Introduction
Coronary artery disease (CAD) is the most common
type of heart disease and cause of heart attacks. Despite the
improvement of pharmacological therapy and coronary
revascularization procedures, there remains a requirement for novel
therapeutic approaches (1,2). Stimulation of vascular and cardiac
repair mechanisms, such as those mediated by stem/progenitor cells,
has become a focus of cardiovascular research (3).
Experimental and clinical studies have suggested the
feasibility and safety of cell-based therapies in patients with
ischemic cardiomyopathy (4,5). To
date, different autologous adult stem and progenitor cells, in
particular several subtypes of bone marrow-derived cells, isolated
adipose tissue-derived and cardiac-derived stem/progenitor cells
have gained attention from researchers. CD133-positive cells have
shown the ability to home to injured myocardium (6) and promote cardiac recovery (7–9). The
molecular mechanisms underlying the cardiac repair process mediated
by CD133-positive cells have also been investigated. Ahmadi et
al (8) suggested that they may
induce myogenesis as well as angiogenesis. Bonanno et al
(10) reported that CD133-positive
cells differentiate into endothelial- and cardiomyocyte-like cells
in vitro. However, several studies have reported that the
numbers of bone marrow-derived progenitor cells with endothelial
differentiation potential are reduced in patients with CAD
(11,12). Therefore, it is necessary to
determine the impact of CAD on the gene expression of
CD133-positive cells, which may advance the understanding regarding
the potential therapeutic mechanism of CD133-positive cells.
Although Liu et al (13) determined certain alterations in the
gene expression of CD133-positive progenitor cells from CAD
patients, the present study aimed to obtain more information via
currently available bioinformatic tools, such as cluster analysis
and functional enrichment analysis. Relevant small molecules were
also predicted, which could provide clues for future drug
development. In addition, the effect of exercise on the gene
expression of CD133-positive cells was also investigated.
Materials and methods
Gene expression data
Gene expression data set (accession number GSE18608)
(13) was downloaded from public
Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/). This dataset
included blood samples from four healthy subjects (H), and from 10
patients with coronary artery disease at baseline (B) and after 3
months of exercise (3M), which was used for isolation of
CD133-positive cells. Gene expression levels were measured using
GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array
(Affymetrix Inc., Santa Clara, CA, USA). Probe annotations were
also acquired.
Pre-treatment and differential
analysis
The 54,675 probes were all mapped to genes. A log2
transformation was applied on the gene expression levels (14). Differential analysis was performed
for H vs. B and H vs. 3M using limma (Linear Models for Microarray
Analysis) (15) package of
Bioconductor R (available at http://www.bioconductor.org/packages/release/bioc/html/limma.html).
Multiple testing correction using the Benjamini-Hochberg method
(16) was performed to adjust the
P-value to the false discovery rate (FDR). FDR<0.25 was set as
the cut-off to screen for the significant DEGs.
DEGs of H vs. B were compared with those of H vs. 3M
to screen out unique genes differentially expressed prior to and
following exercise. In addition, Student's t-test was performed for
unique DEGs between the groups. P<0.05 was set as the
cut-off.
Cluster analysis
Two-way cluster analysis was performed using the
expression levels of the DEGs by package pheatmap (17) in statistical software R (R version,
2.15, pheatmap version, 0.6.1; R Core Team, Vienna, Austria).
Euclidean distance was adopted to cluster the genes and produce the
dendrograms.
Functional enrichment analysis
Functional enrichment analysis was performed for the
DEGs using Database for Annotation, Visualization and Integration
Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (18) online tools. The statistical method
was performed based upon the hypergeometric distribution. P<0.05
was selected as the cut-off.
Prediction of relevant small
molecules
Connectivity map database (CMap; http://broad.mit.edu.rpa.skh.org.tw:81/cmap) is
designed to link gene patterns associated with disease to
corresponding patterns produced by drug candidates (19,20).
Relevant small molecules were predicted using the DEGs of H vs. 3M
and those with |score| >0.8 were retained.
