Introduction
Breast cancer is the most common type of invasive
cancer in females worldwide (1,2). In
addition to conventional prognostic factors, including tumor size,
lymph node status, estrogen receptor (ER)/progesterone receptor
(PR) status, erbB2 receptor (HER2) status, several other biological
markers, including Ki67, tau and topo II, have been investigated in
order to correlate their status with the prognosis of patients with
breast cancer (3,4). Due to a lack of specific receptors,
triple-negative (ER−, PR−, HER2−) breast cancer (TNBC) exhibits
marked resistance to chemotherapy, hormone therapy and targeted
therapy (5–7). The subtype comprises ~15% of all
cases of breast cancer and results in tumors, which are typically
larger in size and higher in grade, compared with other types of
breast cancer.
The identification of intrinsic subtypes of breast
cancer has provided a number of novel therapeutic targets and
enabled the design of novel clinical trials (8). Due to the lack of specific
therapeutic targets with TNBC, previous studies have aimed to
identify novel molecular markers, including phosphoinositide
3-kinase, epidermal growth factor receptor (EGFR), cell-cycle
regulatory proteins, heat shock proteins, epigenetic pathways and
androgen receptors, and clinical trials have been performed
(8,9). In this context, the loss of androgen
receptor expression has been found to predict early recurrence in
triple-negative and basal-like breast cancer (10). However, the therapeutic outcomes
have not reached initial expectations.
Previous genome-wide association investigations have
identified genetic variants, which are associated with an increased
risk of developing breast cancer. Furthermore, gene expression
profile analyses have provided multi-gene signatures associated
with TNBC carcinogenesis (11,12).
Although investigations are increasingly focussing on the
recurrence (5,6,13)
and treatment (7) of TNBC, the
molecular profiles of paired specimens obtained from patients with
and without subsequent recurrence require further investigation.
Therefore, the aim of the present study was to investigate and
compare the gene expression profiles obtained from patients with
TNBC, with and without recurrent status.
Patients and methods
Human TNBC tissue samples
In the present study, 30 patients, who had been
diagnosed with TNBC based on pathological confirmation (ER <1%;
PR <1%; HER2, not amplified) and who had been followed up for ≥5
years were recruited between June 2011 and May 2012. The tumor
stage of these patients was determined using the American Joint
Committee on Cancer TNM system (version 6; Springer, Inc., New
York, NY, USA). The clinicopathological information was
retrospectively reviewed and identified as either recurrent [n=15,
stage II (9), IIIA (2), IIIC (4)] or non-recurrent [n=15, stage II
(6), IIIA (1), IIIC (8)] at the end of the investigation period
(2011–2012). Subsequent to obtaining informed consent from the
patients, tumor specimens were obtained during surgery and were
stored in liquid nitrogen under the regulations of Taipei Veterans
General Hospital (Taipei, China). The present study was approved by
the institutional review board of the hospital (2011-06-001GCF;
Taipei, China) for microarray gene expression profiling analysis.
The tumor tissues were divided into two groups; the recurrent
group, defined as the patients being having a recurrence status
years later, and the non-recurrent group, defined as those without
recurrence at the end of the investigation.
Isolation of RNA from tumor tissues and
oligonucleotide microarray analysis
Total RNA was isolated using a modified single-step
guanidinium thiocyanate method (TRIzol reagent; cat. no. T-9424;
Sigma-Aldrich, St. Louis, MO, USA). The concentration of total RNA
required for oligonucleotide micro-array analysis was ≥0.6 g/l with
a quality ratio ≥1.8 for absorbance at 260/280 nm and 260/230 nm
using a NanoDrop 1000 (Thermo Fisher Scientific, Pittsburgh, PA,
USA). Suitable RNA samples were then sent to Genome Research
Center, National Yang-Ming University (Taiwan, China), for analysis
with the human Affymetrix expression set, Hu-133 2.0 (Affymetrix,
Inc., Santa Clara, CA, USA). The dataset obtained was analyzed
following normalization against the global signal. The relative
expression levels for each gene were obtained using GeneSpring
software (GeneSpring GX 11.5; Agilent Technologies, Inc., Santa
Clara, CA, USA) and further pathway analysis using Ingenuity
Pathway Analysis software (IPA; Qiagen, Redwood City, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Following extraction of tumor total RNA,
complementary DNA (cDNA) was created using a First Strand cDNA
Synthesis kit (Invitrogen Life Technologies, CA, USA).
TaqMan® Gene Expression Assays (Invitrogen Life
Technologies) were then used to validate the differential
expression at the mRNA level of various identified genes, such as
MMP9 and SERPINE1 set in the recurrent and non-recurrent TNBC
samples. The TaqMan system is supported by a well established
primer database, which significantly reduces experimental failure
due to inappropriate primer design (Invitrogen Life Technologies).
