Introduction
Diabetes mellitus (DM) is a common endocrine and
metabolic disease, and presents a major medical problem worldwide.
The World Health Organization has predicted that DM will be the
seventh leading cause of mortality by 2030 (1). In addition, it is the underlying
cause of microvascular disorders, including macrovascular diseases,
diabetic nephropathy and retinopathy (2). With the rapid development of social
economy and continuous improvement of living standards, the
prevalence of DM continues to increase. Currently, >347,000,000
individuals worldwide have beene diagnosed with DM (3). Type 2 DM (T2DM) accounts for 90–95%
of all these cases. It leads to a higher risk of chronic kidney
disease) (4), which may develop
into end-stage renal disease and has serious consequences on
morbidity and the mortality rates in this population. It is known
that effective glycemic control may delay the deterioration in
kidney function (5). T2DM is also
one of the leading causes of cardiovascular disease, as
atherosclerosis is the important pathological mechanism underlying
macrovascular disease in T2DM, resulting in major life-threatening
events, including myocardial infarction and stroke (6). Furthermore, clinical investigations
have suggested that certain intensive therapies are ineffective in
the prevention of cardiovascular-associated mortality in T2DM
(7). Atherosclerosis in diabetic
patients has a complex pathogenesis, which is initiated and
modified by environmental risk factors and a number of genes,
several of which remain to be elucidated (8). The overall genetic contribution is
reported to be ~50–70%, and a substantial number of genes have been
reported to be associated with T2DM (9–11).
To date, a number of the most important mechanisms have been
reported, including lipid metabolism disorders (12), endothelial dysfunction (13), chronic inflammation (14) and imbalance in redox homeostasis
(15). Notably, lipid metabolism
disorders are critical in atherosclerosis in T2DM patients.
MicroRNAs (miRNAs) represent the largest family of
short non-coding RNA molecules, with a length of 19–23 nucleotides
(16). It is well known that
miRNAs regulate the expression of target genes in a
post-transcriptional manner, through incomplete base pairing with
the 3′-untranslated region of target mRNAs, inhibiting translation
and/or causing mRNA degradation (17). These may contribute to the
regulation of a wide variety of cellular functions. It is now well
established that miRNAs account for ~1% of predicted genes in
higher eukaryotic genomes, and that up to 30% of protein-coding
genes may be regulated by miRNAs (18). In addition, previous studies have
demonstrated that miRNAs are critical regulators of cell cycle,
proliferation, differentiation, development and apoptosis during
embryogenesis (16,19,20).
As well as in the cell, miRNAs also exists in the circulatory
system, therefore, circulating miRNAs in the plasma are expected to
become novel biomarkers for molecular diagnostic and therapeutic
agents for various disease.
In a previous study (21), high throughput gene chips were used
to screen differential gene expression profiles in healthy
controls, patients with T2DM and its complication with patients
with unstable angina. The findings indicated that the occurrence of
T2DM is a complicated and dynamic process affected by various
miRNAs. Among the differentially expressed miRNAs, miR-9-3p was one
of the genes with the most significant difference in expression. To
the best of our knowledge, there are no reports evaluating the
association between miR-9-3p and lipid metabolism in T2DM combined
with atherosclerosis. It is well-known that the liver, as the main
organ of energy metabolism, is the most frequently involved organ
in glucose and lipid metabolic disorders (22). HepG2 cell lines have been used to
examine lipid metabolism disorder in several diseases. Therefore,
the aim of the present study was to evaluate the effects of
miRNA-9-3p on lipid metabolism in HepG2 cells, and investigate its
molecular mechanisms by cell transfection, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses.
The present study revealed that miRNA-9-3p has an
important role in development of T2DM, indicating its potential as
a therapeutic target. The future applications of miRNA-based
diagnostics and therapeutics in metabolic diseases requires further
research.
Materials and methods
Cell culture
The HepG2 cells were maintained in high
glucose-Dulbecco's modified Eagle's medium (DMEM) (Gibco Life
Technologies, Carlsbad, CA, USA), supplemented with 10% fetal
bovine serum (FBS; GE Healthcare Life Sciences, Logan, UT, USA) and
1% penicillin/streptomycin (XiangBo Biological Technology Co.,
Ltd., Guangzhou, China) in a humidified atmosphere of 5%
CO2 and 95% air at 37°C. The media was replaced every
2–3 days.
Cell transfection
The miRNA-9-3p mimic, miRNA-9-3p inhibitor
(anti-miRNA-9-3p) and their corresponding negative control (NC)
were purchased from Invitrogen Life Technologies (Carlsbad, CA,
USA). The HepG2 cells were transfected with the miRNA-9-3p mimic
(50 nM), miRNA-9-3p inhibitor (100 nM) or their negative controls
for 48 h using Lipofectamine 2000 (Invitrogen Life Technologies),
according to the manufacturer's instructions. The HepG2 cells were
divided into a normal group, NC group, miRNA-9-3p mimic group and
anti-miRNA-9-3p group.
MTT assay
Cell proliferation was determined using an MTT assay
24, 48 and 72 h following transfection. The HepG2 cells were plated
in 96-well plates at a density of 3×104 per well.
Subsequently, 10 µl 5 mg/ml MTT solution (Sigma-Aldrich, St.
Louis, MO, USA) was added to each well of the 96-well plate, and
the plates were incubated in the dark at 37°C for 2 h. The
absorbance was measured at a wavelength of 490 nm with a microplate
reader (BIORAD680; Bio-Rad Laboratories, Inc., Hercules, CA, USA),
and the results were expressed as a percentage of the control.
Intracellular triglyceride (TG) and total
cholesterol (TC) measurements
The concentrations of TG and TC in the HepG2 cells
were measured using a Total cholesterol Test kit and a Triglyceride
Test kit (BioVision, Inc., Milpitas, CA, USA), according to the
manufacturer's instructions, based on phosphoglycerol
oxidase/peroxidase enzymatic reactions. Briefly, HepG2 cells at 80%
confluence were washed three times with 1 ml cold
phosphate-buffered saline (PBS; Zhongshan Jinqiao Biological
Technology Co., Ltd., Beijing, China). The concentrations of TG and
TC were measured in the HepG2 cell lysates. The cell lysates,
working liquid and distilled water blended at 37°C for 6 min. After
cooling, the absorbent density was tested.