Results
Differentially expressed genes
Normalized gene expression data are shown in
Fig. 1. A total of 131 and 71 DEGs
were identified in patients with CAD prior to and following
exercise, respectively. Fewer DEGs were observed in patients after
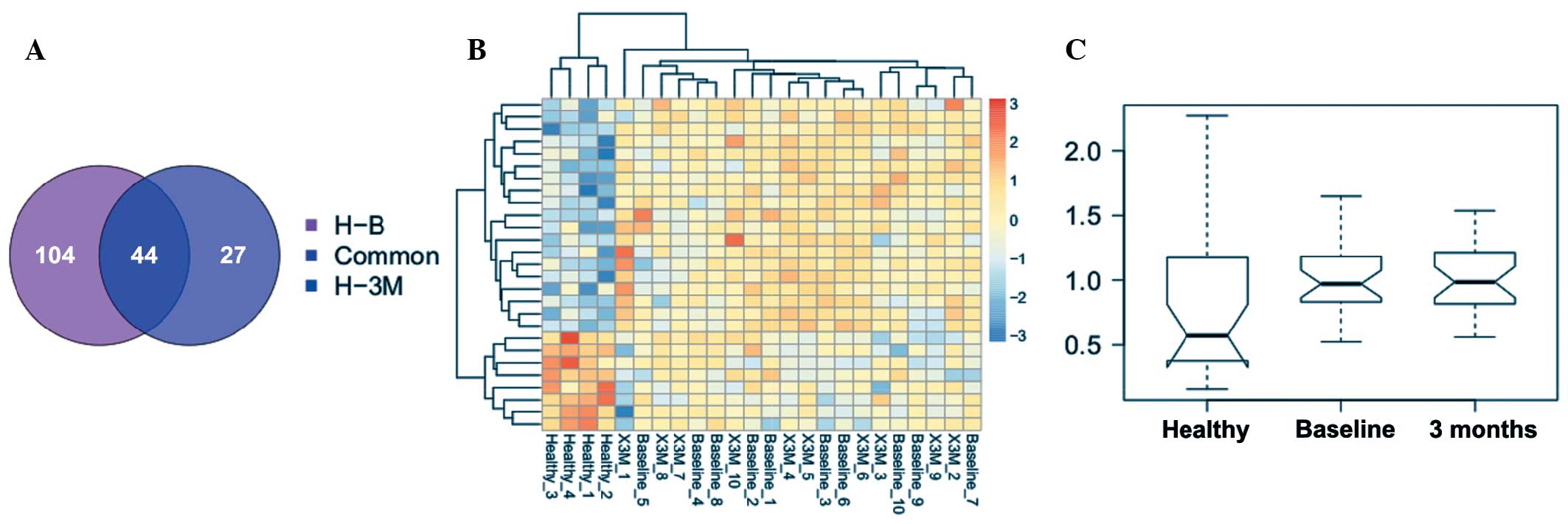
3 months of exercise. The two groups of DEGs were compared and 44
common genes were identified (Fig.
2A).