All samples were analyzed in triplicate.
Statistical analysis
Analysis of the microarray dataset was divided into
three steps. Firstly, the differentially expressed genes were
identified using Student's t-test and a repeated measures one-way
analysis of variance. The purpose of this step was to identify a
gene set associated with recurrent TNBC and with the various cancer
grades. Secondly, clustering analysis was performed to validate the
performance of the identified gene set, through separation of the
different cancer groups, namely recurrent, vs. non-recurrent TNBCs
and grade 1, vs. grade 2, vs. grade 3. The purpose of this step was
to reduce the number of gene sets originally obtained and to obtain
gene sets, which were optimal for cancer subtype classification.
The final step was gene set annotation using GeneSpring software
and IPA. Gene, Gene Ontology and pathway information were
integrated in order to evaluate the biomedical importance of the
identified gene sets.
Results
Clinical and pathological information, which was
obtained by retrospective review completed at the end of 2012,
resulted in two groups of patients, which were divided into those
with subsequent recurrence [n=15, stage II (9), IIIA (2), IIIC (4)] and those without recurrence [n=15,
stage II (6), IIIA (1), IIIC (8)], as shown in Fig. 1.
Hierarchical clustering analysis of 30
patients with triple-negative breast cancer
A microarray dataset of the 30 TNBC samples was
collected and a total 54,675 genes were eligible for analysis. Gene
expression data were normalized against normal tissue controls
using quantile normalization. The molecular characteristics of the
TNBC were investigated at the mRNA level, according to previously
described criteria (8).
Hierarchical clustering analysis of the 30 TNBC samples, of which
15 were non-recurrent and 15 were recurrent, was performed to
identify genes that were upregulated by ≥2-fold. The results
demonstrated no significant clustering among the non-recurrent and
recurrent samples (data not shown).
Principal component analysis of 15
patients with recurrent triple-negative breast cancer
To investigate the level of heterogeneity within the
recurrent tumor samples, principal component analysis was performed
and the results demonstrated a marked difference in distribution
between the stage IIIc recurrent samples and the early stage (stage
IIa, IIb, IIIa) recurrent samples (Fig. 2). Thedr results suggested that the
gene expression profiles of the stage IIIc TNBC tumors were
different from those of the early stage TNBC tumors, and they were
also different from those of the stage IV, metastatic tumors (data
not shown).
A number of significant pathways involving specific
genes were either upregulated or downregulated during the early
stages of recurrent TNBC. For validation of the microarray
information, RT-qPCR was performed on selected genes, including
MMP9 (Fig. 3A) and SERPINE1
(Fig. 3B).
Using bioinformatics software analysis, the 30 TNBC
samples were further stratified into four subgroups depending on
stage, classified as early stage (IIa, IIb, IIIa) or stage IIIc,
and recurrence, classified as non-recurrent or recurrent. In early
stage recurrence, the significant pathways with upregulated genes
in the recurrence samples included the osteoblast and ossification
pathway, the serotonin receptor 2 pathway, various prostate cancer
genes and the epidermal growth factor (EGF)-EGFR pathway (Fig. 4A; Table I); while the significant pathways
with downregulated genes included the actin cytoskeleton pathway,
the G protein pathway, the inflammatory response pathway and the
insulin synthesis pathway (Table
II).
 | Table ISignificant pathways associated with
upregulated genes in early astage recurrent triple-negative breast
cancer. |
Table I
Significant pathways associated with
upregulated genes in early astage recurrent triple-negative breast
cancer.