RNA extraction
Total RNA was isolated from the HepG2 cells using
TRIzol reagent (Invitrogen Life Technologies) and an miRNeasy mini
kit (Invitrogen Life Technologies), according to the manufacturer's
instructions. The concentration and purity of total RNA samples was
measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop
Technologies, Wilmington, DE, USA), with an average 260/280 ratio
of 2.02, and an average 260/230 ratio of 1.79. RNA integrity was
determined by agarose gel electrophoresis. Agarose (0.5 g;
Zhongshan Jinqiao Biological Technology Co., Ltd.) was dissolved in
electrophoresis buffer (50 ml; Zhongshan Jinqiao Biological
Technology Co., Ltd.), and the samples were added to the gel
following blending. The voltage was adjusted to 100 V, to allow RNA
migration from the anode to the cathode. The gel was treated with
EB dyeing liquid (Applygen Technologies Inc. Beijing, China) for 5
min.
RT-qPCR
The concentration, purity and integrity of the RNA
samples in the present study met the criteria (A260/A280 ratio
>2.0 and an A260/A230 ratio >1.7) for evaluation using
RT-qPCR analysis. For RNA analysis, single-strand cDNA synthesis
was performed using a PrimeScript II 1st strand cDNA Synthesis kit
(Takara Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's instructions. The reaction volume was 50 µl:
RNase-DNase free water 32.5 µl, buffer 10 µl, dNTP mix (2.5 mM) 4
µl, forward primer (10 µM) 1 µl, reverse primer (10 µM) 1 µl, cDNA
1 µl and PrimeStar HS DNA polymerase 0.5 µl. RT-qPCR was performed
on an ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad,
CA, USA), with the following conditions: One cycle of initial
denaturation at 95°C for 10 min; followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min. The primers designed in the present
study (Sangon Biotech Co., Ltd., Shanghai, China) were as follows:
sirtuin type 1 (SIRT1), sense 5′-CTACTGGTCTTACTTTGAGGG-3′, and
antisense 5′-CAAGGGATGGTATTTATGCT-3′; and GAPDH, sense
5′-TCCACCACCCTGTTGCTGTA-3′ and anti-sense
5′-ACCACAGTCCATGCCATCAC-3′. GAPDH was used as an internal control.
The threshold cycle (Ct), defined as the cycle at which PCR
amplification reached a significant value, was determined as the
mean value. The relative expression levels for each mRNA were
determined using the 2−ΔΔCt method (23).
Western blot analysis
The cells were washed in fresh PBS, homogenized in
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich)
containing protease inhibitors (Roche Diagnostics, Indianapolis,
IN, USA) and centrifuged at 10,000 x g for 15 min at 4°C. Protein
concentration was determined using a Bio-Rad protein assay reagent
(Bio-Rad Laboratories, Inc.). Equal quantities of total protein (40
µg) were subjected to electrophoreses on 10% SDS-PAGE
(OriGene Technologies, Inc., Beijing, China), and then transferred
onto polyvinylidene difluoride membranes (Roche Diagnostics GmbH,
Mannheim, Germany). The membranes were blocked with 5% non-fat dry
milk in Tris-buffered saline-Tween 20 (0.1%) (TBST; Zhongshan
Jinqiao Biological Technology Co., Ltd.), and incubated with mouse
anti-SIRT1 monoclonal antibody (A-4111-100; 1:200; Abcam,
Cambridge, MA, USA) or mouse anti-β-actin monoclonal antibody
(sc-47778; 1:500; Abcam) at 37°C for 2 h. Subsequently, the
proteins were incubated with the corresponding horseradish
peroxidase-conjugated secondary antibodies (111-035-003; 1:5,000;
Abcam) at 37°C for 1 h and then washed three times with TBST for 15
min. The proteins were detected using an enhanced chemi
luminescence (ECL) system (GE Healthcare Life Sciences).
Densitometric quantification of the bands was performed using
ImageJ software (version 1.41; National Institutes of Health,
Bethesda, MD, USA). The loading volume of each sample was
normalized by the β-actin protein band density.
Statistical analysis
Statistical analysis in the present study was
performed using SPSS (version 18.0; SPSS, Inc., Chicago, IL, USA).
