Introduction
miRNA-206 is known as a muscle-specific miRNA, one
of the so-called myomiRs, and has been consistently found using
northern blotting, microarray, RNase protection and quantitative
polymerase chain reaction (qPCR) analyses to be specifically
expressed in skeletal muscle, but rarely detectable in other adult
tissues (1). Its primary
transcript, contained in a synapse-associated 7H4 non-coding RNA,
was first identified in the motor endplate of the rat skeletal
neuromuscular junction, long before the 'microRNA era' (2). Accumulated evidence has shown that
miR-206 is important in the growth and development of skeletal
muscle through targeting a number of genes, including gap junction
protein α-1 (3), polymerase (DNA
directed) α-1 (4),
follistatin-like 1 (5), utrophin
(5), paired box 7 (6) and histone deacetylase 4 (7). In a model of amyotrophic lateral
sclerosis, a novel pathway for neuromuscular synapse repair
regulated by miR-206 has been identified (8). This indicates that miR-206 may be a
key modulator in establishing muscular function through
coordinating innervation. A previous study reported that sonic
hedgehog signaling inhibited the expression of miR-206, which
increased the protein levels of brain-derived neurotrophic factor
(BDNF), an essential signal for airway smooth muscle innervation
(9). This further supports the
neuronal function of miR-206 in a muscular context.
The expression of miR-206 has also been detected at
significant levels in adult mouse skin using RNase protection
assays (3); however, no further
investigation of miR-206 in skin has been reported. Another two
reports demonstrated that miR-206 is downregulated in human
melanoma biopsies (10) and in
skin from human papillomavirus 8-transgenic mice (11). Our previous study found that the
mouse homolog of hsa-miR-206, miR-206-3p, was often expressed in
embryonic skin at a level, which was one quarter of the level in
skeletal muscle (12). Several
other skin-specific or skin-associated miRNAs have been identified
(13,14) and, in comparison, miR-206 is
expressed at relatively low levels in adult skin. However, its
expression profile in embryonic skin remains to be fully
elucidated. In a previous screen of differentiating keratinocyte
(KC) miRNAs, miR-206 was not identified (14), although it had muscular and
neuronal functions in muscle. Therefore, the present study
hypothesized that miR-206 is involved in skin development, possibly
through an association with neurofunction. To confirm this
hypothesis, the present study investigated the spatial and temporal
expression of miR-206-3p and its target gene, Bdnf, during
mouse skin development, and determined whether the dynamic
expression of miR-206-3p is involved in the KC differentiation
program.
Materials and methods
Mice and tissue samples
Wild-type BALB/c mice were bred and maintained in
accordance with the Principles of Laboratory Animal Care at the
Laboratory Animal Center of Jiangsu University (Zhenjiang, China;
License no. SYXK [SU] 2008-0024) in a temperature and
humidity-controlled room maintained in a 12 h light/dark cycle with
food and water ad libitum. All animal experiments in the
present study were approved by the ethics committee of Jiangsu
University. Mature (6–12 week-old) female mice were housed
separately until natural mating with a 2:1 female to male ratio,
and were assessed for vaginal plug formation on the morning
following mating. Females with vaginal plugs were designated as 0.5
day postcoitum (dpc), and neonatal mice on the day of delivery were
designated as 1 day postpartum (dpp). Full-thickness skin and
subcutaneous muscle were dissected for cryosections and
TRIzol® homogenization (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), respectively. Pregnant females
at various stages of gestation were intraperitoneally anesthetized
with 1% pentobarbital sodium (35–50 mg/kg; Sigma-Aldrich, St.
Louis, MO, USA) and embryos were collected by cesarean section. A
skin tissue sample of 3 × 3 mm and a 3 mm3 subcutaneous
skeletal muscle tissue sample were dissected from the back of the
embryos of at least three different mothers at each time-point.
Following tissue harvest, the anesthetized mice were sacrificed by
cervical dislocation. The harvested tissues were snap-frozen in
liquid nitrogen prior to being stored at −80°C.
Cryosections
The frozen tissues were embedded in
Tissue-Tek® OCT™ (Sakura Finetek, Tokyo, Japan) and cut
at a thickness of 8–10 µm on a CM1900 freezing microtome
(Leica Microsystems GmbH, Wetzlar, Germany). The cryosections were
mounted on poly-L-lysine coated slides (Wuhan Boster Biological
Technology, Ltd., Wuhan, China), following which the slides were
air-dried for 10 min, fixed for 10 min with 4% paraformaldehyde
(Sangon Biotech Co., Ltd., Shanghai, China), rinsed with Dulbecco's
phosphate-buffered saline (PBS) and stored at −20°C following
thorough drying.
In situ hybridization (ISH)
The frozen sections were air-dried for ~15 min,
following which hybridization was performed using an Enhanced
Sensitive ISH Detection kit (cat. no. MK1032; Wuhan Boster
Biological Technology, Ltd.), according to the manufacturer's
protocol. Briefly, the sections were treated with pepsin supplied
in the Enhanced Sensitive ISH Detection kit at 37°C for 5 min,
washed with 0.5 M Tris-buffered salin (TBS; Sangon Biotech Co.,
Ltd.) and then 0.1% diethylpyrocarbonate-treated water (Sangon
Biotech Co., Ltd.), pre-hybridized at 40°C for 1 h, and then
hybridized with 100 nM 5′-digoxigenin (DIG)-labeled LNA™ probe
(Exiqon A/S, Vedbaek, Denmark) against miR-206 at 50°C for 8 h.
