Introduction
A class of small non-coding RNAs, termed miRNAs are
now recognized, which contribute to essential biological processes,
including development, cellular differentiation, proliferation,
stress responses, apoptosis and metabolism, as well as tumor
initiation and progression (1,2).
These miRNAs are 18–24 nucleotides long (3). miRNAs are considered to exhibit tumor
suppressor gene and oncogene regulatory roles, and show complex
patterns of disease-and tissue-specific expression, which are
associated with their ability to regulate several targets that are
crucial to the carcinogenic process (2). In multiple types of cancer, aberrant
patterns of miRNA expression have been observed, which can affect
cancer cell proliferation (4),
apoptosis (5) and metastasis
(6), and potentially define the
cancer stem cell phenotype (7).
Colorectal cancer (CRC), the third most common type
of cancer worldwide, is the fourth leading cause of
cancer-associated mortality, accounting for 90% incidence and
mortality rates (8,9). In 2003, the differential expression
of miRNAs was first reported to be associated with CRC (10). From a clinical point of view,
cancer of the colon and rectum are two distinct entities, which
require different treatment strategies (2,11).
Accordingly, they require separate treatment when analyzing the
genetics and biology of the diseases. For colon cancer, extensive
catalogues of downregulated miRNAs have been identified in the
previous years, whereas only limited data are available for rectal
cancer (2).
In order to understand the function of miRNA in
human rectal cancer, miRNA profiling was performed in a previous
study (1), wherein microRNA-144
was found to show aberrant expression and was identified as rectal
cancer-specific; having not been reported in colon cancer.
Originally, miRNA (miR)-144 was identified as an erythroid-specific
miRNA, which is required for subsequent erythroid lineage survival
and maturation (12,13). In addition, it can increase the
severity of anemia, and decrease glutathione regeneration and
antioxidant capacity (14).
Previous tumor investigations, involving a comprehensive
meta-analysis of miRNA expression microarrays, revealed that
miR-144 is downregulated in lung cancer, prostate cancer and
hepatocellular carcinoma (15),
indicating that it may be a novel human cancer-associated miRNA. To
date, there are no reports that miR-144 has a functional role in
rectal cancer. In the present study, miR-144 was characterized in
rectal cancer and found that the upregulation of miR-144 inhibited
malignant progression (migration and proliferation) of rectal
cancer cells. According to previous reports, Rho-kinase (ROCK) is a
serine/threonine kinase, which functions downstream of the small
GTPase RhoA (16). ROCK isoforms
have been implicated in a variety of cellular functions, including
smooth muscle contraction, actin cytoskeleton organization
(17), cytokinesis (18), cell adhesion and motility (19). In addition, the activation of ROCK1
by RhoA can promote cell invasion and motility in prostate cancer
and colorectal carcinoma cells (20,21).
In order to determine whether the inhibitory effects of miR-144 on
the progression of rectal cancer are associated with ROCKs, the
present study performed a series of relative experiments, and found
that miR-144 inhibits the migration and proliferation of SW837 and
SW1463 rectal cancer cells by targeting the crucial oncogene,
ROCK1.
Materials and methods
Cell lines and cell culture
Human SW837 and SW1463 cell lines were purchased
from American Type Culture Collection (ATCC; Manassas, VA, USA).
These two cell lines originated from rectum tissue belonging to
epithelial cells, and they were selected as rectal cell carcinoma
cells. The SW837 and SW1463 cells were cultured in Leibovitz's 15
(L-15) culture medium (cat. no. 30-2008; ATCC) supplemented with
fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA), to a final
concentration of 10% at 37°C. The cells were passaged 1–2 times per
week and were cultured in free gas exchange with atmospheric air. A
human rectal mucosa epithelium cell was purchased from PriCells
(Wuhan, China); which was derived from normal rectal mucosa
epithelium and was used as a normal control (NC).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA and miR were extracted from the above cell
line (5×105 per well in six-well plates at 80%
confluency) using an RNeasy Mini kit and an miR Neasy Mini kit
(Qiagen, Valencia, CA, USA). Following quantitation, the extracted
total RNA was reverse-transcribed using a High Capacity cDNA
Archive kit (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and a TaqMan microRNA reverse transcription kit
(Applied Biosystems), according to the manufacturer's protocol. The
RT products were mixed with TaqMan universal PCR master mix II, and
qPCR was performed on an Applied Biosystems Prism 7500 Fast
Sequence Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR parameters were as follows: 95°C for 20
sec, 40 cycles at 95°C for 3 sec and 60°C for 30 sec. The primers
for mRNA were as follows: miR-144 (cat. no. 204754; Exiqon A/S,
Vedbaek, Denmark); ROCK-1, forward 5′-AAG AGA GTG ATA TTG AGC AGT
TGC G-3′ and reverse 5′-TTC CTC TAT TTG GTA CAG AAA GCC A-3′; and
ROCK2, forward 5′-TGC GGT CAC AAC TCC AAG C-3′ and reverse 5′-GGA
AAC CCA TCA TCT GCC TCA G-3′. RNU48 or β-actin were used as an
endogenous control. Primers were synthesized by Shanghai Sangon
Biological Engineering and Technology Service (Shanghai, China).
All reactions were performed in triplicate. The mRNA and miRNA
expression levels were determined using the 2−ΔCq method
(22).