All data from the experiments are presented as the mean ± standard
error of the mean. Significant differences in the mean values were
evaluated using Student's t-test, and one-way analysis of variance
was performed to compare the means among groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miRNA-9-3p in HepG2
cells
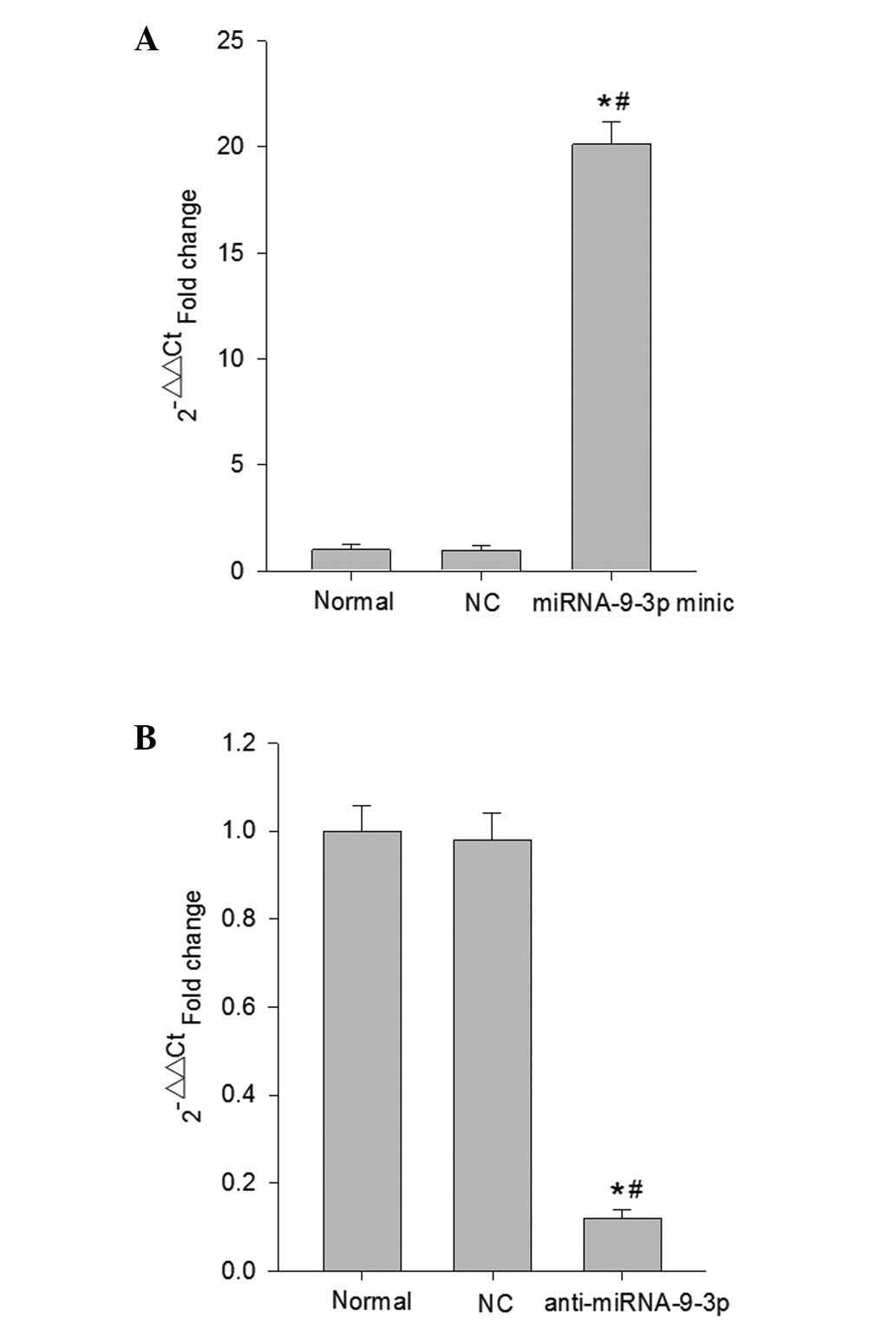
To determine the potential cellular effects of
miRNA-9-3p on HepG2 cells, the HepG2 cells were transfected with
the miRNA-9-3p mimic or miRNA-9-3p inhibitor, and their NCs for 48
h. To confirm whether miR-9-3p was upregulated or downregulated in
the HepG2 cells, the expression levels of miRNA-9-3p in the HepG2
cells were determined using RT-qPCR analysis (Fig. 1). The results revealed that
miRNA-9-3p was expressed at significantly higher levels in the
HepG2 cells transfected with the miRNA-9-3p mimic, compared with
the levels in the normal control group and NC group (P<0.05;
Fig. 1A). By contrast, miRNA-9-3p
was downregulated in the HepG2 cells transfected with the
miRNA-9-3p inhibitor, compared with the normal control group and NC
group (P<0.05;Fig. 1B).
Effects of miRNA-9-3p on HepG2 cell
proliferation
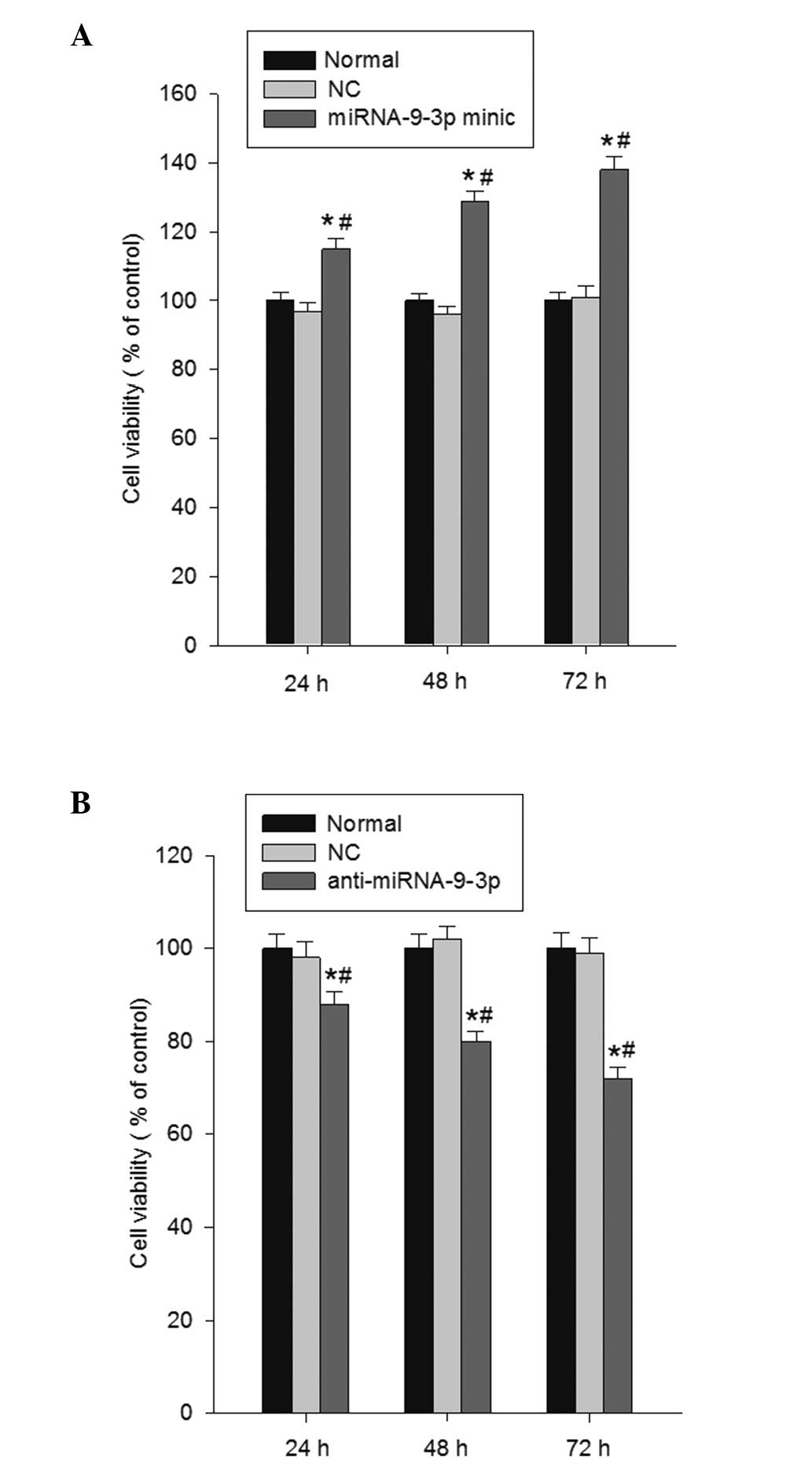
The present study investigated the effect of
miRNA-9-3p on cell proliferation using MTT growth assays at 24, 48
and 72 h post-transfection. The data revealed that miRNA-9-3p
overexpression significantly enhanced cell proliferation, compared
with the normal group and NC group (P<0.05; Fig. 2A). However, in the HepG2 cell
lines, transfection with the miRNA-9-3p inhibitor markedly
inhibited cell proliferation (P<0.05; Fig. 2B). Collectively, these results
demonstrated that the inhibition of miRNA-9-3p reduced the growth
of the HepG2 cells.