Following hybridization, the slides were washed with 2× saline
sodium citrate (SSC; Sangon Biotech Co., Ltd.), 0.5× SSC, 0.2× SSC,
sequentially. Subsequently, the slides were blocked using the
blocking solution supplied in the Enhanced Sensitive ISH Detection
kit for 30 min at 37°C, and incubated with biotinylated anti-DIG
antibody (supplied in the Enhanced Sensitive ISH Detection kit) for
2 h at 37°C. Following washing with 0.5 M TBS, alkaline phosphatase
(AP)-conjugated strep-tavidin (supplied in the Enhanced Sensitive
ISH Detection kit) was applied to the section for 1 h at room
temperature. Immediately following washing of the slides, AP
substrate, containing 0.2 mM levamisol, was prepared and then
applied to the sections in the dark at 30°C for 2 h. When the
desired intensity of chromogenic reaction was reached (strongly
stained target and unstained background), the slides were washed
with water, followed by mounting in aqueous medium and imaging on a
DM LB2 microscope (Leica Microystems GmbH).
Immunohistochemistry (IHC)
The frozen sections were air-dried and incubated for
heat-induced antigen retrieval in 0.01 M citrate buffer (pH 6.0;
Sangon Biotech Co., Ltd.) at 100°C for 10 min. Following cooling to
room temperature, 0.6% H2O2/80% methanol was
applied to the sections for 10 min at room temperature to eliminate
endogenous peroxidation. Following washing with D-PBS, the sections
were blocked with 10% bovine serum albumin (Sangon Biotech Co.,
Ltd.) and 0.2% Triton X-100 (Sangon Biotech Co., Ltd.) in D-PBS for
30 min at room temperature, prior to incubation with anti-mouse
BDNF rabbit polyclonal antibody (cat. no. BA0565; Wuhan Boster
Biological Technology, Ltd.) diluted 100-folds or anti-mouse
ubiquitin carboxy-terminal hydrolase L1 (UCHL1) rabbit polyclonal
antibody (cat. no. BS1293; Bioworld Technology, Inc. St. Louis
Park, MN, USA) diluted 80-folds at 4°C overnight. Following washing
with PBS with 0.1% Tween 20 (PBS-T; Sangon Biotech Co., Ltd.), the
sections were added with horseradish peroxidase (HRP)-conjugated
goat anti-rabbit IgG antibody (cat. no. BA1055; Wuhan Boster
Biological Technology, Ltd.) diluted 500-fold for 2 h at room
temperature. Following washing with PBST, 1%
H2O2 was diluted 100-fold with 0.05%
diaminobenzidine (Sangon Biotech Co., Ltd.) in TBS to prepare an
active substrate solution, which was then applied to the sections
for color development. The reaction was terminated by washing,
following which the sections were counterstained with hematoxylin,
dehydrated with alcohol, mounted with neutral balsam and imaged on
a DM LB2 microscope (Leica Microsystems GmbH).
Cell culture and in vitro
differentiation
To determine cell differentiation, three independent
back skin biopsies (5 mm × 2 cm) from 1 dpp mouse were used to
isolate epidermal keratinocytes. Firstly, the skin biopsies were
washed three times in D-PBS containing high-concentration
antibiotics (200 U/ml penicillin and 200 U/ml streptomycin; Sangon
Biotech Co., Ltd.) for 5 min, and subcutaneous residues were
curetted. Secondly, the epidermis of each skin biopsy was separated
from the dermal compartment using 0.3% Dispase® II
(Roche Diagnostics GmbH, Mannheim, German) digestion overnight at
4°C. Subsequently, the epidermal sheets were trypsinized with 0.25%
trypsin-EDTA (Invitrogen; Thermo Fisher Scientific, Inc.) for 10
min at 37°C, followed by the addition of 0.1% soybean trypsin
inhibitor (Sigma-Aldrich) at a 1:1 volume ratio to trypsin-EDTA,
and were subsequently filtered to yield single cell suspensions.
The keratinocytes were cultured in defined keratinocyte-serum-free
medium (Invitrogen; Thermo Fisher Scientific, Inc.), and seeded at
a density of 105/cm2 on 2% gelatin coated
dishes at 37°C and 5% CO2 to near confluency.
For in vitro differentiation, the primary
keratinocytes cultured under standard conditions to ~70% confluency
were induced by increasing the calcium concentration of the growth
medium to 1 mM for 5 days.
RNA extraction and reverse transcription
(RT)-qPCR
Total RNA extraction was performed according to the
manufacturer's protocol. Total RNA was extracted using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.);
prior to the addition of 0.2 ml chloroform (Sangon Biotech Co.,
Ltd.) per 1 ml TRIzol®. The samples were then
centrifuged at 12,000 × g for 15 min at 4°C, and the aqueous phase
was then placed into another tube with an equal volume of 100%
isopropanol (Sangon Biotech Co., Ltd.). Precipitation was conducted
at −20°C overnight as opposed to at room temperature for 10 min as
previously described (15), and
then centrifuged at 12,000 × g for 10 min at 4°C. The RNA pellet
was washed twice with 1 ml of 80% ethanol (Sangon Biotech Co.,
Ltd.) at 7,500 × g for 5 min at 4°C, prior to being air dried for
5–10 min, and dissolved in RNase-free water (Sangon Biotech Co.,
Ltd.). RNA concentration was measured using a NanoDrop 1000 UV/Vis
spectrophotometer (Thermo Fisher Scientific, Inc.). Prior to the RT
reaction, the RNAs were treated with DNase (NEB, Ipswich, MA, USA),
according to the manufacturer's protocol.