| Pathway | P-value |
|---|
|
Hs_Osteoblast_Signaling_WP322_53892 | 0.00118489 |
|
Hs_Endochondral_Ossification_WP474_45241 | 0.00172066 |
|
Hs_Serotonin_Receptor_2_and_ELK-SRF-GATA4_signaling_WP732_49572 | 0.00175804 |
|
Hs_GPCRs,_Other_WP117_45343 | 0.00466699 |
|
Hs_Spinal_Cord_Injury_WP2431_56064 | 0.00606436 |
|
Hs_Monoamine_GPCRs_WP58_48221 | 0.00656731 |
|
Hs_Integrated_Cancer_pathway_WP1971_44858 | 0.00777973 |
|
Hs_Prostate_Cancer_WP2263_54728 | 0.00807849 |
|
Hs_Serotonin_Receptor_2_and_STAT3_Signaling_WP733_45035 | 0.01460303 |
|
Hs_EGF-EGFR_Signaling_Pathway_WP437_47973 | 0.01537961 |
|
Hs_RANKL-RANK_Signaling_Pathway_WP2018_48442 | 0.01752249 |
|
Hs_MAP_kinase_cascade_WP1844_44888 | 0.01822052 |
|
Hs_Myometrial_Relaxation_and_Contraction_Pathways_WP289_45373 | 0.01972625 |
|
Hs_SIDS_Susceptibility_Pathways_WP706_55364 | 0.02248320 |
|
Hs_Oncostatin_M_Signaling_Pathway_WP2374_54418 | 0.02260932 |
|
Hs_Glial_Cell_Differentiation_WP2276_53125 | 0.02541592 |
|
Hs_Heme_Biosynthesis_WP561_45350 | 0.03255886 |
|
Hs_Apoptosis_Modulation_and_Signaling_WP1772_44957 | 0.03513467 |
|
Hs_APC-C-mediated_degradation_of_cell_cycle_proteins_WP1782_44955 | 0.03611077 |
 | Table IISignificant pathways associated with
downregulated genes in early stagea recurrent triple-negative breast
cancer. |
Table II
Significant pathways associated with
downregulated genes in early stagea recurrent triple-negative breast
cancer.
| Pathway | P-value |
|---|
|
Hs_Calcium_Regulation_in_the_Cardiac_Cell_WP536_44983 | 7.37E-05 |
|
Hs_Biogenic_Amine_Synthesis_WP550_44978 | 3.17E-04 |
|
Hs_Regulation_of_Actin_Cytoskeleton_WP51_49262 | 3.96E-04 |
|
Hs_Regulation_of_beta-cell_development_WP1897_45053 | 5.57E-04 |
|
Hs_Myometrial_Relaxation_and_Contraction_Pathways_WP289_45373 | 5.90E-04 |
|
Hs_G_Protein_Signaling_Pathways_WP35_45294 | 0.00159171 |
|
Hs_Neural_Crest_Differentiation_WP2064_47071 | 0.00229661 |
|
Hs_Hypothetical_Network_for_Drug_Addiction_WP666_45365 | 0.00281034 |
|
Hs_Benzo(a)pyrene_metabolism_WP696_44980 | 0.00286947 |
|
Hs_Inflammatory_Response_Pathway_WP453_41201 | 0.00308023 |
|
Hs_Selenium_Pathway_WP15_56489 | 0.00656218 |
|
Hs_Osteoblast_Signaling_WP322_53892 | 0.00703770 |
|
Hs_Insulin_Synthesis_and_Processing_WP1830_44856 | 0.00703770 |
|
Hs_Tryptophan_metabolism_WP465_45056 | 0.00858096 |
|
Hs_Estrogen_signaling_pathway_WP712_48214 | 0.01417347 |
|
Hs_SIDS_Susceptibility_Pathways_WP706_55364 | 0.01731102 |
|
Hs_EPO_Receptor_Signaling_WP581_41162 | 0.02338902 |
|
Hs_miRs_in_Muscle_Cell_Differentiation_WP2012_45377 | 0.03248199 |
|
Hs_Metabolism_of_amino_acids_and_derivatives_WP1847_52373 | 0.03248199 |
|
Hs_Endothelin_WP2197_56443 | 0.03644705 |
Metastasis-associated gene expression in
early stage recurrent TNBC
Since metastasis is a mechanism associated with
tumor recurrence in TNBC, a subset of overexpressed genes were
identified in tumor samples obtained from the patients with early
stage (IIa, IIb or IIIa) TNBC, who had undergone subsequent tumor
recurrence. These included genes encoding MMPs, genes involved in
cancer cell migration (CDH2) and genes involved in cell
adhesion/motility (KRAS, CDC42, RAC1, ICAM and SRGAP2) (Fig. 4B). Notably, genes associated with
tumor stemness and angiogenesis were not overexpressed in these
tumor samples.
Significant pathways in stage IIIc
recurrent TNBC associated with upregulated and downregulated
genes
In contrast to the observations for early
recurrence, the significant pathways involving upregulated genes
present in stage IIIc recurrence samples included the WNT signaling
pathway, glycogen metabolism pathway, integrated pancreatic cancer
pathway, mitogen-activated protein kinase (MAPK) cascade pathway,
brain-derived neurotrophic factor (BDNF) signaling pathway and
prostaglandin synthesis pathway (Table III), while the significant
pathways involving downregulated genes in the stage IIIc recurrence
samples included the AMPK signaling pathway, interleukin (IL)-2
signaling pathway and BDNF signaling pathway (Table IV).
 | Table IIISignificant pathways associated with
upregulated genes in stage IIIc recurrent triple-negative breast
cancer. |
Table III
Significant pathways associated with
upregulated genes in stage IIIc recurrent triple-negative breast
cancer.