Cell transfection and immunoblotting
The SW837 and SW1463 cells (5×105 per
well in six-well plates, 80% confluency) were transfected with pre
miR-144 or a scrambled pre miR-control (Ambion; Thermo Fisher
Scientific, Inc.) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Following transfection for 72 h, for miR-144 or mRNA (ROCK1 and
ROCK2) analysis, total RNA was extracted using previously described
methods (23). The cells were
lysed in Triton X-100 containing 1% phosphate-buffered saline (PBS)
with a cocktail of protease inhibitors (Roche Diagnostics, Basal,
Switzerland) for protein analysis. The lysates were then
centrifuged at 16,000 g for 15 min, and the protein concentrations
of the supernatants were determined using a bicinchoninic acid
assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). The
ROCK1 and ROCK2 proteins (40 µg/lane) were analyzed using
immunoblotting following transfer onto polyvinylidene fluoride
membranes (Sigma-Aldrich) from a 10% SDS-PAGE gel (Sigma-Aldrich).
Subsequently, followingblocking with 0.5% skimmed milk powder
(Sigma-Aldrich) in 1X PBS-Tween 20 (Sigma-Aldrich), the membranes
were incubated with primary monoclonal rabbit anti-human ROCK1
antibody (cat. no. ab45171) or monoclonal rabbit anti-human ROCK2
antibody (cat. no. ab125025), and monoclonal rabbit anti-human
β-actin antibody (cat. no. ab115777), at a dilution of 1:4,000
overnight at 4°C. Following washing three times (10 min each) with
1X PBS-Tween 20, the membranes were incubated with horseradish
peroxidase (HRP)-conjugated polyclonal goat anti-rabbit IgG (cat.
no. ab6721) secondary antibody (1:10,000) at room temperature for 1
h, and washed again. The reactive bands were detected using
enhanced chemiluminescence (GE Healthcare Life Sciences, Chalfont,
UK), according to the manufacturer's protocol. The relative levels
of each protein to β-actin were analyzed. All primary and secondary
antibodies were purchased from Abcam (Cambridge, MA, USA). The
resulting bands of the blot were analyzed using Gel-Pro Analyzer
4.0 software (Media Cybernetics Inc., Silver Spring, MD, USA).
Cell viability
Following transfection with pre-miR-144, the
viabilities of the SW837 or SW1463 cells were determined using an
MTT assay (Sigma-Aldrich). Briefly, 5×103 transfected
cells were plated in 96-well plates. Following incubation at 37°C
for different periods of time (24, 48 and 72 h), the culture medium
was removed and MTT (10 µl; 0.5%) was added. Following
incubation for another 4 h at 37°C, the culture medium was replaced
with dimethyl sulfoxide (10 µl; 4%; Sigma-Aldrich), and the
optical density (O)570 was measured using a microplate reader
(Multiskan MK2; Molecular Devices; Thermo Fisher Scientific,
Inc.).
Cell migration analysis using a Transwell
assay
Migration assays were performed in Transwells
(8.0-µm pore size). For the migration assay, the
miR-144-transfected SW837 and SW1463 cells (~2×104 cells
per well), in L-15 culture medium containing 10% FBS, were added to
the upper wells. L-15 media, containing 10% FBS was added to the
lower wells. The miR-control-transfected SW837 and SW1463 cells
were used as a control. The cells, which migrated through the
filter after 24, 48 or 72 h were stained with 0.1% crystal violet
(Sigma-Aldrich) and counted using phase contrast microscopy
(Axiovert 200M; Carl Zeiss, Jena, Germany) (24). The same experiments were performed
to detect the migration abilities of the miR-144 and ROCK1
co-transfected SW837 and SW1463 cells.
Bromodeoxyuridine (BrdU) assay
A BrdU Cell Proliferation Assay kit (cat. no. 2752;
EMD Millipore, Bedford, MA, USA) was used to detect cell
proliferation ability. The miR-144-transfected SW837 and SW1463
cells were synchronized and plated in 96 wells (3×103
cells/well) in L-15 culture medium containing 10% FBS, following
which 10 µl BrdU solution was added and the cells were
incubated at 37°C for 24 h, 48 h and 72 h, respectively. Following
incubation, 100 µl/well fixing solution (2752b; 4%
paraformaldehyde in PBS; EMD Millipore) was added and the cells
were incubated at 37°C for 15 min. Subsequently, 100 µl/well
of pre-prepared detection antibody solution (anti-BrdU mouse
monoclonal antibody; 2752c) was added and incubated at 37°C for 1
h, following which 50X plate wash concentrate (2752h) was diluted
and used to wash the plates. Subsequently, 100 µl/well of
prepared HRP-conjugated goat anti-mouse IgG secondary antibody
(2752e) was added and incubated at 37° for 30 min. The plates were
washed with diluted plate wash concentrate. Finally, 100 µl
TMB substrate was added prior to incubation for 30 min. The
quantity of BrdU, which was incorporated into the cells was
determined at 450 nm using a microplate reader. The same
experiments were performed to detect the proliferation ability of
the miR-144 and ROCK1 co-transfected SW837 or SW1463 cells.
miR-144 and ROCK1 co-transfection
assays
The SW837 and SW1463 cells were first transfected
with pre miR-144 using Lipofectamine 2000, according to the
manufacturer's protocol. Following transfection, the cDNA of ROCK1
was sub-cloned using pyrobest DNA polymerase (Takara Bio, Inc.
Otsu, Japan) and inserted into a pEGFP-N1 vector (Clontech
Laboratories, Inc., Mountain View, CA, USA). These vectors were
transfected into the miR-144-transfected SW837 and SW1463 cells
using Lipofectamine 2000. Mock-pEGFP-N1 (mock-ROCK1) was used as a
control. The cells were used for subsequent experiments following
transfection for 24, 48 or 72 h.