For amplification of β-actin (Actb) and Bdnf, cDNAs
were generated using a ReverTra Ace-α-® Reverse
Transcription kit (Toboyo Co., Ltd., Osaka, Japan). According to
the manufacturer's instructions, the reaction was performed using
500 ng total RNA in a 10 µl total volume, as follows: 30°C
for 10 min, 42°C for 30 min and 99°C for 5 min, and stored at
−20°C. The miR-206-3p and Snord68 primers for RT and qPCR were
designed according to a previous report (12). A pulsed gene-specific RT reaction
(16) in a 10 µl total
volume containing 500 ng RNA and stem-loop primer (4 nM for
miR-206-3p, 1 nM for Snord68) was applied, as follows: 16°C for 30
min, followed by 60 cycles at 20°C for 30 sec, 42°C for 30 sec and
50°C for 1 sec, with a final step at 99°C for 5 min and storage at
−20°C.
A DyNAmo™ ColorFlash SYBR® Green qPCR kit
(Thermo Fisher Scientific, Inc.) was used in a modified protocol
regarding the volumes of reagents and cDNA in the 10 µl
total volume: 5 µl 2X master mix, 1 µl RT product,
forward and reverse primer pairs (180 nM for Actb, Bdnf and
miR-206-3p; 100 nM for Snord68). A three-step qPCR was performed
for Actb and Bdnf, as follows: 95°C for 7 min; 40 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 20 sec and
extension at 72°C for 15 sec. A two-step qPCR was performed for
Snord68 and miR-206-3p, as follows: 95°C for 7 min; 40 cycles of
denaturation at 95°C for 10 sec and extension (Snord68 at 66°C;
miR-206-3p at 59°C) for 30 sec. All primers (Table I) were synthesized by Sangon
Biotech Co., Ltd.
 | Table IPrimers used in the RT-qPCR assay. |
Table I
Primers used in the RT-qPCR assay.
| Target | Accession no. | Reaction
(RT/qPCR) | Primer sequence
(5′-3′) | Amplicon size
(bp) |
|---|
| Snord68 | NR_028128.1 | RT |
CTCAACTGGTGTCGTGGAGTCGGCAA | 93 |
| | |
TTCAGTTGAGCATCAGAT | |
| | qPCR | Forward:
ACGACTAGGGCTGTACTGACTTGATG | |
| | qPCR | Reverse:
CTCAACTGGTGTCGTGGAGTCGG | |
| miR-206-3p | MIMAT0000239 | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTC
AGTTGAGCCACACAC | 71 |
| | qPCR | Forward:
AGCTCGATTAAGGTGGAATGTAAGGAAGT | |
| | qPCR | Reverse:
CTCAACTGGTGTCGTGGAGTCGG | |
| Actb | NM_007393.3 | qPCR | Forward:
GGCTGTATTCCCCTCCATCG | 154 |
| | qPCR | Reverse:
CCAGTTGGTAACAATGCCATGT | |
| Bdnf | NM_007540.4 | qPCR | Forward:
TCATACTTCGGTTGCATGAAGGC | 145 |
| | qPCR | Reverse:
TCGTCGTCAGACCTCTCGAAC | |
All RT-qPCR reactions, including the RT minus
controls and no-template controls, were run in triplicate on an
CFX™ 96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The quantification cycle (Cq), defined as the
fractional cycle number at which the fluorescence passes the fixed
threshold, was determined using CFX Manager™ Software version 1.6
(Bio-Rad Laboratories, Inc.). The expression levels of miR-206-3p
and Bdnf were calculated using the ∆Cq method (17) using Snord68 and Actb as a
reference, respectively.
Western blotting
Following removal of the RNA aqueous phase from the
TRIzol® homogenate, the remaining phenol-chloroform
layer was used for protein isolation, according to manufacturer's
protocol. A total of 1.5 ml isopropanol was added per of 1 ml
TRIzol® for 10 min at room temperature, prior to being
centrifuged at 12,000 × g for 10 min at 4°C. The protein pellet was
washed three times with 2 ml of 0.3 M guanidine hydrochloride
(Sangon Biotech Co., Ltd.) in 95% ethanol at 7,500 × g for 5 min at
4°C, and once with 2 ml of 100% ethanol. The protein pellet was
subsequently air dried for 5–10 min, and dissolved in 0.5% SDS/3 M
urea (Sangon Biotech Co., Ltd.). The proteins were quantified using
a bicinchoninic acid assay kit (CWBio, Beijing, China).
Subsequently, the samples were denatured at 95°C for 5 min, and 30
µg proteins per lane were separated using 12% SDS-PAGE and
transferred onto Immobilon®-P membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked in 5% defatted milk
for 1 h at room temperature, and were then probed with anti-mouse
ACTB rabbit polyclonal antibody (cat. no. 20536-1-AP; ProteinTech,
USA) diluted 2,000-fold, anti-BDNF rabbit polyclonal antibody (cat.