Effects of miRNA-9-3p on the levels of TG
and TC in HepG2 cells
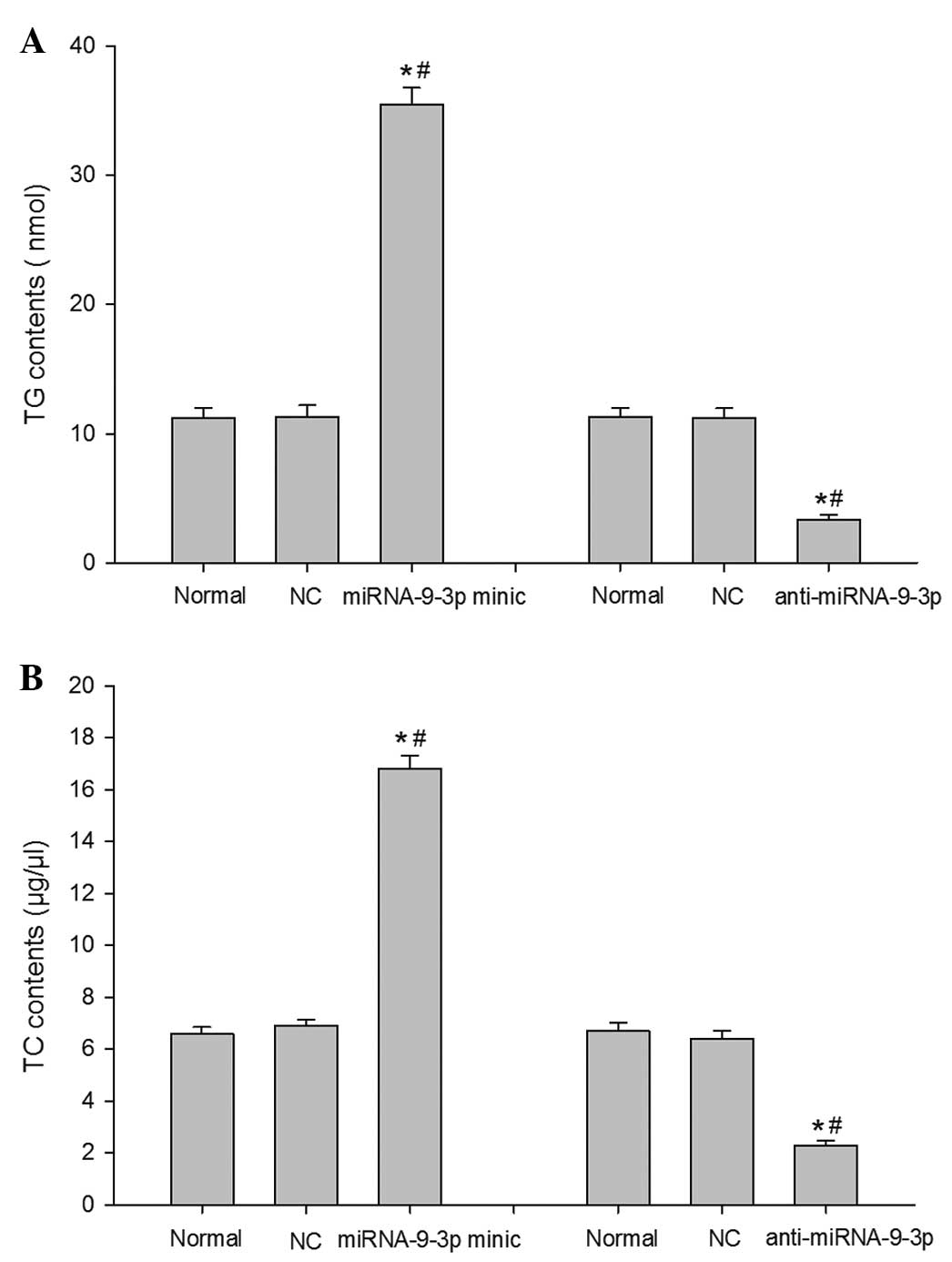
Subsequently, the present study analyzed the effect
of miRNA-9-3p on lipid accumulation in the HepG2 cells. The TG and
TC contents in the cells were measured using phospho-glycerol
oxidase/peroxidase enzymatic reactions. As shown in Fig. 3A and B, the levels of TG and TC in
the miRNA-9-3p group were significantly increased, compared with
the normal group an NC group (P<0.05). By contrast, transfection
with the miRNA-9-3p inhibitor caused downregulation in the levels
of TG and TC (P<0.05). These results indicated that inhibition
of the expression of miRNA-9-3p significantly decreased the TG and
TC contents, and relieved lipid deposition in the HepG2 cells.