Statistical analysis
Each data point was obtained from three repeated
experiments. Data are expressed as the mean ± standard deviation,
and differences were analyzed using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference. The
western blotting experiments were performed several times with
similar results, of which the optimal image was selected to
present.
Results
Expression of miR-144 in rectal carcinoma
cell lines
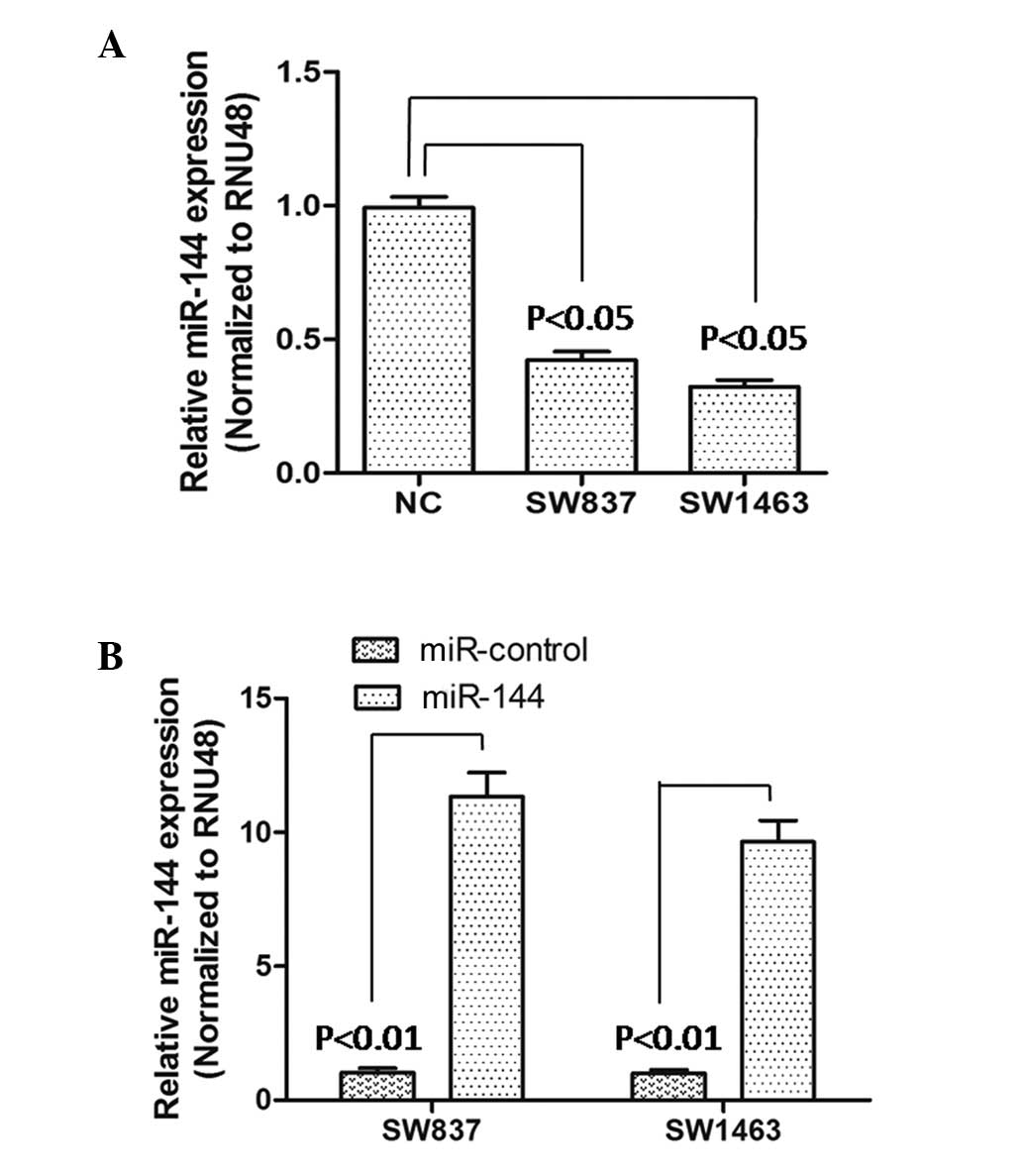
To identify miR-144 in rectal carcinoma, SW837 and
SW1463 cell lines were selected as model cells, and the expression
levels of miR-144 were determined in these cells. Following RT-qPCR
analysis, the results showed that the expression of miR-144 was
significantly downregulated (P<0.05), compared with that of the
NC cells (Fig. 1A). Following
this, the effect of miR-144 on rectal carcinoma cells was examined
by constructing miR-144-overexpressing SW837 and SW1463 cells. The
transfection effectiveness was verified using RT-qPCR. As shown in
Fig. 1B, the results suggested
that the expression levels of miR-144 were significantly higher
than those of the miR-control-transfected SW837 and SW1463 cells,
which indicated that the miR-144-overexpressing SW837 and SW1463
cells constructed were suitable for subsequent experiments.
Cell viability, migration and
proliferation in miR-144-overexpressing rectal carcinoma cells
Subsequently, the viability, migration and
proliferation abilities of the SW837 and SW1463 cells were detected
using the methods, described above. As shown in Fig. 2A, the percentage cell viability was
downregulated (P<0.05) in the miR-144-SW837 and the
miR-144-SW1463 cells, compared with the same cells transfected with
the miR-control. As shown in Fig.