| Pathway | P-value |
|---|
|
Hs_Factors_involved_in_megakaryocyte_development_and_platelet_production_WP1815_42038 | 8.41E-05 |
|
Hs_Wnt_Signaling_Pathway_and_Pluripotency_WP399_54212 | 0.00149959 |
|
Hs_Wnt_Signaling_Pathway_WP428_45008 | 0.00162369 |
|
Hs_Nuclear_receptors_in_lipid_metabolism_and_toxicity_WP299_45336 | 0.00262814 |
|
Hs_Glycogen_Metabolism_WP500_45329 | 0.00337628 |
|
Hs_G13_Signaling_Pathway_WP524_45304 | 0.00365162 |
|
Hs_Integrated_Pancreatic_Cancer_Pathway_WP2256_54274 | 0.00683842 |
|
Hs_Integrated_Pancreatic_Cancer_Pathway_WP2377_56677 | 0.00683842 |
|
Hs_G_Protein_Signaling_Pathways_WP35_45294 | 0.00755248 |
|
Hs_Calcium_Regulation_in_the_Cardiac_Cell_WP536_44983 | 0.00855703 |
|
Hs_Synaptic_Vesicle_Pathway_WP2267_56444 | 0.00898066 |
|
Hs_Eicosanoid_Synthesis_WP167_45234 | 0.01199396 |
|
Hs_Muscle_contraction_WP1864_44927 | 0.01665074 |
|
Hs_MAPK_Cascade_WP422_44889 | 0.02439614 |
|
Hs_Prostaglandin_Synthesis_and_Regulation_WP98_45273 | 0.02763810 |
|
Hs_BDNF_signaling_pathway_WP2380_54595 | 0.03042269 |
|
Hs_Endothelin_WP2197_56443 | 0.03104245 |
|
Hs_Vitamin_A_and_carotenoid_metabolism_WP716_52902 | 0.04216790 |
|
Hs_DNA_damage_response_(only_ATM_dependent)_WP710_46091 | 0.04453868 |
 | Table IVSignificant pathways associated with
downregulated genes in stage IIIc recurrent triple-negative breast
cancer. |
Table IV
Significant pathways associated with
downregulated genes in stage IIIc recurrent triple-negative breast
cancer.
| Pathway | P-value |
|---|
|
Hs_Inflammatory_Response_Pathway_WP453_41201 | 2.37E-04 |
|
Hs_Synaptic_Vesicle_Pathway_WP2267_56444 | 9.45E-04 |
|
Hs_GPCRs,_Class_C_Metabotropic_glutamate,_pheromone_WP501_45341 | 0.00142081 |
|
Hs_AMPK_signaling_WP1403_44950 | 0.00217159 |
|
Hs_Transport_of_inorganic_cations-anions_and_amino_acids-oligopeptides_WP1936_45061Hs_Binding_of_RNA_by_Insulin-like_Growth_Factor-2_mRNA_Binding_Proteins_f(IGF2BPs-IMPs-VICKZs)_WP1789_44977 |
0.00530772
0.01120121 |
|
Hs_IL-2_Signaling_pathway_WP49_48400 | 0.01140687 |
|
Hs_BDNF_signaling_pathway_WP2380_54595 | 0.01594213 |
|
Hs_Regulation_of_Actin_Cytoskeleton_WP51_49262 | 0.01748728 |
|
Hs_Calcium_Regulation_in_the_Cardiac_Cell_WP536_44983 | 0.01878150 |
|
Hs_GPCR_downstream_signaling_WP1824_42047 | 0.02959318 |
|
Hs_Arrhythmogenic_right_ventricular_cardiomyopathy_WP2118_47057 | 0.03163952 |
|
Hs_Focal_Adhesion_WP306_45270 | 0.03231586 |
|
Hs_Peroxisomal_lipid_metabolism_WP1878_45246 | 0.03323053 |
|
Hs_RalA_downstream_regulated_genes_WP2290_53118 | 0.03323053 |
|
Hs_Integrated_Pancreatic_Cancer_Pathway_WP2256_54274 | 0.03932082 |
|
Hs_Integrated_Pancreatic_Cancer_Pathway_WP2377_56677 | 0.03932082 |
|
Hs_BMP_signalling_and_regulation_WP1425_44981 | 0.04406138 |
|
Hs_Neurotransmitter_Release_Cycle_WP1871_42089 | 0.04406138 |
Stemness-associated gene expression in
stage IIIc recurrent TNBC
Since cancer stemness involves self-renewal ability
and aggressive behavior, it was not unexpected that a subset of
overexpressed stemness genes, including CD44, WNT 4 and WNT 16,
were identified in tumor samples obtained from patients with late
stage (IIIc) TNBC and subsequent recurrence (Fig. 5A). In addition, a gene set
associated with angiogenesis (Fig.