Effects of miRNA-9-3p on the expression
of SIRT1 mRNA in HepG2 cells
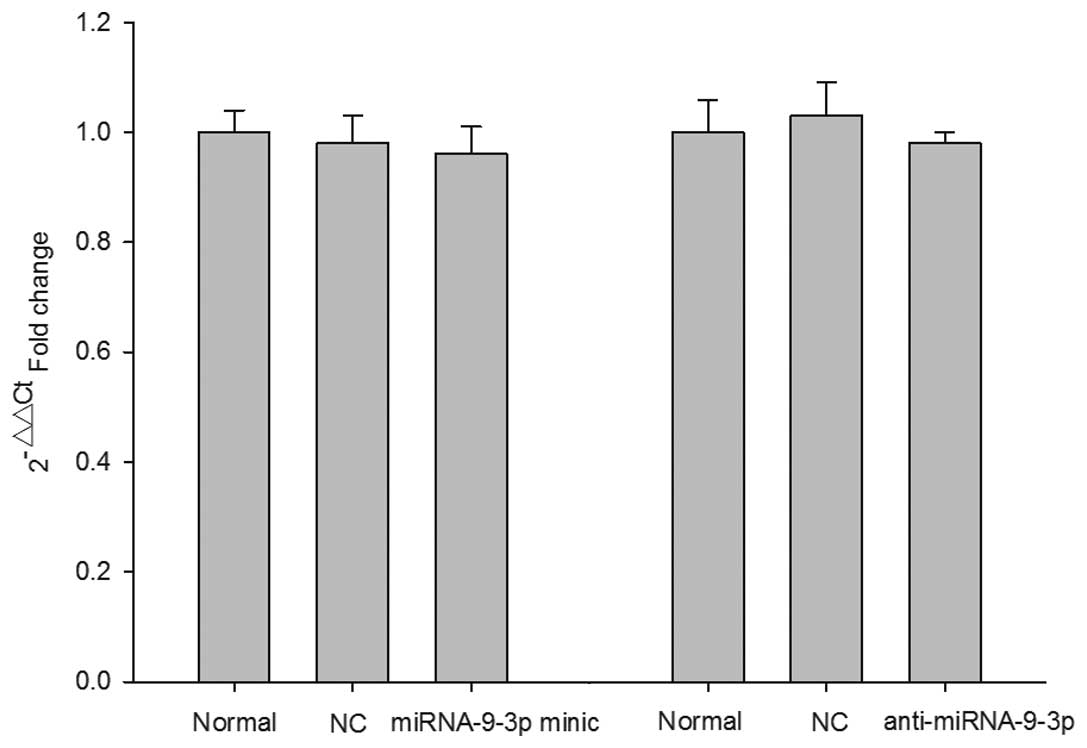
RT-qPCR analysis of the mRNA levels of SIRT1 was
performed 48 h post-transfection. As shown in Fig. 4, the RT-qPCR results demonstrated
no significant difference in the mRNA expression levels of SIRT1
relative to the mRNA expression of GAPDH in any group.
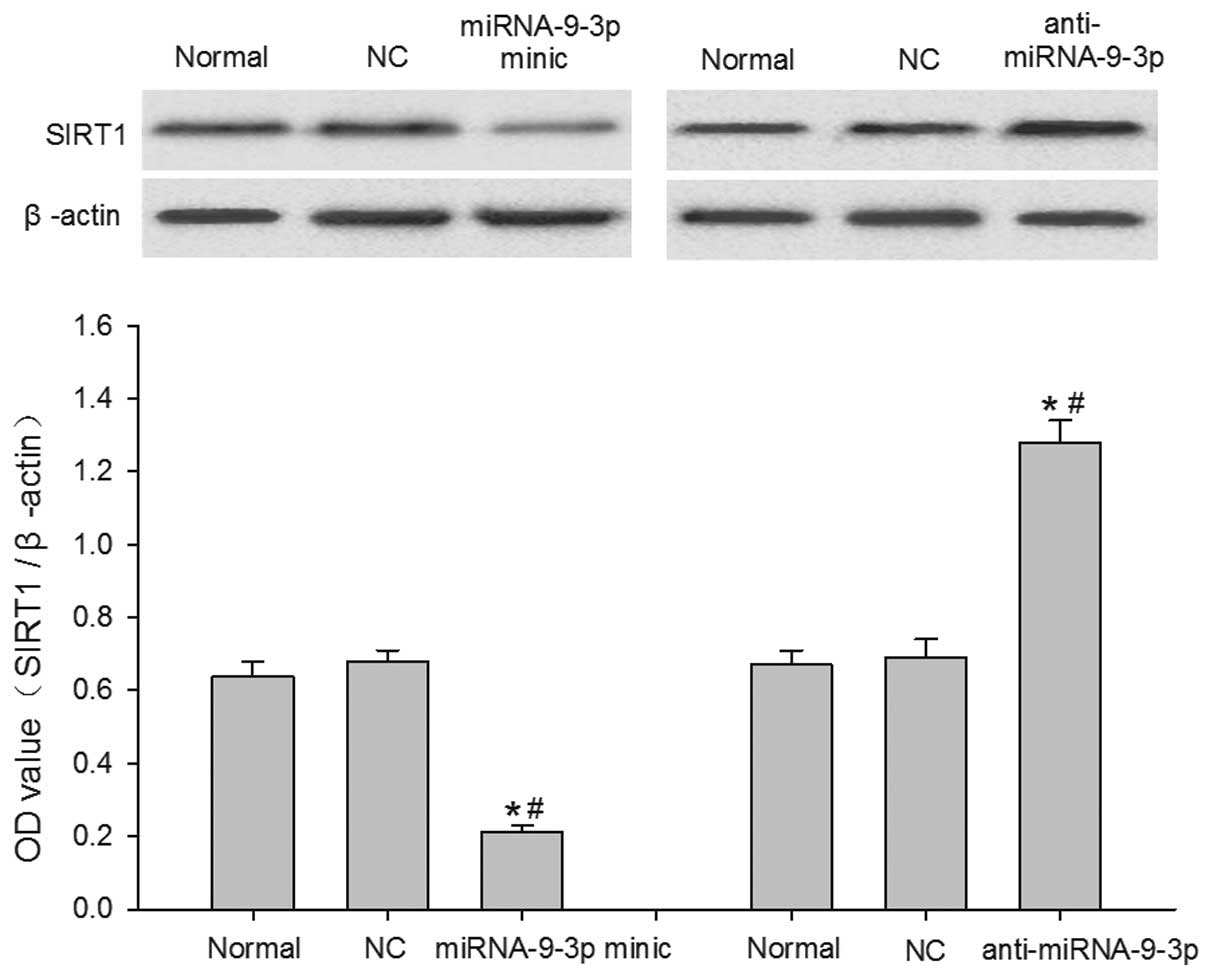
Effects of miRNA-9-3p on the protein
expression of SIRT1 in HepG2 cells
The protein expression of SIRT1 was analyzed using
western blot analysis 48 h post-transfection. The data were
normalized to the expression of β-actin. As shown in Fig. 5, the protein levels of SIRT1 were
significantly decreased in the miRNA-9-3p-transfected HepG2 cells,
compared with the cells in the normal group and NC group
(P<0.05). However, the protein levels of SIRT1 were
significantly increased when miRNA-9-3p was inhibited via
transfection with anti-miRNA-9-3p, compared with the normal group
and NC group (P<0.05). These results indicated that the protein
expression of SIRT1 is endogenously regulated by miRNA-9-3p in
HepG2 cells, suggesting that the expression of SIRT1 was
post-transcriptionally regulated by miRNA-9-3p in the HepG2
cells.