2B, the number of migrating SW837 and SW1463 cells increased
with increasing duration (P<0.05; 24 h, 48 h and 72 h). However,
the numbers of migrated miR-144-SW837 (blue line) or miR-144-SW1463
cells (red line), were lower, compared with those in the
miR-control-SW837 or miR-control-SW1463 groups at each time point.
As shown in Fig. 2C, the relative
proliferation of the miR-144-SW837 cells was lower than that of the
miR-control-SW837 cells following culture for 24, 48 and 72 h. For
the proliferation of the SW1463 cells, similar results were
observed (Fig. 2D). In addition,
from these four experiments, the decreases in cell viability,
migration and proliferation abilities was more marked in the
miR-144-SW837 cells, compared with the miR-144-SW1463 cells, which
may be associated with the primary levels of miR-144 in the
corresponding cells. Taken together, these results in the SW837 and
SW1463 cells indicated that miR-144 was involved in inhibiting the
progression of rectal carcinoma.
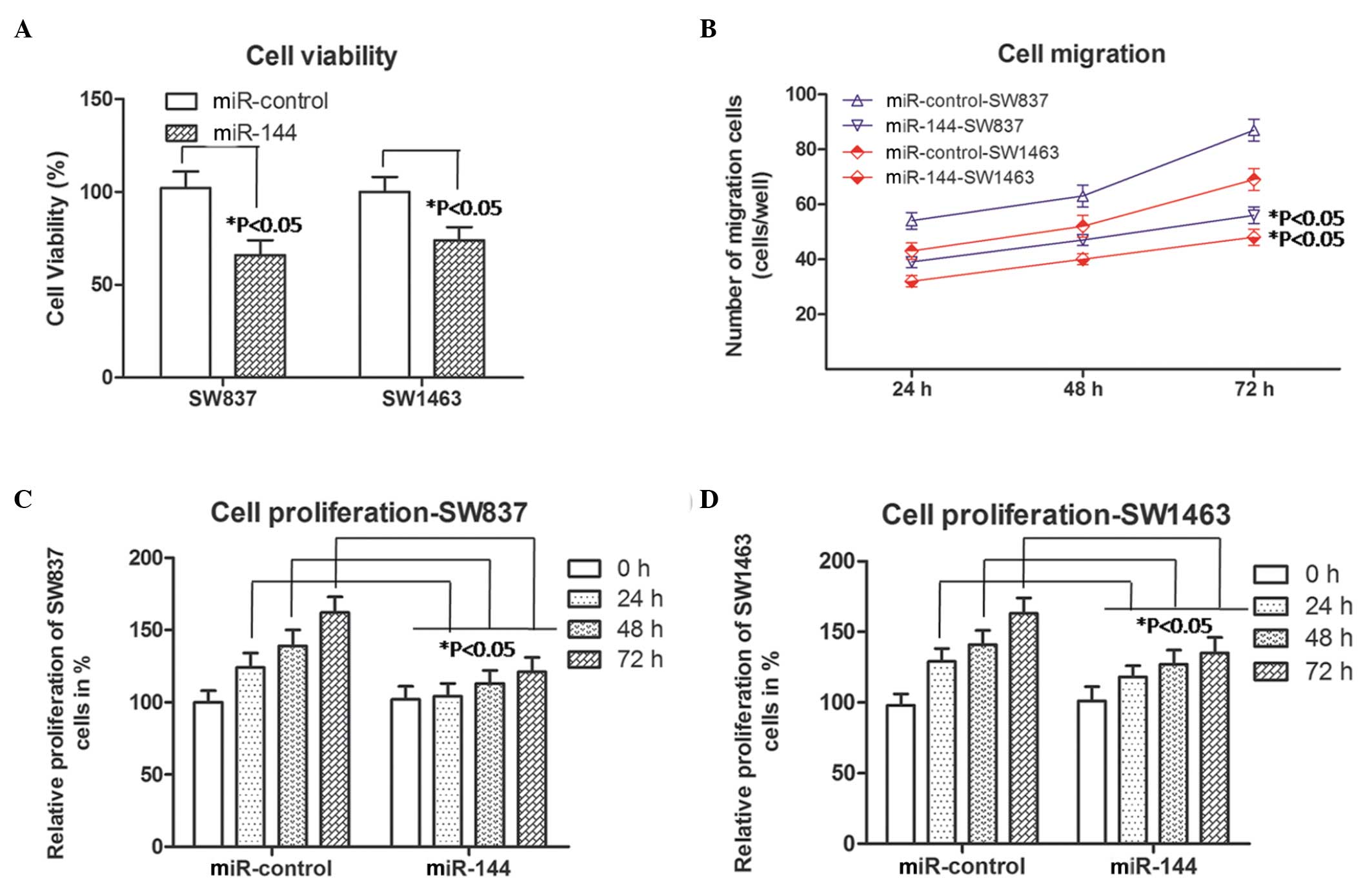 | Figure 2Cell viability, migration and
proliferation in miR-144 overexpressed rectal carcinoma cells
(SW837 and SW1463). (A) Cell viability was analyzed using an MTT
assay following transient transfection for different durations.
*P<0.05, compared with the corresponding miR-control
(Student's t-test). (B) Numbers of SW837 (blue lines) and SW1463
(red lines) cells, which migrated into the lower wells. Data were
obtained 24, 48 and 72 h following transfection.