5B), but not metastasis, was to be overexpressed in these tumor
samples.
Functional pathway analysis
In order to elucidate the possible networks
involving the pathways identified in the present study, the
above-mentioned upregulated and downregulated genes in the early
stage and stage IIIc tumor samples were investigated using
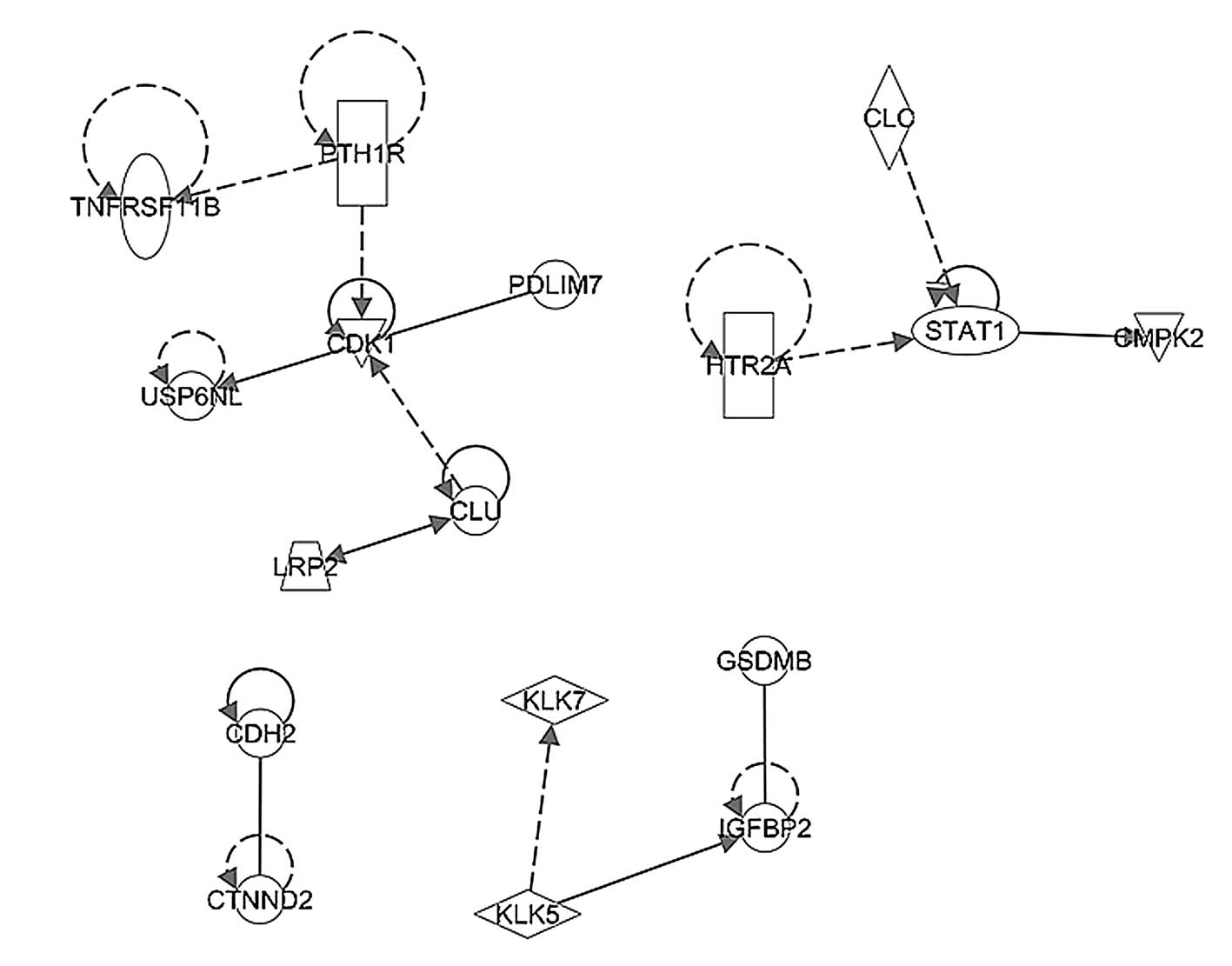
ingenuity pathway analysis. The network shown in Fig 6 represents the significant pathway
network incorporating CDK1 and STAT1, which regulated cell growth
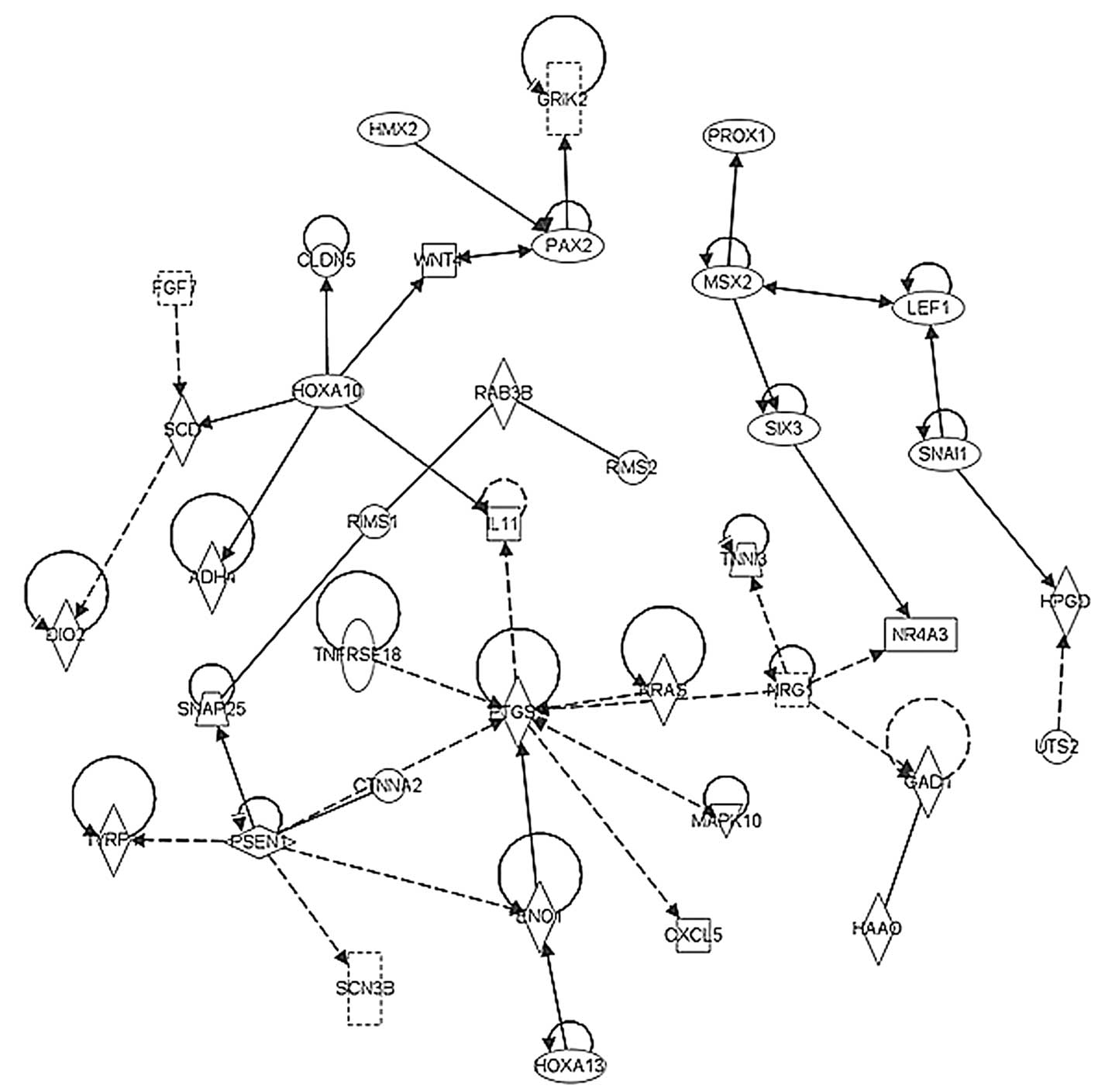
in the early stage TNBC tumor samples. By contrast, Fig. 7 shows the significant pathway
network incorporating PTGS, RAS and WNT, which promoted
angiogenesis and stemness in the late stage TNBC tumors.
Discussion
It has been suggested that TNBC is not a complete
proxy for basal-like breast cancer. Although an appreciation of the
significance of basal-like breast cancers predates gene-expression
investigations by a number of years, this term was not in
widespread use until later (14,15).
Several of the most well-known genes associated with basal-like
breast cancer, including keratin 5 and keratin 17, do not differ
significantly different between caucasian and Asian populations
(16). This observation is
contradictory to the fact that the epidemiology and prognosis of
breast cancer between different ethnicities is generally reported
to be different (17). There is no
internationally accepted definition of these tumors. However, the
majority of basal-like cancers are also TNBCs, and the majority of
cases of TNBC (~80%) are also cases of basal-like breast cancer;
therefore, it has been suggested that the triple-negative phenotype
and the basal-like phenotype are effectively synonymous (15,16).