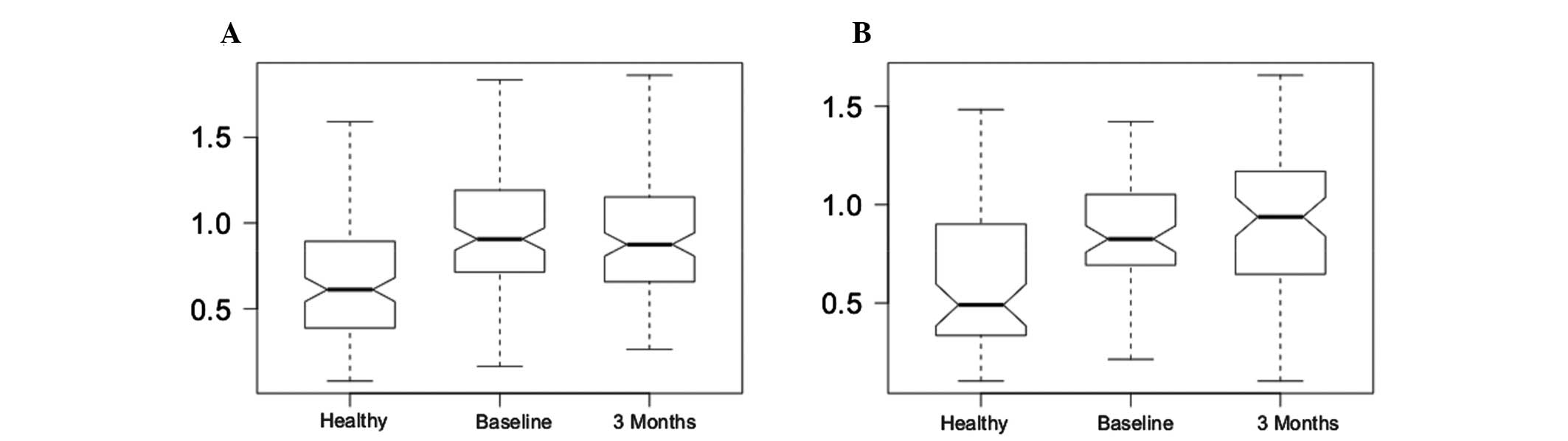
A t-test was performed using the expression levels
of the DEGs for H vs. B and H vs. 3M (Fig. 3 and Table I). The difference was significant,
which confirmed the reliability of the DEGs.
 | Table IStudent's t-test result based upon the
expression levels of differentially expressed genes. |
Table I
Student's t-test result based upon the
expression levels of differentially expressed genes.
| Comparison | Baseline
specific | 3M specific | Common |
|---|
| H vs. B | 8.82E-05 | 4.81–04 | 4.21E-03 |
| H vs. 3M | 6.15E-04 | 1.70E-04 | 5.36E-02 |
Cluster analysis result
The cluster analysis result is shown in Fig. 2B. CAD samples could be well
separated from healthy controls, confirming the reliability of the
DEGs. In addition, CAD samples prior to and following exercise were
clustered into one group with expression levels of common DEGs,
suggesting that 3 months of exercise would not bring significant
improvements in patients and further drug treatment may be
required.
Functional enrichment analysis
Functional enrichment analysis showed that response
to peptide hormone stimulus was most significantly enriched in the
common DEGs (Table II), including
protein kinase C α (PRKCA), protein kinase C ι (PRKCI) and
phosphodiesterase 3B (PDE3B). In addition, anti-apoptosis and
regulation of glucose transport pathways were also significantly
over-represented by the common DEGs.
 | Table IIFunctional enrichment analysis result
for the common differentially expressed genes. |
Table II
Functional enrichment analysis result
for the common differentially expressed genes.
| Term | Count | P-value | Genes |
|---|
| GO:0043434~response
to peptide hormone stimulus | 3 | 1.57E-02 | PRKCA, PRKCI,
PDE3B |
|
GO:0006916~anti-apoptosis | 3 | 2.70E-02 | SERPINB9, PRKCI,
POLB |
| GO:0046324~regulation
of glucose import | 2 | 4.07E-02 | PRKCA, PRKCI |
| GO:0010827~regulation
of glucose transport | 2 | 4.19E-02 | PRKCA, PRKCI |
| GO:0009187~cyclic
nucleotide metabolic process | 2 | 4.67E-02 | PDE3B, GUCY1A3 |
| GO:0019899~enzyme
binding | 4 | 1.37E-02 | PDE3B, NBEA, POLB,
GGA2 |
Relevant small molecules
A total of 12 relevant small molecules were revealed
by CMap based upon the expression levels of common DEGs (Table III). 5252917 [corresponding to
N-(2-benzooxazol-2-yl-phenyl) -4-methyl-benzenesulfon-amide] was
the most negatively correlated small molecule.
 | Table IIIRelevant small molecules. |
Table III
Relevant small molecules.