no. PB0013; Wuhan Boster Biological Technology, Ltd.) diluted
250-fold and rabbit anti-mouse involucrin (IVL) polyclonal antibody
(cat. no. 55328-1-AP; ProteinTech, USA) diluted 300-fold,
respectively, at 4°C overnight. Following washing with PBST, the
membranes were then incubated with HRP-conjugated goat anti-rabbit
IgG antibody (cat. no. BA1055; Wuhan Boster Biological Technology,
Ltd.) diluted 4,000-fold for 1 h at room temperature. Following
washing, the immunolabeling proteins were reacted with
chemiluminescent HRP substrate (EMD Millipore) and visualized using
a ChemiDoc™ XRS system (Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analyses were performed using Stata/SE
11.2 for Windows (StataCorp, College Station, TX, USA). Where
appropriate, data are presented as the mean ± standard deviation of
at least three independent samples. Two sample's mean comparison
was performed using Student's t-test. One-way analysis of variance
was used to determine differences among at least three groups, and
a Bonferroni multiple comparison was performed to test variances
within groups. Two-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-206-3p and BDNF during
mouse skin development
The present study first examined the expression
levels of miR-206-3p in full-thickness skin at different
developmental phases. The RT-qPCR analysis indicated that the
expression of miR-206-3p was low at 13.5 dpc, increased gradually
by ~4.5-fold up to 17.5 dpc (13.5 dpc, vs. 17.5 dpc: P<0.001),
and subsequently decreased to a level similar to that observed at
13.5 dpc (13.5 dpc, vs. 16 weeks: P=1.000; Fig. 1A). The mRNA expression level of
Bdnf decreased until 17.5 dpc (13.5 dpc, vs. 17.5 dpc:
P=0.001) and remained at a steady level (Fig. 1B). The change in the level of BDNF
precursor (pro-BDNF) was inversely correlated with that of
miR-206-3p (Fig. 1C and D). The
anti-BDNF antibody used in the present study was raised against a
peptide mapping at the N-terminal end of the mature form of BDNF;
therefore, it reacted with multiple forms of BDNF, including
pro-BDNF and a minor truncated form of the pro-BDNF (28 kDa
(18) in the adult skin (Fig. 1D). The temporal expression pattern
of mature BDNF closely resembled that of its precursor, pro-BDNF
(Fig. 1C). The levels of
miR-206-3p expressed in the muscle tissue was at a comparable level
to that in the skin at 13.5 dpc; however, its level increased in
the muscle by >100-fold during the neonatal period (13.5 dpc,
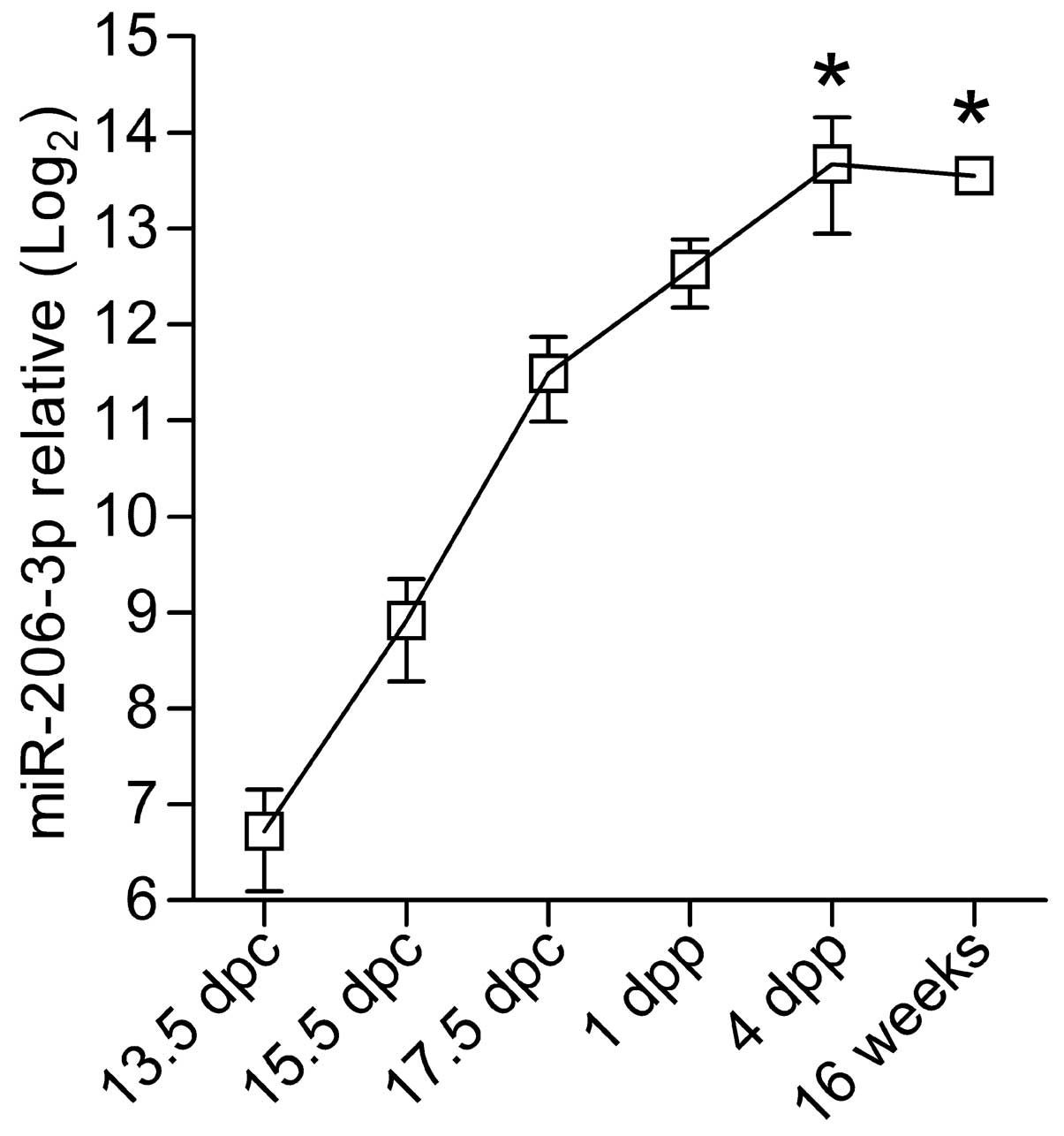
vs. 16 weeks: P=0.001; Fig. 2),
confirming the muscle-specificity of miR-206-3p.