However, clinical, microarray and immunohistochemical observations
have demonstrated that this is not the case (18). The present study enrolled TNBC
cases with ER (<1%), PR (<1%) and non amplified HER2.
Furthermore, in the present study, the gene expression levels of
basal-like breast cancer specific genes, including keratin 5 and
keratin 17, were not uniquely upregulated, which was in agreement
with those previously reported in Asian TNBC patients (16), indicating that these types of
cancer do not possess basal-like breast cancer characteristics.
Compared with previous study designs that used tumor
and non-tumor samples for TNBC analysis, the present study was the
first, to the best of out knowledge, to focus on the analysis of
paired recurrent, vs. non-recurrent TNBC samples. Therefore, the
design of the present study was likely to identify potentially
important biomarkers and therapeutic targets for the treatment and
prevention of TNBC recurrence. It was hypothesized that the TNBC
population is heterogeneous and that various subgroups will exhibit
different, and possibly specific, expression signatures. The
identification of such signatures offers potential perspective into
the individualized treatment protocols that are available. These
signaling pathways are also likely to be involved in the
upregulation of metastatic and/or cancer cell self-renewal genes,
leading to higher levels of metastatic activity and resulting in
the recurrence of TNBC.
TNBC is a highly diverse type of cancer, and
subtyping of TNBC tumors is necessary in order to identify
appropriate molecular-based therapies. Evolving technologies are
permitting increasing quantities of molecular data to be obtained
from tumor tissues, which enable the development of more
personalized treatment strategies (19). In this context, whether all
basal-like cancers are enriched with cancer stem cells or whether
they have a disproportionately high content of cells undergoing
epithelial-to-mesenchymal transition (EMT) remains to be elucidated
(20). In the present study, which
focused on the molecular mechanisms in paired specimens obtained
from patients with and without recurrence, the results suggested
that the significant pathways involving upregulated genes that are
present in stage IIIc recurrent samples included the WNT, glycogen
metabolism, integrated pancreatic cancer, MAPK cascade, BDNF and
prostaglandin signaling pathways. In addition, the presence of
overexpressed stemness genes, including CD44, WNT 4 and WNT 16,
were also identified. By contrast, a different subset of
overexpressed genes, which included genes encoding MMPs, and genes
involved in the cancer cell migration (CDH2) pathway, cell adhesion
or motility (KRAS, CDC42, RAC1, ICAM and SRGAP2) and EMT (TWIST1),
were identified in the tumor samples obtained from patients with
early (stage IIa, IIb, IIIa) tumors. These observations may be
useful for biomarker selection, drug discovery and clinical trial
design, all of which may assist in the identification of
appropriate targeted therapies for patients with TNBC (13).
Kuo et al (13) reported that deregulated genes
within the transforming growth factor (TGF)-β signaling pathway
were markedly involved in the distant recurrence of TNBC (13). The overexpression of TGF-β1 has
been observed to be mediated by two upstream regulators, tumor
necrosis factor and IL-1β, which are known mediators of the
immune/inflammatory response; furthermore, TGF-β1 itself is crucial
to the regulation of T cell-mediated immunity (21). In the present study, similar
deregulation of the inflammatory response pathway and the IL-2
signaling pathway were observed in the samples from patients with
stage IIIc TNBC recurrence. Taken together, these findings
suggested that the distant metastatic invasion of TNBC may be
induced by immune/inflammatory deregulation.
According to the St Gallen consensus for
chemotherapy guidelines (22), all
triple-negative patients are recommended to receive adjuvant
systemic chemotherapy in combination of anthracycline-based
regimens with taxanes, however, this approach often results in
serious side effects in patients. A number of pathway-targeted
agents, including EGFR inhibitors, DNA repair pathway inhibitors
and anti-angiogenic agents, have been used in clinical trials as
targeted therapies for TNBC (7,23).
These may be used, alongside traditional chemotherapy treatments,
to treat triple-negative patients with an unfavorable prognosis.
The gene profiling in the current study may provide a prognostic
predictor and, thus may become a clinically useful tool for the
identification of triple-negative patients who are at low risk of
recurrence. The subsequent provision of moderate doses of combined
regimens, or the anthracycline-based regimens alone, in these
patients can be offered to reduce patient side effects. Among the
stage IIIc recurrence group, the prostaglandin synthesis and
regulation signaling pathway exhibited significant alterations in
expression. COX-2, an inducible form of cyclooxygenase, is the rate
limiting step in the production of prostaglandins, which has been
suggested to be involved in long-term inflammation and the
promotion of cancer growth. Therefore, the results of the present
study suggest that this pathway is likely to be important in the
late stages of tumor growth and metastasis.