*P<0.05, compared with the corresponding miR-control
group (Student's t-test). (C) Relative percentage proliferation of
the SW837 cells percent. (D) Relative proliferation of SW1463
cells. Data were obtained 0, 24, 48 and 72 h following
transfection,. *P<0.05, compared with the miR-control
group at the corresponding time point. The data are presented as
the mean ± standard deviation of three independent experiments.
miR, microRNA. |
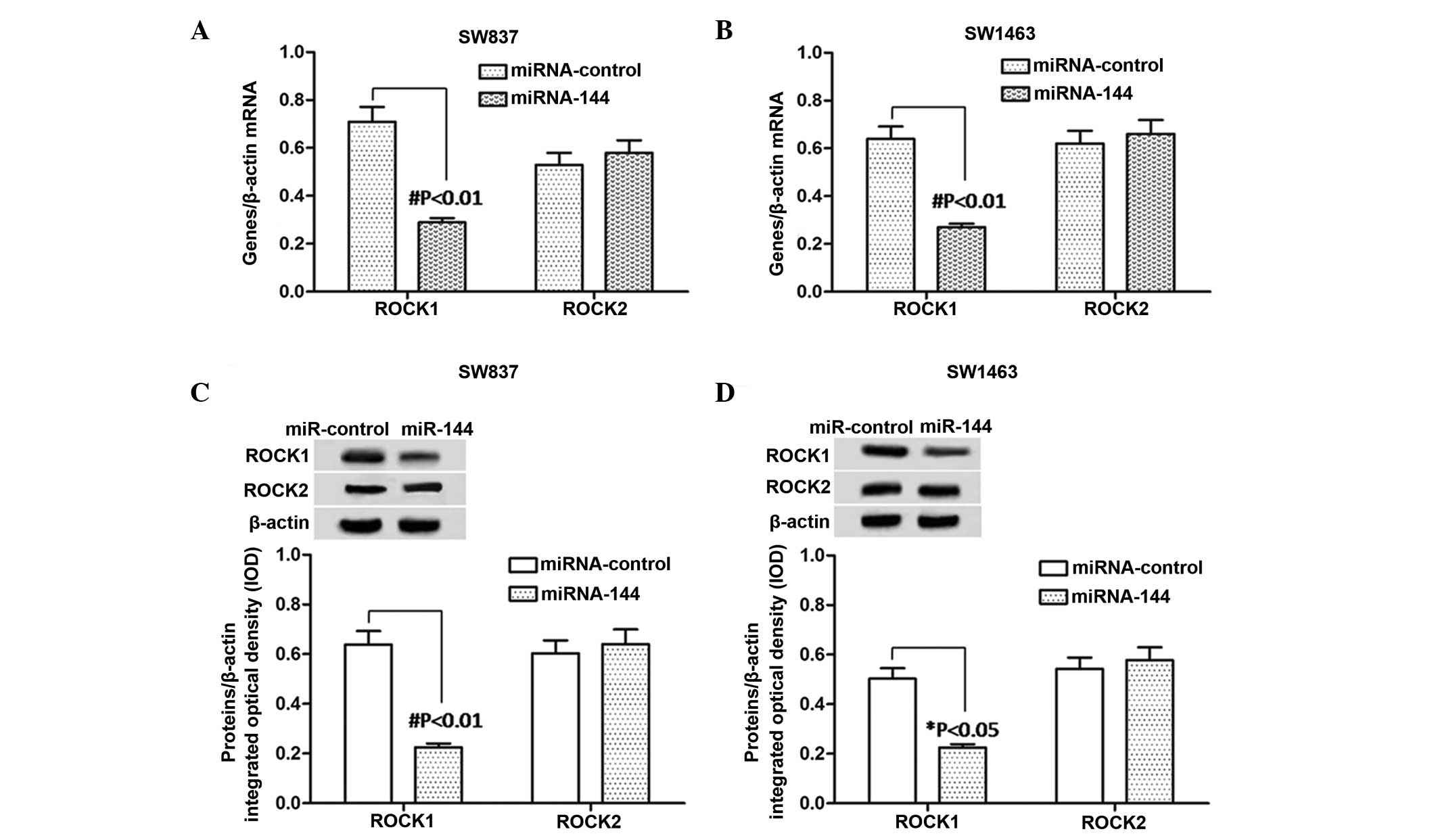
Expression levels of ROCK1 and ROCK2 in
miR-144-overexpressing rectal carcinoma cells
According to the above results, miR-144 inhibited
SW837 and SW1463 cell proliferation and migration. In order to
examine the underlying mechanism, the present study examined the
effects of miR-144 on ROCK1 and ROCK2 in the miR-144-SW837 and
miR-144-SW1463 cells. The mRNA and protein expression levels of
ROCK1 and ROCK2 were detected using RT-qPCR and western blot
analyses, respectively. The bands of the western blot were analyzed
using Gel-Pro Analyzer 4.0 software. As shown in Fig. 3A and B, the mRNA expression level
of ROCK1 was significantly downregulated (P<0.01) in the
miR-144-trans-fected cells, compared with the
miR-control-transfected cells, in the SW837 and SW1463 cells.
However no change in the mRNA levels of ROCK2 were observed.
Similar changes were observed in the protein expression levels of
ROCK1 in the SW837 cells (P<0.01) and SW1463 cells (P<0.05)
and ROCK2 (Fig. 3C and D). These
results suggested that the expression of ROCK1 was reduced by the
overexpression of miR-144 in the SW837 and SW1463 cells.