Discussion
miRNAs are a group of small non-coding RNAs, which
modulate the expression of target genes (24). Increasing evidence has revealed
that miRNAs are critical in cell cycle regulation, cell
differentiation, proliferation and apoptosis (25,26).
miRNAs control these cellular biological functions by targeting the
expression of genes. Therefore, the understanding and elucidation
of functional targeted genes is a focus of investigations.
Additionally, miRNAs are involved in several physiological and
pathological processes as negative regulators. It has been reported
that miRNAs are important role in insulin signaling and insulin
secretion, however, the association between miRNAs and insulin
resistance remains to be fully elucidated (27). In HepG2 cells lines, a previous
study demonstrated that the overexpression of miRNA-145 inhibits
glucose uptake, decreases the phosphorylation of protein kinase B
(Akt) and insulin receptor substrate-1 (IRS-1), and induces
resistin-induced insulin resistance (28). The IRS-1/phosphoinositide 3-kinase
(PI3K)/AKT pathway is the predominant insulin signaling pathway
involved in the regulation of blood glucose in T2DM. Another study
revealed that miRNA-126 overexpression inhibited the PI3K/AKT
pathway by regulating the expression of IRS-1, suggesting that
miRNA-126 may be associated with glucose metabolism (29). The knockout of miRNA-375 in mice
reduces the secreting ability of pancreatic islet B cells, causing
insulin resistance and severe clinical symptoms (30), whereas the overexpression of
miRNA-29a and miRNA-29b may inhibit insulin induced glucose uptake
(31). In previous years, studies
have reported that miRNA not only directly regulates insulin
secretion, pancreatic development and pancreatic islet cell
differentiation, but also indirectly regulates lipid metabolism and
is involved in diabetes and its complications (32,33,34).
However, whether miRNAs are involved in lipid metabolism in
diabetes remain to be elucidated. Therefore, the purpose of the
present study was to investigated whether miRNA-9-3p is involved in
the process of lipid metabolism.
The human miRNA-9 gene locates on chromosome 15, and
includes miRNA-9-3p and miRNA-9-5p. miRNA-9, with a brain source,
is highly expressed in the nervous system and is conserved among
species, exhibiting 100% similarity between Drosophila
melanogaster and vertebrates (35). miRNA-9 has been demonstrated to be
crucial in the repair of the myelin sheath following
hypoxic-ischemic brain damage (36). Several studies have demonstrated
that miRNA-9 regulates the differentiation of oligodendrocyte
progenitor cells by regulating serum response factor (37), and maintain myelinogenesis
(38). Importantly, the expression
of miR-9-3p in islet β cells is involved in the physiological
processes of insulin secretion and glucose metabolism (39). A study by Plaisance demonstrated
that miR-9-3p overexpression increased the secretion of
granuphilin-a protein, and reduced the exocrine abilities of the
islet β cells (40). In addition,
miRNA-9-3p in the plasma may be involved in cholesterol metabolism
and angiogenesis, adjusting the biological function of the
myocardium (41). However, whether
there is a regulatory association between miRNA-9-3p and lipid
metabolism disorder in T2DM remains to be elucidated.
In the present study, the effect of miRNA-9-3p on
lipid accumulation in the HepG2 cells was examined by transfection
of miRNA-9-3p minic, miRNA-9-3p inhibitor and their corresponding
NCs into HepG2 cells. For the first time, to the best of our
knowledge, the present study demonstrated that the inhibition of
miRNA-9-3p suppressed the proliferation of HepG2 cells and
significantly decreased TG and TC contents, reducing lipid
accumulation in the HepG2 cells. These results indicated that
miRNA-9-3p may offer potential as a therapeutic target for diseases
associated with lipid metabolism. Subsequent investigations aim to
identify biological or molecular tools to alter the expression of
miRNA-9-3p for further elucidation of its function and therapeutic
implications.
To examine the molecular mechanisms underlying the
inhibition of lipid deposition, the present study assessed the
expression of SIRT1, at mRNA and protein levels, in the HepG2
cells. The results of the RT-qPCR and western blot analyses
revealed that the inhibition of miRNA-9-3p upregulated the protein
expression of SIRT1 protein, but had no effect on the mRNA
expression of SIRT1. SIRT1 is an NAD+-dependent
deacetylase, which has been linked to the regulation of cell aging,
life span and energy metabolism (42). SIRT1 function has been reported to
regulate a wide range of cellular functions affecting metabolic
homeostasis. SIRT1 not only exerts anti-oxidative, anti-apoptotic,
and anti-inflammatory effects against cellular injury, it also
inhibits sugar dysplasia effects and improves insulin sensitivity
in liver, muscle and adipose tissue (43-45).
