|
1
|
National Spinal Cord Injury Statistical
Center: Spinal cord injury facts and figures at a glance. J Spinal
Cord Med. 36:1–2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu J: China spinal cord injury network:
changes from within. Lancet Neurol. 8:606–607. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thuret S, Moon LD and Gage FH: Therapeutic
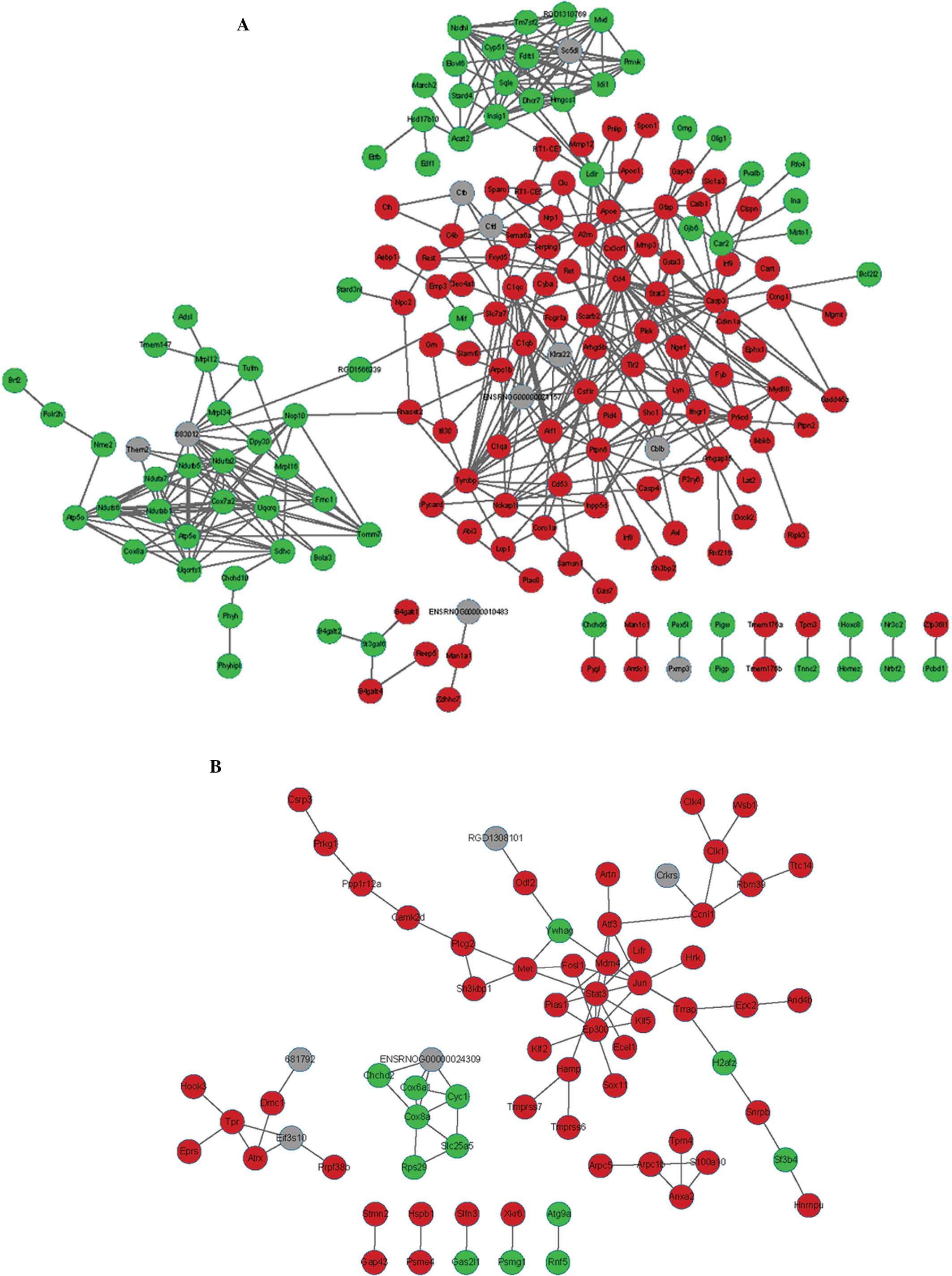
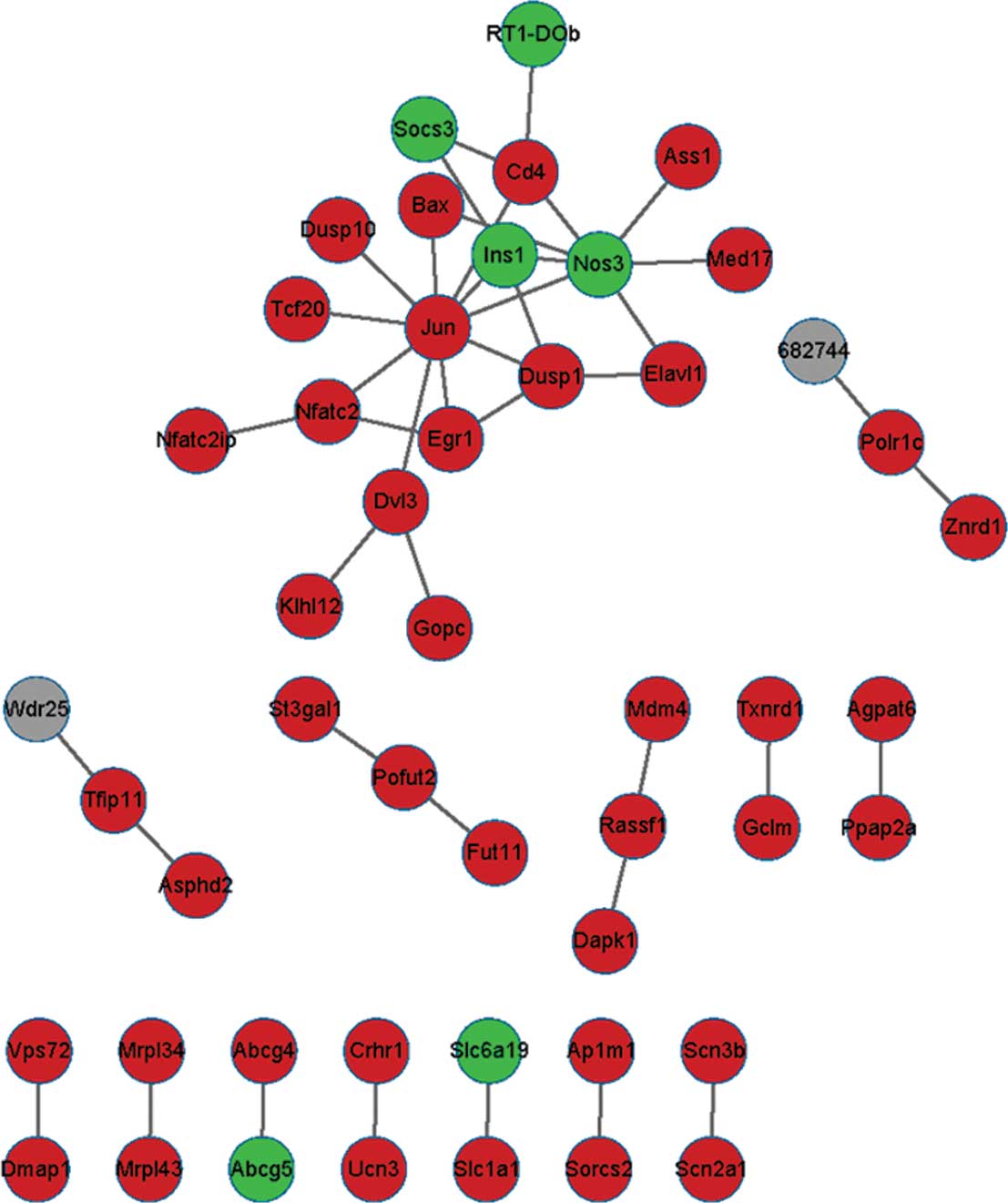
interventions after spinal cord injury. Nat Rev Neurosci.
7:628–643. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Freund P, Weiskopf N, Ashburner J, et al:
MRI investigation of the sensorimotor cortex and the corticospinal
tract after acute spinal cord injury: a prospective longitudinal
study. Lancet Neurol. 12:873–881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McDonald JW and Sadowsky C: Spinal-cord
injury. Lancet. 359:417–425. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mariano ED, Batista CM, Barbosa BJ, et al:
Current perspectives in stem cell therapy for spinal cord repair in
humans: a review of work from the past 10 years. Arq
Neuropsiquiatr. 72:451–456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Young W: Spinal cord regeneration. Cell
Transplant. 23:573–611. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu NK and Xu XM: Phospholipase A2 and its
molecular mechanism after spinal cord injury. Mol Neurobiol.
41:197–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Donnelly DJ and Popovich PG: Inflammation
and its role in neuroprotection, axonal regeneration and functional
recovery after spinal cord injury. Exp Neurol. 209:378–388. 2008.
View Article : Google Scholar
|
|
10
|
Lee K, Na W, Lee JY, et al: Molecular
mechanism of Jmjd3-mediated interleukin-6 gene regulation in
endothelial cells underlying spinal cord injury. J Neurochem.
122:272–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kerschensteiner M, Gallmeier E, Behrens L,
et al: Activated human T cells, B cells and monocytes produce
brain-derived neurotrophic factor in vitro and in inflammatory
brain lesions: a neuroprotective role of inflammation? J Exp Med.
189:865–870. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yekutiel M, Brooks M, Ohry A, Yarom J and
Carel R: The prevalence of hypertension, ischaemic heart disease
and diabetes in traumatic spinal cord injured patients and
amputees. Paraplegia. 27:58–62. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Myers J, Lee M and Kiratli J:
Cardiovascular disease in spinal cord injury: an overview of
prevalence, risk, evaluation and management. Am J Phys Med Rehabil.