| CMap name | Enrichment | P-value |
|---|
| 5252917 | −0.98 | 6.80E-04 |
| MG-262 | −0.91 | 1.30E-03 |
| DL-thiorphan | −0.90 | 1.91E-02 |
| Biotin | −0.90 | 2.08E-03 |
| CP-944629 | −0.86 | 6.80E-04 |
| AH-6809 | −0.86 | 4.15E-02 |
| 5230742 | −0.85 | 4.58E-02 |
| Alsterpaullone | −0.82 | 1.17E-02 |
| PNU-0251126 | −0.80 | 1.40E-04 |
| Alfadolone | 0.82 | 1.20E-02 |
| Ketoconazole | 0.82 | 1.93E-03 |
| 5211181 | 0.92 | 1.37E-02 |
Discussion
In the present study, a total of 131 and 71 DEGs
were identified in patients with CAD prior to and following 3
months of exercise. The fewer DEGs identified after 3 months of
exercise suggested that exercise may normalize the expression of
certain genes in CD133-positive cells collected from patients with
CAD. However, further comparative study indicated 44 common genes
prior to and following 3 months of exercise, illustrating that
these genes were not altered by exercise and further treatment for
these genes is necessary.
Functional enrichment analysis showed that response
to peptide hormone stimulus was the most significant functional
term enriched in the common DEGs, including PRKCA, PRKCI, and
PDE3B. Protein kinase C (PKC) is a family of serine- and
threonine-specific protein kinases that can be activated by calcium
and the second messenger diacylglycerol. PKC family members
phosphorylate a wide variety of protein targets and are known to be
involved in diverse cellular signaling pathways. Braz et al
(21) first reported that PRKCA
regulates cardiac contractility and propensity towards heart
failure. Hambleton et al (22) further indicated that inducible and
myocyte-specific inhibition of PRKCA enhances cardiac contractility
and protects against infarction-induced heart failure. Hambleton
(23) drew a similar conclusion.
PRKCI is involved in the regulation of myocardial coherence and
remodeling during cardiac morphogenesis (24). Phosphodiesterase type 3 (PDE3) is
an important regulator of cAMP-mediated responses within the
cardiovascular system. Sun et al (25) indicated that PDE3A is the main
subtype of PDE3 expressed in platelets and cardiac ventricular
myocytes, and is responsible for the functional changes caused by
PDE3 inhibition. PDE3B may exhibit a different role in the
cardiovascular system, however, further studies are required to
determine this. Apoptosis is important in cardiovascular diseases,
such as atherosclerosis, ischemic heart disease and congestive
heart failure. It was demonstrated that three DEGs were involved in
anti-apoptosis pathways: Serpin B9 (SERPINB9), PRKCI and DNA
polymerase β. SERPINB9 is a member of the serpin family that has
been found to be an intrinsic inhibitor of granzyme B, which is
associated with apoptosis and thus is involved in cardiovascular
diseases (26). A protective role
of SERPINB9 against apoptosis has been reported in in vivo
and in vitro models (27,28).
All of these findings suggest that the above genes involved in
hormone response and apoptosis may serve as crucial therapeutic
targets for CAD.
To further examine the underlying treatment
approaches for the 44 genes, which were not altered by exercise,
the CMap database was used to predict the small molecules. As a
result, 12 small molecules were obtained, such as 5252917 and
MG-262. 5252917 was previously described as a mitotic inhibitor and
thus induces cell apoptosis (29),
and MG-262 is a proteasome inhibitor (30). Zheng et al (31) indicated that pharmacologically
induced proteasome inhibition is sufficient to activate autophagy
in cardiomyocytes. Stangl et al (32) reported that inhibition of the
ubiquitin-proteasome pathway induces differential heat shock
protein responses in cardio-myocytes and results in early cardiac
protection. These studies suggest that proteasome inhibitors may be
used in the treatment of cardiovascular diseases.
In conclusion, a number of DEGs were revealed in
CD133-positive cells obtained from patients with CAD. These
findings may advance the understanding of the molecular mechanisms
underlying cardiac repair and thus benefit cell-based therapy
development. Exercise showed a positive effect on the gene
expression of CD133-positive cells, but did not completely reverse
it. The non-altered genes may require treatment with small
molecules drugs..