Furthermore, the activation and upregulation of SIRT1 may be
beneficial in age-associated disorders, including T2DM and
neurodegenerative diseases (46).
Similarly, down-regulation in the expression of SIRT1 has been
observed in insulin-resistant subjects (47). In liver tissue, SIRT1 is key in the
regulation of lipid metabolism balance. Lovis et al
(48) found that SIRT1-knockout in
the liver increases the level of glucose in the plasma, reduces
insulin sensitivity and upregulates the levels of free fatty acids
and cholesterol. These effects may be associated with the
downregulation of ABCA1 and low density lipoprotein receptor
(LDLR), reported by Balasubramanyam et al (49). In addition, Adlakha et al
(50) revealed that miRNA-128
regulated lipid metabolism by targeting the expression of SIRT1 in
HepG2 cells. These findings indicate that the expression SIRT1
regulates lipid metabolism. Therefore, maintaining SIRT1 levels or
stimulating upregulation in the expression of SIRT1 may be
beneficial in the treatment of a variety of diseases with lipid
metabolic disorders.
In conclusion, the findings of the present study
indicated that the inhibition of miRNA-9-3p exerted a regulatory
effect on lipid accumulation, reducing TG and TC contents, by
regulating the protein expression of SIRT1 in the HepG2 cells. This
novel evidence may assist in understanding the potential roles of
miRNA-9-3p in T2DM, and supports the possibility of a synthetic
inhibitor of miRNA-9-3p as a novel therapeutic strategy for lipid
metabolism disorder in T2DM.
Acknowledgments
The present study was supported by a grant from the
Scientific Research Fund Project of Hebei Province (grant. no.
20130310).
Abbreviations:
|
DM
|
diabetes mellitus
|
|
T2DM
|
type 2 diabetes mellitus
|
|
miRNAs
|
microRNAs
|
|
SIRT1
|
sirtuin type 1
|
|
TG
|
triglycerides
|
|
TC
|
total cholesterol
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Ludovico O, Carella M, Bisceglia L, Basile
G, Mastroianno S, Palena A, De Cosmo S, Copetti M, Prudente S and
Trischitta V: Identification and clinical characterization of adult
patients with multigenerational diabetes mellitus. PLoS One.
10:e01358552015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou Y, Fang R, Liu LH, Chen SD and Tang
HD: Clinical characteristics for the relationship between type-2
diabetes mellitus and cognitive impairment: A cross-sectional
study. Aging Dis. 6:236–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
García ALM, Villarreal RE, Galicia RL,
Martínez GL and Vargas DER: The cost of polypharmacy in patients
with type 2 diabetes mellitus. Rev Med Chil. 143:606–611. 2015.In
Spanish. View Article : Google Scholar
|
|
4
|
National Kidney Foundation: KDOQI Clinical
Practice Guideline for Diabetes and CKD: 2012 Update. Am J Kidney
Dis. 60:850–886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perkovic V, Heerspink HL, Chalmers J,
Woodward M, Jun M, Li Q, MacMahon S, Cooper ME, Hamet P, Marre M,
et al: Intensive glucose control improves kidney outcomes in
patients with type 2 diabetes. Kidney Int. 83:517–523. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Madonna R and De Caterina R: Cellular and
molecular mechanisms of vascular injury in diabetes-part I:
Pathways of vascular disease in diabetes. Vasc Pharmacol. 54:68–74.
2011. View Article : Google Scholar
|
|
7
|
Duckworth W, Abraira C, Moritz T, Reda D,
Emanuele N, Reaven PD, Zieve FJ, Marks J, Davis SN, Hayward R, et
al: Glucose control and vascular complications in veterans with
type 2 diabetes. New Engl J Med. 360:129–139. 2009. View Article : Google Scholar
|
|
8
|
Li H, Feng SJ, Su LL, Wang W, Zhang XD and
Wang SX: Serum hepcidin predicts uremic accelerated atherosclerosis
in chronic hemodialysis patients with diabetic nephropathy. Chin
Med J (Engl). 128:1351–1357. 2015. View Article : Google Scholar
|
|
9
|
Talmud PJ, Hingorani AD, Cooper JA, Marmot
MG, Brunner EJ, Kumari M, Kivimäki M and Humphries SE: Utility of
genetic and non-genetic risk factors in prediction of type 2
diabetes: Whitehall II prospective cohort study. BMJ.
340:b48382010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Gan W, Lu L, Dong X, Han X, Hu C,
Yang Z, Sun L, Bao W, Li P, et al: A genome-wide association study
identifies GRK5 and RASGRP1 as type 2 diabetes loci in Chinese
Hans. Diabetes. 62:291–298. 2013. View Article : Google Scholar
|
|
11
|
Wang C, Li L, Wang L, Ping Z, Flory MT,
Wang G, Xi Y and Li W: Evaluating the risk of type 2 diabetes
mellitus using artificial neural network: An effective
classification approach. Diabetes Res Clin Pract. 100:111–118.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Watanabe T, Hirata M, Yoshikawa Y,
Nagafuchi Y, Toyoshima H and Watanabe T: Role of macrophages in
atherosclerosis. Sequential observations of cholesterol-induced
rabbit aortic lesion by the immunoperoxidase technique using
monoclonal antimacrophage antibody. Lab Invest. 53:80–90.
1985.PubMed/NCBI
|
|
13
|
Werns SW, Walton JA, Hsia HH, Nabel EG,
Sanz ML and Pitt B: Evidence of endothelial dysfunction in
angiographically normal coronary arteries of patients with coronary
artery disease. Circulation. 79:287–291. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansson GK: Immune and inflammatory
mechanisms in the development of atherosclerosis. Br Heart J.