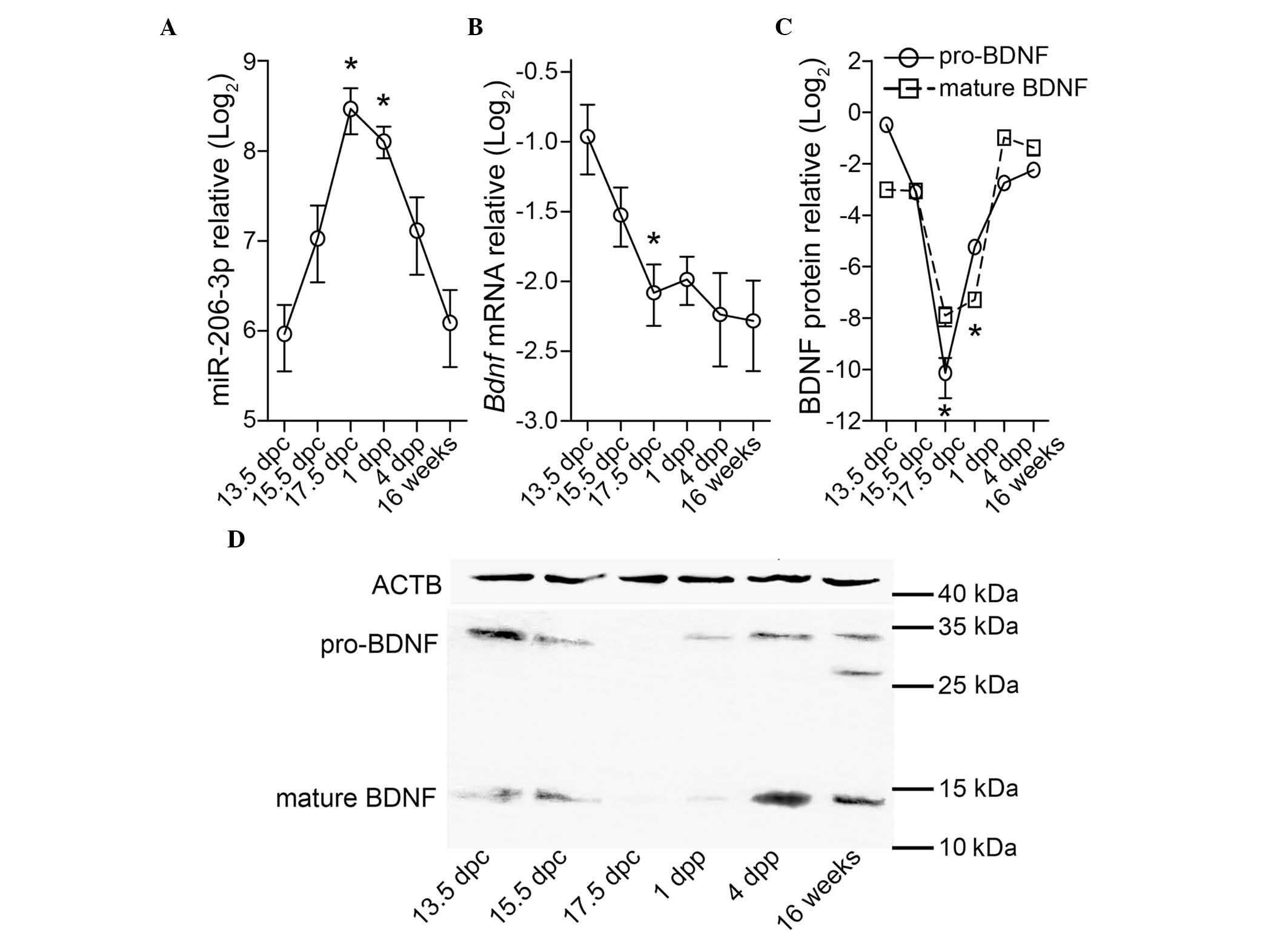 | Figure 1Full-thickness skin tissue from the
backs of wide-type BALB/c mice at 13.5 dpc, 15.5 dpc, 17.5 dpc, 1
dpp, 4 dpp and 16 weeks postpartum were measured for the expression
of (A) miR-206-3p, normalized to Snord68 and the (B) mRNA
expression of Bdnf, normalized to Actb using reverse
transcription-quantitative polymerase chain reaction analyses. (C
and D) Protein expression of BDNF, normalized to ACTB was
determined using western blotting. All relative expression data are
plotted in Log2 scale using the mean standard deviation
from three independent samples. *P<0.05, vs. 13.5
dpc. dpc, days postcoitum; dpp, days postpartum; miR, microRNA;
BDNF, brain-derived neurotrophic factor; ACTB; β-actin. |
Due to its decline in expression following birth,
miR-206-3p was not be considered skin-specific, as tissue-specific
miRNAs have been defined to be expressed at a level >20-fold
higher in a specific tissue, compared with the mean level of
expression in all other tissues (19). However, as its expression level was
inversely correlated with BDNF in the present study, the tissue
distribution of miR-206-3p and BDNF were assessed. ISH showed that
miR-206-3p was regionalized during skin development. The expression
of miR-206-3p was widespread at 13.5 dpc, and was evident in the
suprabasal layer between 17.5 dpc and 1 dpp, following which it was
confined to the epidermal tissues at 4 dpp. The expression of
miR-206-3p was weakened in the adult (Fig. 3). By contrast, BDNF immuno-labeling
was scattered and weak at 13.5 dpc, and was barely visible at 1
dpp. Subsequently, it was expressed in the supra-basal layer and
inside hair follicles, where its expression was located adjacent
to, but not overlapping, that of miR-206-3p.