Acknowledgments
The authors wish to thank Fenghe (Shanghai)
Information Technology Co., Ltd (Shanghai, China) for their ideas
and help, which gave a valuable added dimension to our
research.
References
|
1
|
Landmesser U and Drexler H: Chronic heart
failure: An overview of conventional treatment versus novel
approaches. Nat Clin Pract Cardiovasc Med. 2:628–638. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ford ES, Ajani UA, Croft JB, Critchley JA,
Labarthe DR, Kottke TE, Giles WH and Capewell S: Explaining the
decrease in U.S. deaths from coronary disease, 1980–2000. N Engl J
Med. 356:2388–2398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Landmesser U, Wollert KC and Drexler H:
Potential novel pharmacological therapies for myocardial
remodeling. Cardiovasc Res. 81:519–527. 2009. View Article : Google Scholar
|
|
4
|
Schächinger V, Erbs S, Elsässer A,
Haberbosch W, Hambrecht R, Hölschermann H, Yu J, Corti R, Mathey
DG, Hamm CW, et al: Improved clinical outcome after intracoronary
administration of bone-marrow-derived progenitor cells in acute
myocardial infarction: Final 1-year results of the REPAIR-AMI
trial. Eur Heart J. 27:2775–2783. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Segers VF and Lee RT: Stem-cell therapy
for cardiac disease. Nature. 451:937–942. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schots R, De Keulenaer G, Schoors D,
Caveliers V, Dujardin M, Verheye S, Van Camp G, Franken PR, Roland
J, Van Riet I and Everaert H: Evidence that intracoronary-injected
CD13+ peripheral blood progenitor cells home to the myocardium in
chronic postinfarction heart failure. Exp Hematol. 35:1884–1890.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartunek J, Vanderheyden M, Vandekerckhove
B, Mansour S, De Bruyne B, De Bondt P, Van Haute I, Lootens N,
Heyndrickx G and Wijns W: Intracoronary injection of CD133-positive
enriched bone marrow progenitor cells promotes cardiac recovery
after recent myocardial infarction: Feasibility and safety.
Circulation. 112:I178–I183. 2005.PubMed/NCBI
|
|
8
|
Ahmadi H, Baharvand H, Ashtiani SK,
Soleimani M, Sadeghian H, Ardekani JM, Mehrjerdi NZ, Kouhkan A,
Namiri M, Madani-Civi M, et al: Safety analysis and improved
cardiac function following local autologous transplantation of
CD133+ enriched bone marrow cells after myocardial infarction. Curr
Neurovasc Res. 4:153–160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klein H, Ghodsizad A, Marktanner R, Poll
L, Voelkel T, Mohammad Hasani MR, Piechaczek C, Feifel N,
Stockschlaeder M, Burchardt ER, et al: Intramyocardial implantation
of CD133+ stem cells improved cardiac function without bypass
surgery. Heart Surg Forum. 10:E66–E69. 2007. View Article : Google Scholar
|
|
10
|
Bonanno G, Mariotti A, Procoli A, Corallo
M, Rutella S, Pessina G, Scambia G, Mancuso S and Pierelli L: Human
cord blood CD133+ cells immunoselected by a clinical-grade
apparatus differentiate in vitro into endothelial- and
cardio-myocyte-like cells. Transfusion. 47:280–289. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heeschen C, Lehmann R, Honold J, Assmus B,
Aicher A, Walter DH, Martin H, Zeiher AM and Dimmeler S: Profoundly
reduced neovascularization capacity of bone marrow mononuclear
cells derived from patients with chronic ischemic heart disease.