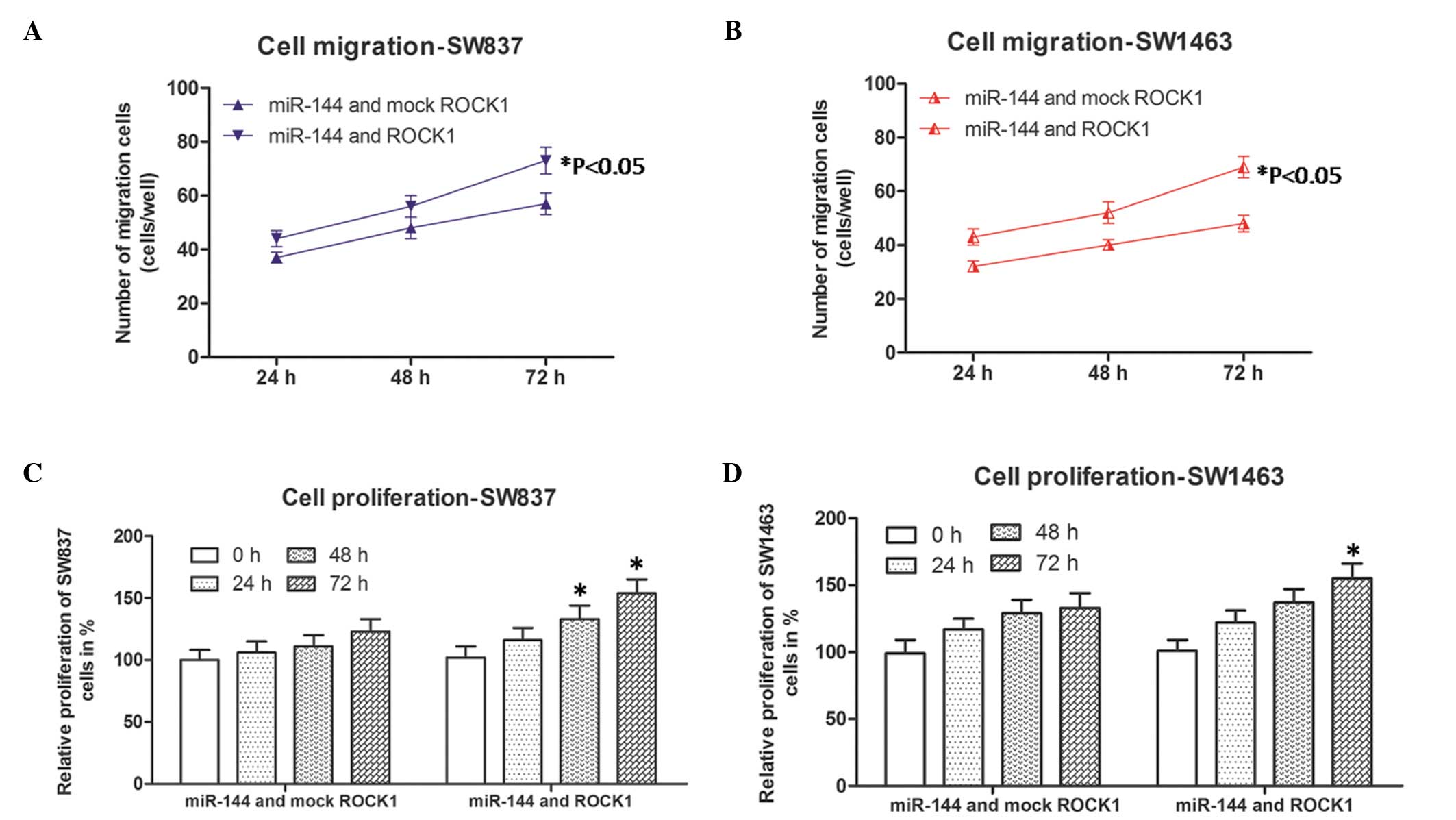
Cell migration and proliferation in
miR-144 and ROCK1-co-transfected rectal carcinoma cells
As the results revealed that only the expression of
ROCK1 was affected by miR-144, co-transfection experiments were
performed to further confirm the role of ROCK1, as described above.
Subsequently, the cell migration and proliferation abilities of the
cells were detected again by corresponding methods, described
above. As shown in Fig. 4A and B,
the numbers of migrated miR-144 and ROCK1 co-transfected SW837
(blue line) and SW1463 cells (red lines) were higher, compared with
those of the miR-144- and mock-ROCK1 co-transfected SW837 and
SW1463 cells at each time point. In addition, the relative
proliferation rates of the miR-144 and ROCK1 co-transfected SW837
and SW1463 cells were more marked, compared with those of the
miR-144 and mock-ROCK1 co-transfected SW837 or SW1463 cells
following culture for 48 and 72 h (SW837), and 72 h (SW1463). These
results suggested that the inhibitory effects of overexpressed
miR-144 on the SW837 and SW1463 cells was by the overexpression of
ROCK1. Taken together, it was concluded that miR-144 inhibited
migration and proliferation in rectal cancer through the
downregulation of ROCK1.
Discussion
Although the expression of several miRNAs are
aberrantly altered in rectal cancer (1), their underlying molecular mechanisms
in the development and progression of rectal cancer remain to be
fully elucidated. Thus, investigating the function of miRNAs, which
are specifically involved in the progression of rectal cancer is
required to improve current knowledge of rectal cancer, and offer
novel insights into its diagnosis and therapy.