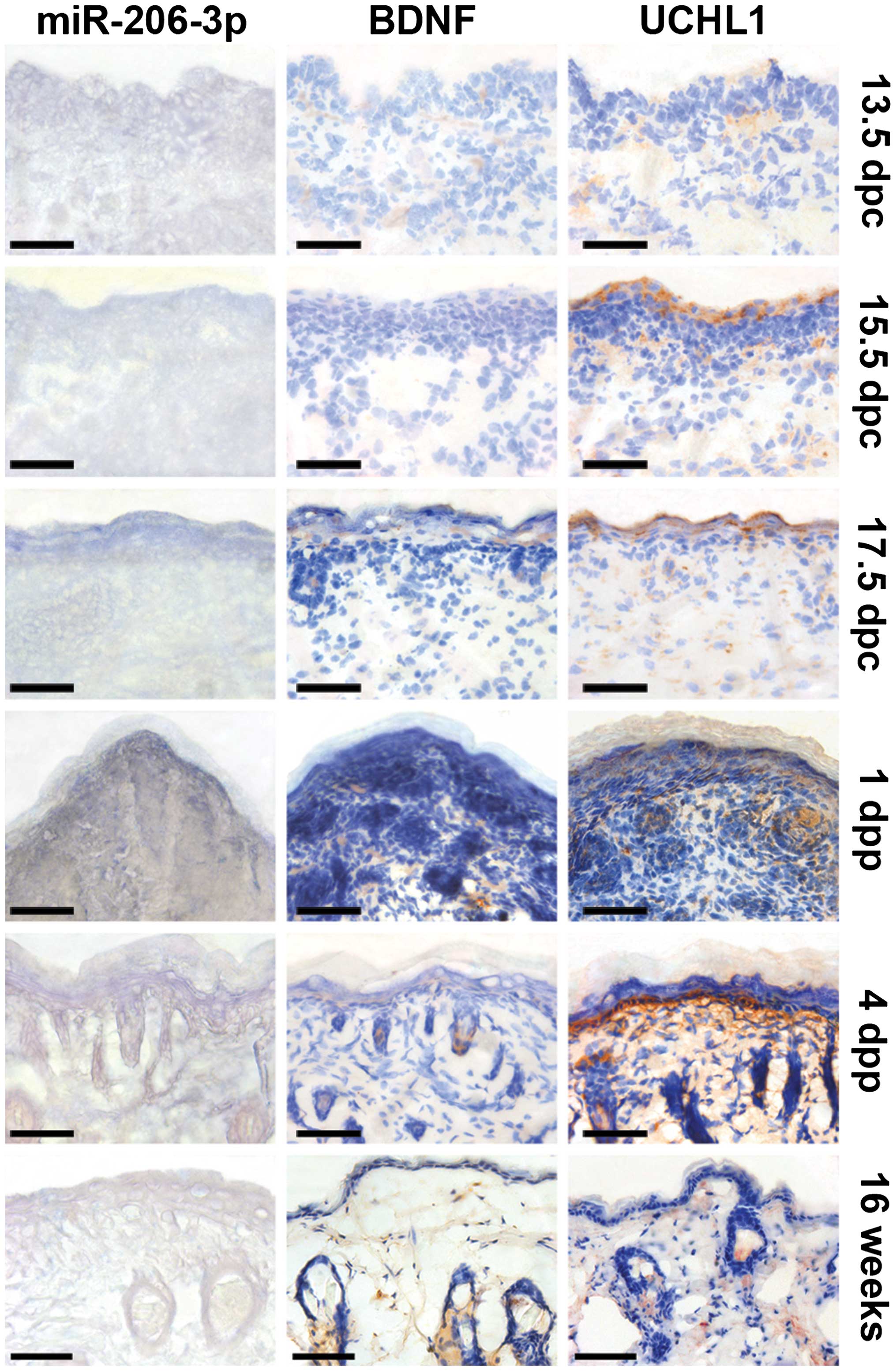 | Figure 3Tissue cryosections of skin from the
backs of wide-type BALB/c mice were examined for the expression of
miR-206-3p by in situ hybridization with 4-nitro-blue
tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate staining (dark
blue). The expression levels of BDNF and UCHL1 were examined using
immunohistochemistry with diaminobenzidine staining (brown) and
counterstained with hematoxylin (blue). Tissues were examined at
13.5 dpc, 15.5 dpc, 17.5 dpc, 1 dpp, 4 dpp and 16 weeks postpartum.