Acknowledgments
The present study was supported, in part, by the
Division of Experimental Surgery of the Department of Surgery,
Taipei Veterans General Hospital (Taiwan, China; grant no.
101DHA0100010) and the Ministry of Health and Welfare (Center of
Excellence for Cancer Research at Taipei Veterans General Hospital
Phase II; M OHW103-TD-B-111-02).
References
|
1
|
Lin YS, Jen YM, Wang BB, Lee JC and Kang
BH: Epidemiology of oral cavity cancer in taiwan with emphasis on
the role of betel nut chewing. ORL J Otorhinolaryngol Relat Spec.
67:230–236. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Siegel R, Bandi P and Jemal A:
Breast cancer statistics, 2011. CA Cancer J Clin. 61:409–418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gradishar WJ: Tamoxifen-what next?
Oncologist. 9:378–384. 2004. View Article : Google Scholar
|
|
5
|
Yamamoto Y and Iwase H:
Clinicopathological features and treatment strategy for
triple-negative breast cancer. Int J Clin Oncol. 15:341–351. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Podo F, Buydens LM, Degani H, Hilhorst R,
Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J,
Monleon D, et al: Triple-negative breast cancer: Present challenges
and new perspectives. Mol Oncol. 4:209–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Shaughnessy J, Osborne C, Pippen JE,
Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM and Bradley C:
Iniparib plus chemotherapy in metastatic triple-negative breast
cancer. N Engl J Med. 364:205–214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohamed A, Krajewski K, Cakar B and Ma CX:
Targeted therapy for breast cancer. Am J Pathol. 183:1096–1112.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thike AA, Yong-Zheng Chong L, Cheok PY, Li
HH, Wai-Cheong Yip G, Huat Bay B, Tse GM, Iqbal J and Tan PH: Loss
of androgen receptor expression predicts early recurrence in
triple-negative and basal-like breast cancer. Mod Pathol.
27:352–360. 2014.
|
|
11
|
Chen LH, Kuo WH, Tsai MH, Chen PC, Hsiao
CK, Chuang EY, Chang LY, Hsieh FJ, Lai LC and Chang KJ:
Identification of prognostic genes for recurrent risk prediction in
triple negative breast cancer patients in Taiwan. PLoS One.
6:e282222011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hicks C, Kumar R, Pannuti A, Backus K,
Brown A, Monico J and Miele L: An integrative genomics approach for
associating GWAS information with triple-negative breast cancer.
Cancer Inform. 12:1–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuo WH, Chang YY, Lai LC, Tsai MH, Hsiao
CK, Chang KJ and Chuang EY: Molecular characteristics and
metastasis predictor genes of triple-negative breast cancer: A
clinical study of triple-negative breast carcinomas. PLoS One.
7:e458312012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kreike B, van Kouwenhove M, Horlings H,
Weigelt B, Peterse H, Bartelink H and van de Vijver MJ: Gene
expression profiling and histopathological characterization of
triple-negative/basal-like breast carcinomas. Breast Cancer Res.
9:R652007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peppercorn J, Perou CM and Carey LA:
Molecular subtypes in breast cancer evaluation and management:
divide and conquer. Cancer Invest. 26:1–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kurebayashi J, Moriya T, Ishida T,
Hirakawa H, Kurosumi M, Akiyama F, Kinoshita T, Takei H, Takahashi
K and Ikeda M: The prevalence of intrinsic subtypes and prognosis
in breast cancer patients of different races. Breast. 16(Suppl 2):
S72–S77. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weigelt B, Baehner FL and Reis-Filho JS:
The contribution of gene expression profiling to breast cancer
classification, prognostication and prediction: A retrospective of
the last decade. J Pathol. 220:263–280. 2010.
|
|
19
|
Rodenhiser DI, Andrews JD, Vandenberg TA
and Chambers AF: Gene signatures of breast cancer progression and
metastasis. Breast Cancer Res. 13:2012011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Y, Teige I, Birnir B and
Issazadeh-Navikas S: Neuron-mediated generation of regulatory T
cells from encephalitogenic T cells suppresses EAE. Nat Med.
12:518–525. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Untch M, Gerber B, Harbeck N, et al: 13th
st Gallen international breast cancer conference 2013: primary
therapy of early breast cancer evidence, controversies, consensus -
opinion of a german team of experts (zurich 2013). Breast Care
(Basel). 8:221–229. 2013.
|
|
23
|
Hudis CA and Gianni L: Triple-negative
breast cancer: An unmet medical need. Oncologist. 16(Suppl 1):
1–11. 2011. View Article : Google Scholar : PubMed/NCBI
|