The present study focused on the role of miR-144 in
rectal cancer, which has been extensively investigated and reported
in other types of cancer, including lung cancer (25), prostate cancer (26) and hepatocellular carcinoma
(27). Previous microRNA
microarray analyses have indicated that miR-144 shows aberrant
expression and appears to be specific to rectal cancer (1). In order to determine the role of
miR-144 in rectal cancer, the present study selected SW837 and
SW1463 cells as rectal cell carcinoma cells, which originated from
rectal tissue belonging to epithelial cells. The results
demonstrated that the expression of miR-144 was significantly
reduced in rectal cell carcinoma cells, and overexpression of
miR-144 repressed the viability, migration and proliferation of the
SW837 and SW1463 cells in vitro by targeting ROCK1.
Previously, miR-144 was identified as an
erythroid-specific miRNA, which is essential for the subsequent
maturation and survival of the erythroid lineage (12,28).
miR-144 can decrease glutathione regeneration and antioxidant
capacity by directly regulating a central regulator of the cellular
response to oxidative stress (29). In addition, a previous study showed
that miR-144 can increase cell growth in HeLa cells (30). Zhao et al reported that the
downregulation of miR-144 is associated with the growth and
invasion of osteosarcoma cells through the regulation of the
expression of TAGLN (31). Cao
et al reported that miR-144 suppresses the proliferation and
metastasis of hepatocellular carcinoma by targeting E2F
transcription factor 3 (E2F3) (27), and Guan et al reported that
the downregulation of miR-144 promotes thyroid cancer cell invasion
by targeting zinc finger E box binding homeobox (ZEB)1 and ZEB2
(32). In the present study, ROCK1
was identified as a novel target of miR-144, by which miR-144
inhibited the migration and proliferation of rectal cancer
cells.
ROCK1 is one of the members of the ROCK family,
which facilitates reorganization of the actin cytoskeleton during
motion (33). Previous studies
have demonstrated that ROCK1 functions as an oncogene and possesses
a wide range of functions, including invasion, migration and
metastasis (34–37). The expression of ROCK1 has also
been found to be increased in several types of cancer, including
glioma, prostate cancer, osteosarcoma and gastric cancer (38,39),
and ROCK1 is targeted by several miRNAs, including miR-584
(40), miR-340 (39), and miR-124 (41). In the present study, ROCK1 was
identified as a novel target of miR-144 in rectal cancer cells.
ROCK1 was significantly downregulated in miR-144-overexpressing
SW837 and SW1463 cells. In addition, the inhibitory effects on the
migration and proliferation of the miR-144 on SW837 and SW1463
cells was controlled by the overexpression of ROCK1.
In conclusion, the present study demonstrated that
miR-144 suppressed the progression of rectal cancer by targeting
ROCK1, suggesting that miR-144 may be a novel biomarker and
therapeutic target for rectal cancer treatment.
Abbreviations:
|
miR-144
|
microRNA-144
|
|
ROCK1
|
Rho-associated coiled-coil containing
protein kinase 1
|
|
CRC
|
colorectal cancer
|
|
miRNAs
|
microRNAs
|
|
E2F3
|
E2F transcription factor 3
|
|
ZEB1
|
zinc finger E box binding homeobox
1
|
|
ZEB2
|
zinc finger E box binding homeobox
2
|
References
|
1
|
Gaedcke J, Grade M, Camps J, Søkilde R,
Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi
BM, Møller S, et al: The rectal cancer microRNAome-microRNA
expression in rectal cancer and matched normal mucosa. Clin Cancer
Res. 18:4919–4930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Slattery ML, Wolff E, Hoffman MD, Pellatt
DF, Milash B and Wolff RK: MicroRNAs and colon and rectal cancer:
Differential expression by tumor location and subtype. Gene
Chromosomes Cancer. 50:196–206. 2011. View Article : Google Scholar
|
|
3
|
Smalheiser NR and Torvik VI: A
population-based statistical approach identifies parameters
characteristic of human microRNA-mRNA interactions. BMC
Bioinformatics. 5:1392004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li P, He QY and Luo CQ: Overexpression of
miR-200b inhibits the cell proliferation and promotes apoptosis of
human hypertrophic scar fibroblasts in vitro. J Dermatol.
41:903–911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao J, Li X, Zou M, He J, Han Y, Wu D,
Yang H and Wu J: miR-135a inhibition protects A549 cells from
LPS-induced apoptosis by targeting Bcl-2. Biochem Bioph Res Commun.
452:951–957. 2014. View Article : Google Scholar
|
|
6
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
10
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dore LC, Amigo JD, Dos Santos CO, Zhang Z,
Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al: A
GATA-1-regulated microRNA locus essential for erythropoiesis. P
Natl Acad Sci USA. 105:3333–3338. 2008. View Article : Google Scholar
|
|
13
|
Lawrie CH: microRNA expression in
erythropoiesis and erythroid disorders. Brit J Haematol.
150:144–151. 2010.
|
|
14
|
Sangokoya C, Telen MJ and Chi JT: microRNA
miR-144 modulates oxidative stress tolerance and associates with
anemia severity in sickle cell disease. Blood. 116:4338–4348. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang W, Peng B, Wang D, Ma X, Jiang D,
Zhao J and Yu L: Human tumor microRNA signatures derived from
large-scale oligonucleotide microarray datasets. Int J Cancer.