The expression of miR-206-3p was widespread at 13.5 dpc, and was
evident in the suprabasal layer between 17.5 dpc and 1 dpp,
following which it was confined to the epidermal tissues at 4 dpp,
which was located near BDNF and UCHL1. Scale bar=50 µm. dpc,
days postcoitum; dpp, days postpartum; miR, microRNA; BDNF,
brain-derived neurotrophic factor; UCHL1, ubiquitin
carboxy-terminal hydrolase L1. |
The pan-neuronal marker, UCHL1, also termed PGP9.5,
revealed a pattern of cutaneous innervation made by nerve fibers.
The nerve fibers innervated and reached the epidermal surface at
high density by 17.5 dpc, however they subsequently became
predominantly subepidermal, which resembled the distribution of
miR-206-3p, until 1 dpp. Following this, the localizations of UCHL1
and miR-206-3p were separate, but remained near each other
(Fig. 3).
Expression of miR-206-3p and BDNF during
keratinocyte differentiation in vitro
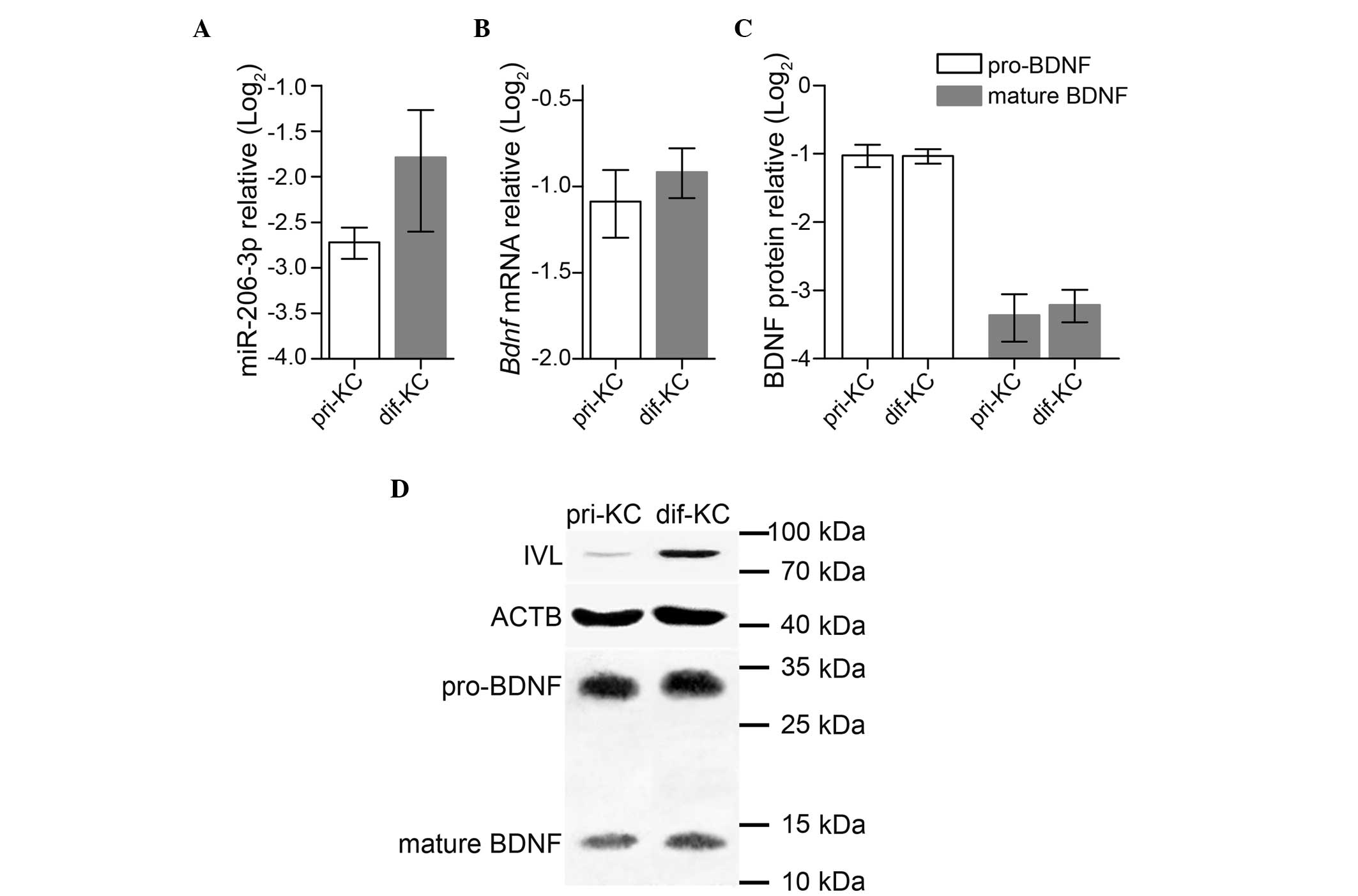
To determine whether the expression of miR-206-3p
was dependent on epidermal differentiation, in vitro
calcium-induced differentiation of primary keratinocytes was
performed. No significant differences were found in the expression
of either miR-206-3p (P=0.227) or Bdnf (P=0.118); mature
BDNF (P=0.106) or pro-BDNF (P=0.905) between the primary and
differentiated keratinocytes (Fig.
4A–C), suggesting that they were not involved in keratinocyte
differentiation. However, the elevated expression of IVL in the
differentiated keratinocytes verified the in vitro
differentiation model (Fig.
4D).
