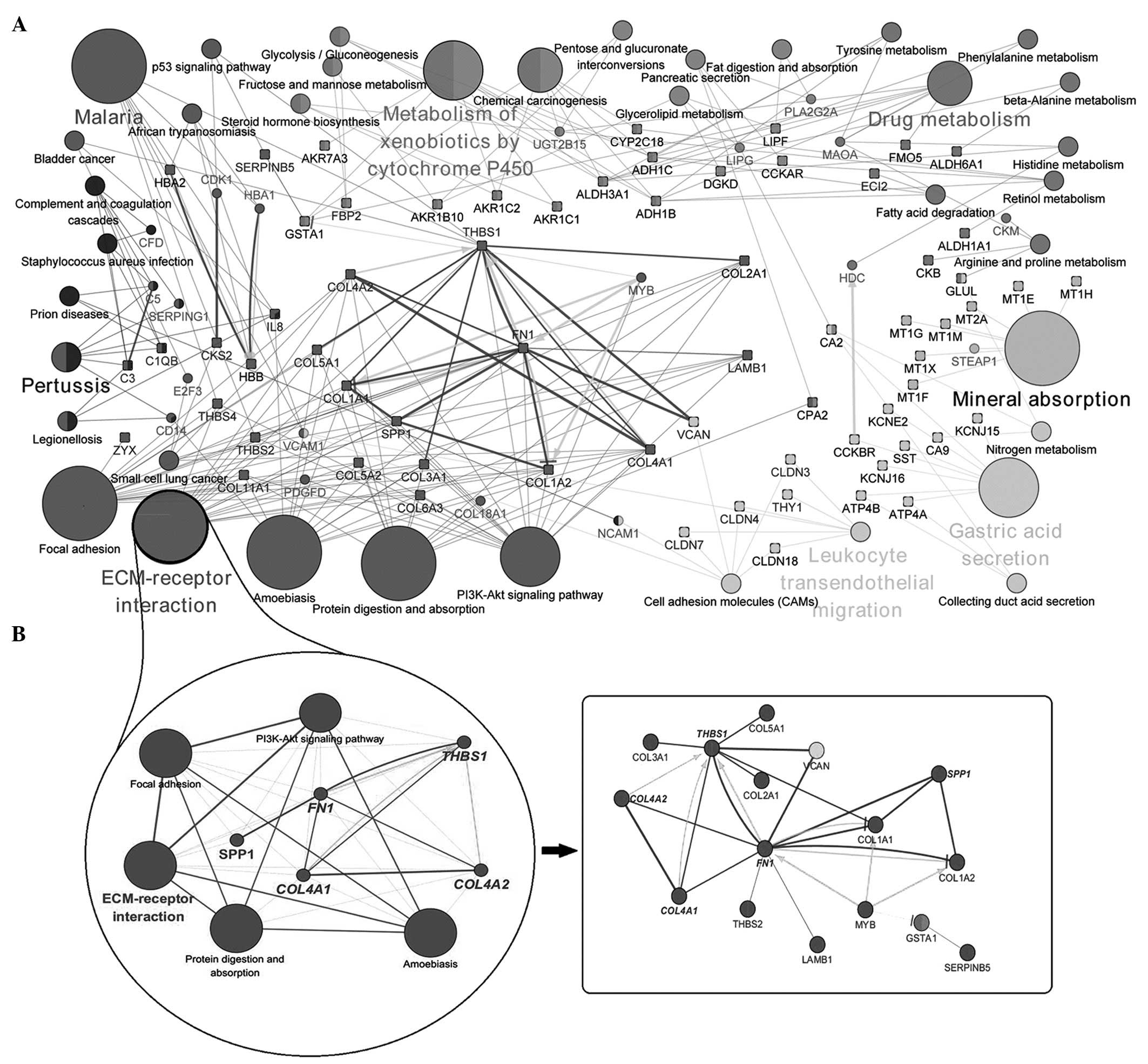
|
1
|
Catalano V, Labianca R, Beretta GD, Gatta
G, de Braud F and Van Cutsem E: GC. Crit Rev Oncol Hematol.
71:127–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006.PubMed/NCBI
|
|
3
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Loscalzo J and Barabasi AL: Systems
biology and the future of medicine. Wiley Interdiscip Rev Syst Biol
Med. 3:619–627. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
D'Souza M, Zhu X and Frisina RD: Novel
approach to select genes from RMA normalized microarray data using
functional hearing tests in aging mice. J Neurosci Methods.
171:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davis JW: Bioinformatics and computational
biology solutions using R and bioconductor. J Am Statistical
Association. 102:388–389. 2007. View Article : Google Scholar
|
|
8
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maere S, Heymans K and Kuiper M: BiNGO: A
cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Winterhalter C, Widera P and Krasnogor N:
JEPETTO: A Cytoscape plugin for gene set enrichment and topological
analysis based on interaction networks. Bioinformatics.