69(Suppl 1): S38–S41. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guzik B, Sagan A, Ludew D, Mrowiecki W,
Chwała M, Bujak-Gizycka B, Filip G, Grudzien G, Kapelak B, Zmudka
K, et al: Mechanisms of oxidative stress in human aortic
aneurysms-association with clinical risk factors for
atherosclerosis and disease severity. Int J Cardiol. 168:2389–2396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim YK: Extracellular microRNAs as
biomarkers in human disease. Chonnam Med J. 51:51–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nature reviews Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ross JS, Carlson JA and Brock G: miRNA:
The new gene silencer. Am J Clin Pathol. 128:830–836. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rui W, Bing F, Hai-Zhu S, Wei D and
Long-Bang C: Identification of microRNA profiles in
docetaxel-resistant human non-small cell lung carcinoma cells
(SPC-A1). J Cell Mol Med. 14:206–214. 2010. View Article : Google Scholar
|
|
21
|
Ortega FJ, Mercader JM, Moreno-Navarrete
JM, Rovira O, Guerra E, Esteve E, Xifra G, Martínez C, Ricart W,
Rieusset J, et al: Profiling of circulating microRNAs reveals
common microRNAs linked to type 2 diabetes that change with insulin
ensitization. Diabetes Care. 37:1375–1383. 2014. View Article : Google Scholar
|
|
22
|
Stachowiak G, Pertyński T and
Pertyńska-Marczewska M: Metabolic disorders in menopause. Prz
Menopauzalny. 14:59–64. 2015.PubMed/NCBI
|
|
23
|
Bubner B and Baldwin IT: Use of real-time
PCR for determining copy number and zygosity in transgenic plants.
Plant Cell Rep. 23:263–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
O'Connell RM, Rao DS, Chaudhuri AA and
Baltimore D: Physiological and pathological roles for microRNAs in
the immune system. Nat Rev Immunol. 10:111–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. P Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar
|
|
27
|
Yang WM, Jeong HJ, Park SY and Lee W:
Saturated fatty acid-induced miR-195 impairs insulin signaling and
glycogen metabolism in HepG2 cells. FEBS Lett. 588:3939–3946. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kent OA, McCall MN, Cornish TC and
Halushka MK: Lessons from miR-143/145: The importance of cell-type
localization of miRNAs. Nucleic Acids Res. 42:7528–7538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elmen J, Lindow M, Schütz S, Lawrence M,
Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et
al: LNA-mediated microRNA silencing in non-human primates. Nature.
452:896–899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Bio. 6:376–385. 2005.
View Article : Google Scholar
|
|
32
|
Gong W, Xiao D, Ming G, Yin J, Zhou H and
Liu Z: Type 2 diabetes mellitus-related genetic polymorphisms in
microRNAs and microRNA target sites. J Diabetes. 6:279–289. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rome S: Are extracellular microRNAs
involved in type 2 diabetes and related pathologies? Clin Biochem.
46:937–945. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hamar P: Role of regulatory micro RNAs in
type 2 diabetes mellitus-related inflammation. Nucleic Acid Ther.
22:289–294. 2012.PubMed/NCBI
|
|
35
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, He X, Han X, Yu Y, Ye F, Chen Y,
Hoang T, Xu X, Mi QS, Xin M, et al: MicroRNA-mediated control of
oligodendrocyte differentiation. Neuron. 65:612–626. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Buller B, Chopp M, Ueno Y, Zhang L, Zhang
RL, Morris D, Zhang Y and Zhang ZG: Regulation of serum response
factor by miRNA-200 and miRNA-9 modulates oligodendrocyte
progenitor cell differentiation. Glia. 60:1906–1914. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li JS and Yao ZX: MicroRNAs: Novel
regulators of oligodendrocyte differentiation and potential
therapeutic targets in demyelination-related diseases. Mol
Neurobiol. 45:200–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramachandran D, Roy U, Garg S, Ghosh S,
Pathak S and Kolthur-Seetharam U: Sirt1 and mir-9 expression is
regulated during glucose-stimulated insulin secretion in pancreatic
β-islets. FEBS J. 278:1167–1174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Plaisance V, Abderrahmani A, Perret-Menoud
V, Jacquemin P, Lemaigre F and Regazzi R: MicroRNA-9 controls the
expression of Granuphilin/Slp4 and the secretory response of
insulin-producing cells. J Biol Chem. 281:26932–26942. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuva-Aydemir Y, Simkin A, Gascon E and Gao
FB: MicroRNA-9: Functional evolution of a conserved small
regulatory RNA. RNA Biol. 8:557–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guarente L and Picard F: Calorie
restriction-the SIR2 connection. Cell. 120:473–482. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roggli E, Britan A, Gattesco S, Lin-Marq
N, Abderrahmani A, Meda P and Regazzi R: Involvement of microRNAs
in the cytotoxic effects exerted by proinflammatory cytokines on
pancreatic beta-cells. Diabetes. 59:978–986. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stefanowicz M, Strączkowski M and
Karczewska-Kupczewska M: The role of SIRT1 in the pathogenesis of
insulin resistance in skeletal muscle. Postepy Hig Med Dosw
(Online). 69:63–68. 2015. View Article : Google Scholar
|
|
45
|
Kitada M and Koya D: SIRT1 in type 2
diabetes: Mechanisms and therapeutic potential. Diabetes Metab J.
37:315–325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lavu S, Boss O, Elliott PJ and Lambert PD:
Sirtuins-novel therapeutic targets to treat age-associated
diseases. Nat Rev Drug Discov. 7:841–853. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
de Kreutzenberg SV, Ceolotto G, Papparella
I, Bortoluzzi A, Semplicini A, Dalla Man C, Cobelli C, Fadini GP
and Avogaro A: Downregulation of the longevity-associated protein
sirtuin 1 in insulin resistance and metabolic syndrome: Potential
biochemical mechanisms. Diabetes. 59:1006–1015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lovis P, Gattesco S and Regazzi R:
Regulation of the expression of components of the exocytotic
machinery of insulin-secreting cells by microRNAs. Biol Chem.
389:305–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Balasubramanyam M, Aravind S,
Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjan H and Mohan V:
Impaired miR-146a expression links subclinical inflammation and
insulin resistance in Type 2 diabetes. Mol Cell Biochem.
351:197–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Adlakha YK, Khanna S, Singh R, Singh VP,
Agrawal A and Saini N: Pro-apoptotic miRNA-128-2 modulates ABCA1,
ABCG1 and RXRα expression and cholesterol homeostasis. Cell Death
Dis. 4:e7802013. View Article : Google Scholar
|