86:142–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Groah SL, Weitzenkamp DA, Lammertse DP,
Whiteneck GG, Lezotte DC and Hamman RF: Excess risk of bladder
cancer in spinal cord injury: evidence for an association between
indwelling catheter use and bladder cancer. Arch Phys Med Rehabil.
83:346–351. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalisvaart JF, Katsumi HK, Ronningen LD
and Hovey R: Bladder cancer in spinal cord injury patients. Spinal
Cord. 48:257–261. 2010. View Article : Google Scholar
|
|
16
|
Siebert JR, Middelton FA and Stelzner DJ:
Intrinsic response of thoracic propriospinal neurons to axotomy.
BMC Neurosci. 11:692010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai J, He X, Wang F, et al: Gene
expression signature analysis and protein-protein interaction
network construction of spinal cord injury. Eur Rev Med Pharmacol
Sci. 17:2941–2948. 2013.PubMed/NCBI
|
|
18
|
Jin L, Wu Z, Xu W, et al: Identifying gene
expression profile of spinal cord injury in rat by bioinformatics
strategy. Mol Biol Rep. 41:3169–3177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gentleman RC, Carey VJ, Bates DM, et al:
Bioconductor: open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smyth GK: Limma: Linear Models for
Microarray Data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R
and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
23
|
Warnes GR, Bolker B, Bonebakker L, et al:
gplots: Various R programming tools for plotting data. R package
version 2.7.4. 2009
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
27
|
Szklarczyk D, Franceschini A, Kuhn M, et
al: The STRING database in 2011: functional interaction networks of
proteins, globally integrated and scored. Nucleic Acids Res.
39(Database Issue): D561–D568. 2011. View Article : Google Scholar :
|
|
28
|
Shannon P, Markiel A, Ozier O, et al:
Cytoscape: a software environment for integrated models of
biomolecular interaction networks. Genome Res. 13:2498–2504. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matys V, Fricke E, Geffers R, et al:
TRANSFAC: transcriptional regulation, from patterns to profiles.
Nucleic Acids Res. 31:374–378. 2003. View Article : Google Scholar :
|
|
30
|
Courtine G, van den Brand R and Musienko
P: Spinal cord injury: Time to move. Lancet. 377:1896–1898. 2011.
View Article : Google Scholar
|
|
31
|
Othman A, Frim DM, Polak P, Vujicic S,
Arnason BG and Boullerne AI: Olig1 is expressed in human
oligodendrocytes during maturation and regeneration. Glia.
59:914–926. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arnett HA, Fancy SP, Alberta JA, et al:
bHLH transcription factor Olig1 is required to repair demyelinated
lesions in the CNS. Science. 306:2111–2115. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lindå H, Sköld MK and Ochsmann T:
Activating transcription factor 3, a useful marker for regenerative
response after nerve root injury. Front Neurol. 2:302011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Flatters S: ATF3: novel signpost for nerve
injury. Neuroreport. 11:A72000. View Article : Google Scholar
|
|
35
|
Koh IU, Lim JH, Joe MK, et al: AdipoR2 is
transcriptionally regulated by ER stress-inducible ATF3 in HepG2
human hepatocyte cells. Febs J. 277:2304–2317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakagomi S, Suzuki Y, Namikawa K,
Kiryu-Seo S and Kiyama H: Expression of the activating
transcription factor 3 prevents c-Jun N-terminal kinase-induced
neuronal death by promoting heat shock protein 27 expression and
Akt activation. J Neurosci. 23:5187–5196. 2003.PubMed/NCBI
|
|
37
|
Kohm AP, Carpentier PA, Anger HA and
Miller SD: Cutting edge: CD4+ CD25+ regulatory T cells suppress
antigen-specific autoreactive immune responses and central nervous
system inflammation during active experimental autoimmune
encephalomyelitis. J Immunol. 169:4712–4716. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liblau RS, Gonzalez-Dunia D, Wiendl H and
Zipp F: Neurons as targets for T cells in the nervous system.
Trends Neurosci. 36:315–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Natarajan R, Singal V, Benes R, et al:
STAT3 modulation to enhance motor neuron differentiation in human
neural stem cells. PLoS One. 9:e1004052014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ambruso DR, Knall C, Abell AN, et al:
Human neutrophil immunodeficiency syndrome is associated with an
inhibitory Rac2 mutation. Proc Natl Acad Sci USA. 97:4654–4659.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ziv Y, Avidan H, Pluchino S, Martino G and
Schwartz M: Synergy between immune cells and adult neural
stem/progenitor cells promotes functional recovery from spinal cord
injury. Proc Natl Acad Sci. 103:13174–13179. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
van den Berg ME, Castellote JM, de
Pedro-Cuesta J and Mahillo-Fernandez I: Survival after spinal cord
injury: A systematic review. J Neurotrauma. 27:1517–1528. 2010.
View Article : Google Scholar : PubMed/NCBI
|