30:1029–1030. 2014. View Article : Google Scholar :
|
|
12
|
Glaab E, Baudot A, Krasnogor N, Schneider
R and Valencia A: EnrichNet: Network-based gene set enrichment
analysis. Bioinformatics. 28:i451–i457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Glaab E, Baudot A, Krasnogor N and
Valencia A: Extending pathways and processes using molecular
interaction networks to analyse cancer genome data. BMC
Bioinformatics. 11:5972010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glaab E, Baudot A, Krasnogor N and
Valencia A: TopoGSA: Network topological gene set analysis.
Bioinformatics. 26:1271–1272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bindea G, Galon J and Mlecnik B: CluePedia
cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu X and Sun S: Comparing a few SNP
calling algorithms using low-coverage sequencing data. BMC
Bioinformatics. 14:2742013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alakwaa FM, Solouma NH and Kadah YM:
Construction of gene regulatory networks using biclustering and
Bayesian networks. Theor Biol Med Model. 8:392011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitra S, Das S and Chakrabarti J: Systems
biology of cancer biomarker detection. Cancer Biomark. 13:201–213.
2013.PubMed/NCBI
|
|
20
|
Zhang L, Sung JJ, Yu J, Ng SC, Wong SH,
Cho CH, Ng SS, Chan FK and Wu WK: Xenophagy in Helicobacter pylori-
and Epstein-Barr virus-induced gastric cancer. J Pathol.
233:103–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang MR, Kim MS, Oh JE, Kim YR, Song SY,
Kim SS, Ahn CH, Yoo NJ and Lee SH: Frameshift mutations of
autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and
colorectal cancers with microsatellite instability. J Pathol.
217:702–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bijian K, Takano T, Papillon J, Khadir A
and Cybulsky AV: Extracellular matrix regulates glomerular
epithelial cell survival and proliferation. Am J Physiol Renal
Physiol. 286:F255–F266. 2004. View Article : Google Scholar
|
|
23
|
Yin Y, Zhao Y, Li AQ and Si LM: Collagen:
A possible prediction mark for gastric cancer. Medical Hypotheses.
72:163–165. 2009. View Article : Google Scholar
|
|
24
|
Yam JW, Tse EY and Ng IO: Role and
significance of focal adhesion proteins in hepatocellular
carcinoma. J Gastroenterol Hepatol. 24:520–530. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ramos DM, But M, Regezi J, Schmidt BL,
Atakilit A, Dang D, Ellis D, Jordan R and Li X: Expression of
integrin beta 6 enhances invasive behavior in oral squamous cell
carcinoma. Matrix Biol. 21:297–307. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen Z, Ye Y, Dong L, Vainionpää S,
Mustonen H, Puolakkainen P and Wang S: Kindlin-2: A novel adhesion
protein related to tumor invasion, lymph node metastasis and
patient outcome in gastric cancer. Am J Surg. 203:222–229. 2012.
View Article : Google Scholar
|
|
27
|
Colacci E, Pasquali A and Severi C:
Exocrine gastric secretion and gastritis: Pathophysiological and
clinical relationships. Clin Ter. 162:e19–25. 2011.PubMed/NCBI
|
|
28
|
Jinawath N, Furukawa Y, Hasegawa S, Li M,
Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H and
Nakamura Y: Comparison of gene-expression profiles between diffuse-
and intestinal-type gastric cancers using a genome-wide cDNA
microarray. Oncogene. 23:6830–6844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ossandon FJ, Villarroel C, Aguayo F,
Santibanez E, Oue N, Yasui W and Corvalan AH: In silico analysis of
gastric carcinoma serial analysis of gene expression libraries
reveals different profiles associated with ethnicity. Mol Cancer.
7:222008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ramaswamy S, Ross KN, Lander ES and Golub
TR: A molecular signature of metastasis in primary solid tumors.
Nat Genet. 33:49–54. 2003. View
Article : Google Scholar
|
|
31
|
Wiedemann S, Wessela T, Schwarz K, Joachim
D, Jercke M, Strasser RH, Ebner B and Simonis G: Inhibition of
anti-apoptotic signals by Wortmannin induces apoptosis in the
remote myocardium after LAD ligation: Evidence for a protein kinase
C-delta-dependent pathway. Mol Cell Biochem. 372:275–283. 2013.
View Article : Google Scholar
|
|
32
|
Adams JC: Functions of the conserved
thrombospondin carboxy-terminal cassette in cell-extracellular
matrix inter-actions and signaling. Int J Biochem Cell Biol.
36:1102–1114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin XD, Chen SQ, Qi YL, Zhu JW, Tang Y and
Lin JY: Polymorphism of THBS1 rs1478604 A>G in 5-untranslated
region is associated with lymph node metastasis of gastric cancer
in a Southeast Chinese population. DNA Cell Biol. 31:511–519. 2012.
View Article : Google Scholar
|
|
34
|
Rosman DS, Phukan S, Huang CC and Pasche
B: TGFBR1* 6A enhances the migration and invasion of
MCF-7 breast cancer cells through RhoA activation. Cancer Res.
68:1319–1328. 2008. View Article : Google Scholar : PubMed/NCBI
|