Circulation. 109:1615–1622. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scheubel RJ, Zorn H, Silber RE, Kuss O,
Morawietz H, Holtz J and Simm A: Age-dependent depression in
circulating endothelial progenitor cells in patients undergoing
coronary artery bypass grafting. J Am Coll Cardiol. 42:2073–2080.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu D, Glaser AP, Patibandla S, Blum A,
Munson PJ, McCoy JP, Raghavachari N and Cannon RO: Transcriptional
profiling of CD133(+) cells in coronary artery disease and effects
of exercise on gene expression. Cytotherapy. 13:227–236. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fujita A, Sato JR, Rodrigues Lde O,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
16
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society. Series
B (Methodological). 289–300. 1995.
|
|
17
|
Kolde R: Pheatmap: Pretty heatmaps. R
package version 0.6. 1:2012.
|
|
18
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Catania A, Urban S, Yan E, Hao C and
Barron Gand Allalunis-Turner J: Expression and localization of
cyclin-dependent kinase 5 in apoptotic human glioma cells. Neuro
Oncol. 3:89–98. 2001.PubMed/NCBI
|
|
21
|
Braz JC, Gregory K, Pathak A, Zhao W,
Sahin B, Klevitsky R, Kimball TF, Lorenz JN, Nairn AC, Liggett SB,
et al: PKC-alpha regulates cardiac contractility and propensity
toward heart failure. Nat Med. 10:248–254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hambleton M, York A, Sargent MA, Kaiser
RA, Lorenz JN, Robbins J and Molkentin JD: Inducible and
myocyte-specific inhibition of PKCalpha enhances cardiac
contractility and protects against infarction-induced heart
failure. Am J Physiol Heart Circ Physiol. 293:H3768–H3771. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hambleton M, Hahn H, Pleger ST, Kuhn MC,
Klevitsky R, Carr AN, Kimball TF, Hewett TE, Dorn GW 2nd, Koch WJ
and Molkentin JD: Pharmacological- and gene therapy-based
inhibition of protein kinase Calpha/beta enhances cardiac
contractility and attenuates heart failure. Circulation.
114:574–582. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rohr S, Bit-Avragim N and
Abdelilah-Seyfried S: Heart and soul/PRKCi and nagie oko/Mpp5
regulate myocardial coherence and remodeling during cardiac
morphogenesis. Development. 133:107–115. 2006. View Article : Google Scholar
|
|
25
|
Sun B, Li H, Shakur Y, Hensley J, Hockman
S, Kambayashi J, Manganiello VC and Liu Y: Role of
phosphodiesterase type 3A and 3B in regulating platelet and cardiac
function using subtype-selective knockout mice. Cell Signal.
19:1765–1771. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saito Y, Kondo H and Hojo Y: Granzyme B as
a novel factor involved in cardiovascular diseases. J Cardiol.
57:141–147. 2011. View Article : Google Scholar
|
|
27
|
Stout-Delgado HW, Getachew Y, Rogers TE,
Miller BC and Thiele DL: The role of serpinb9/serine protease
inhibitor 6 in preventing granzyme B-dependent hepatotoxicity.
Hepatology. 46:1530–1540. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bird CH, Blink EJ, Hirst CE, Buzza MS,
Steele PM, Sun J, Jans DA and Bird PI: Nucleocytoplasmic
distribution of the ovalbumin serpin PI-9 requires a
nonconventional nuclear import pathway and the export factor Crm1.
Mol Cell Biol. 21:5396–5407. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fayad W, Rickardson L, Haglund C, Olofsson
MH, D'Arcy P, Larsson R, Linder S and Fryknäs M: Identification of
agents that induce apoptosis of multicellular tumour spheroids:
Enrichment for mitotic inhibitors with hydrophobic properties. Chem
Biol Drug Des. 78:547–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kisselev AF, van der Linden WA and
Overkleeft HS: Proteasome inhibitors: An expanding army attacking a
unique target. Chem Biol. 19:99–115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng Q, Su H, Tian Z and Wang X:
Proteasome malfunction activates macroautophagy in the heart. Am J
Cardiovasc Dis. 1:214–226. 2011.PubMed/NCBI
|
|
32
|
Stangl K, Günther C, Frank T, Lorenz M,
Meiners S, Röpke T, Stelter L, Moobed M, Baumann G, Kloetzel PM and
Stangl V: Inhibition of the ubiquitin-proteasome pathway induces
differential heat-shock protein response in cardiomyocytes and
renders early cardiac protection. Biochem Biophys Res Commun.
291:542–549. 2002. View Article : Google Scholar : PubMed/NCBI
|