129:1624–1634. 2011. View Article : Google Scholar
|
|
16
|
Lock FE, Ryan KR, Poulter NS, Parsons M
and Hotchin NA: Differential regulation of adhesion complex
turnover by ROCK1 and ROCK2. PloS One. 7:e314232012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmandke A, Schmandke A and Strittmatter
SM: ROCK and Rho: Biochemistry and neuronal functions of
Rho-associated protein kinases. Neuroscientist. 13:454–469. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Madaule P, Furuyashiki T, Eda M, Bito H,
Ishizaki T and Narumiya S: Citron, a Rho target that affects
contractility during cytokinesis. Microsc Res Tech. 49:123–126.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Narumiya S, Tanji M and Ishizaki T: Rho
signaling, ROCK and mDia1, in transformation, metastasis and
invasion. Cancer Metast Rev. 28:65–76. 2009. View Article : Google Scholar
|
|
20
|
Wilkinson S, Paterson HF and Marshall CJ:
Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin
phosphorylation and cell invasion. Nat Cell Biol. 7:255–261. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of mir-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Simionescu N, Niculescu LS, Sanda GM,
Margina D and Sima AV: Serum microRNA profiling of hyperlipidemic
and/or hyperglycemic patients reveals specifically increased levels
of miR-122, miR-125a, miR-486 and miR-92a. Ann Rom Soc Cell Biol.
19:53–64. 2014.
|
|
23
|
Zheng LH, Zhang DZ, Zhang YF, Wen YH and
Wang YC: mTOR signal transduction pathways contribute to TN-C FNIII
A1 overexpression by mechanical stress in osteosarcoma cells. Mol
Cells. 37:118–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han L, Liang XH, Chen LX, Bao SM and Yan
ZQ: SIRT1 is highly expressed in brain metastasis tissues of
non-small cell lung cancer (NSCLC) and in positive regulation of
NSCLC cell migration. Int J Clin Exp Patho. 6:23572013.
|
|
25
|
Zha W, Cao L, Shen Y and Huang M: Roles of
Mir-144-ZFX pathway in growth regulation of non-small-cell lung
cancer. PLoS One. 8:e741752013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA profiling of prostate cancer. J Cancer.
4:350–357. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumor Biol. 35:10759–10764. 2014.
View Article : Google Scholar
|
|
28
|
Fu YF, Du TT, Dong M, Zhu KY, Jing CB,
Zhang Y, Wang L, Fan HB, Chen Y, Jin Y, et al: Mir-144 selectively
regulates embryonic alplha-hemoglobin synthesis during primitive
erythropoiesis. Blood. 113:1340–1349. 2009. View Article : Google Scholar
|
|
29
|
Sangokoya C, Telen MJ and Chi JT: microRNA
miR-144 modulates oxidative stress tolerance and associates with
anemia severity in sickle cell disease. Blood. 116:4338–4348. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu
J, Wang K, Liu D, Zhang X and Yin W: The downregulation of miR-144
is associated with the growth and invasion of osteosarcoma cells
through the regulation of TAGLN expression. Int J Mol Med.
34:1565–1572. 2014.PubMed/NCBI
|
|
32
|
Guan H, Liang W, Xie Z, Li H, Liu J, Liu
L, Xiu L and Li Y: Down-regulation of miR-144 promotes thyroid
cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine.
48:566–574. 2015. View Article : Google Scholar
|
|
33
|
Wen X, Ding L, Wang JJ, Qi M, Hammonds J,
Chu H, Chen X, Hunter E and Spearman P: ROCK1 and LIM Kinase
modulate retrovirus particle release and cell-cell transmission
events. J Virol. 88:6906–6921. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC,
Lee JH, Lee SS, Ashktorab H, Smoot DT, Ryu KW, et al: MicroRNA 135a
suppresses lymph node metastasis through down-regulation of ROCK1
in early gastric cancer. PLoS One. 9:e852052014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rath N and Olson MF: Rho-associated
kinases in tumorigenesis: re-considering ROCK inhibition for cancer
therapy. EMBO Rep. 13:900–908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Choy E, Hornicek FJ, Yang S, Yang
C, Harmon D, Mankin H and Duan Z: ROCK1 as a potential therapeutic
target in osteosarcoma. J Orthop Res. 29:1259–1266. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vigil D, Kim TY, Plachco A, Garton AJ,
Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL, et
al: ROCK1 and ROCK2 are required for non-small cell lung cancer
anchorage-independent growth and invasion. Cancer Res.
72:5338–5347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang C, Zhang S, Zhang Z, He J, Xu Y and
Liu S: ROCK has a crucial role in regulating prostate tumor growth
through interaction with c-Myc. Oncogene. 33:5582–5591. 2014.
View Article : Google Scholar
|
|
39
|
Zhou X, Wei M and Wang W: MicroRNA-340
suppresses osteosarcoma tumor growth and metastasis by directly
targeting ROCK1. Biochem Biophys Res Commun. 437:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ueno K, Hirata H, Shahryari V, Chen Y,
Zaman MS, Singh K, Tabatabai ZL, Hinoda Y and Dahiya R: Tumour
suppressor microRNA-584 directly targets oncogene Rock-1 and
decreases invasion ability in human clear cell renal cell
carcinoma. Br J Cancer. 104:308–315. 2011. View Article : Google Scholar :
|
|
41
|
Hu CB, Li QL, Hu JF, Zhang Q, Xie JP and
Deng L: miR-124 inhibits growth and invasion of gastric cancer by
targeting ROCK1. Asian Pac J Cancer Prev. 15:6543–6546. 2014.
View Article : Google Scholar : PubMed/NCBI
|