Discussion
During mouse development, miR-206-3p is first
detected at low levels, as early as 9.5 dpc, using northern
blotting and the cloning frequency of the whole embryo (20). It is then primarily restricted to
skeletal muscle in the adult (21). miR-206-3p has also been detected in
adult skin (3,10,11),
where it is expressed at a quarter of the level expressed in 12.5
dpc skeletal muscle (12). For the
first time, to the best of our knowledge, the present study
demonstrated the spatiotemporal expression of miR-206-3p in skin.
The results of the present study revealed that miR-206-3p was
expressed dynamically during mouse skin development, with its level
increasing from 13.5 dpc, peaking at 17.5 dpc and declining
following birth. The fluctuation in the expression of miR-206-3p
was accompanied by an inverse change in the protein level of its
Bdnf target gene However, the mRNA expression levels of
Bdnf did not parallel with its protein expression levels.
The tissue distribution of miR-206-3p was similar or located
adjacent to that of UCHL1 during skin development, suggesting the
potential involvement of miR-206-3p in cutaneous innervation.
It is commonly observed that the majority of miRNAs
are not essential for tissue establishment, but are important for
late tissue differentiation and maintenance (22); however, the declining expression
level of miR-206-3p in postnatal skin in the present study
suggested that it may not be associated with cutaneous maturation.
In addition, the pattern of miR-206-3p distribution, which did not
indicate skin-specificity, also supports this assumption. The in
vitro keratinocyte model demonstrated that miR-206-3p was
independent of keratinocyte differentiation. These findings,
together with previous evidence indicating miR-206-3p involvement
in muscle innervation (2,9), led the present study to investigate
the possible role of miR-206-3p during skin innervation. A previous
study clearly demonstrated that Bdnf is directly suppressed
by miR-206 during myogenic differentiation, underlining a
retrograde regulatory role of miR-206 at the neuromuscular junction
(23). Of note, BDNF is capable of
mediating neuronal differentiation and growth, synapse formation
and plasticity, and higher cognitive functions in the mammalian
brain (24). It has been shown
that keratinocyte-derived BDNF is essential for the innervation of
hair follicles and development of Meissner corpuscles (25,26).
Therefore, the bell-shaped expression pattern of miR-206-3p during
skin development is likely attributed to two consecutive and
overlapping processes of innervation: i) all sensory terminals
transiently hyperinnervate the skin and penetrate to the epidermal
surface prior to retracting subepidermally at a late embryonic
stage (27); ii) the hair follicle
keratinocytes begin to concentrate in newly developed follicular
innervation sites when intra-epidermal nerve fiber endings are
present (28), during which
miR-206-3p may exert its neuromodulation through the
post-transcriptional suppression of Bdnf. Although the
production of mature BDNF was almost parallel to that of its
precursor during this time frame, its level of expression was lower
at 13.5 dpc and higher in the adult stage, compared with that of
pro-BDNF. This suggested that other mechanisms, including
post-translational processing or endocytic uptake, may involved in
regulating the level of mature BDNF, which is available for the
innervation and early postnatal survival of cutaneous sensory
organs (25,29).
The results of the present study suggested that
upregulation in the expression of miR-206-3p at late embryonic
stages suppressed the expression of BDNF and induced the
hyperinnervated fibers to retract. The subsequent decline in the
expression levels of miR-206-3p then enabled the levels of BDNF to
increase to meet the requirements of the newly developed follicular
innervations. In conclusion, these findings indicated a potential
role of miR-206-3p in cutaneous innervation, which is largely
mediated by the neurotrophic support of BDNF. These findings may
revise current understanding of this muscle-specific miRNA, and
adds support to the possibility that miRNAs have functions in
addition to those, which are predominant. Cutaneous innervation
requires examination from the perspective of miRNAs, and this
mechanism may be amenable for the development of possible
therapeutic approaches.
Acknowledgments
This study was partially supported by a grant from
the Postgraduate's Innovation Project of Jiangsu Province, China to
Dr Yuan Mu (grant. no. CXLX11_0606).
References
|
1
|
McCarthy JJ: MicroRNA-206: The skeletal
muscle-specific myomiR. Biochim Biophys Acta. 1779:682–691. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Velleca MA, Wallace MC and Merlie JP: A
novel synapse-associated noncoding RNA. Mol Cell Biol.
14:7095–7104. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anderson C, Catoe H and Werner R: MIR-206
regulates connexin43 expression during skeletal muscle development.
Nucleic Acids Res. 34:5863–5871. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim HK, Lee YS, Sivaprasad U, Malhotra A
and Dutta A: Muscle-specific microRNA miR-206 promotes muscle
differentiation. J Cell Biol. 174:677–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosenberg MI, Georges SA, Asawachaicharn
A, Analau E and Tapscott SJ: MyoD inhibits Fstl1 and Utrn
expression by inducing transcription of miR-206. J Cell Biol.
175:77–85. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao
X and Wang DZ: MicroRNA-1 and microRNA-206 regulate skeletal muscle
satellite cell proliferation and differentiation by repressing
Pax7. J Cell Biol. 190:867–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winbanks CE, Wang B, Beyer C, Koh P, White
L, Kantharidis P and Gregorevic P: TGF-beta regulates miR-206 and
miR-29 to control myogenic differentiation through regulation of
HDAC4. J Biol Chem. 286:13805–13814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Williams AH, Valdez G, Moresi V, Qi X,
McAnally J, Elliott JL, Bassel-Duby R, Sanes JR and Olson EN:
MicroRNA-206 delays ALS progression and promotes regeneration of
neuromuscular synapses in mice. Science. 326:1549–1554. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Radzikinas K, Aven L, Jiang Z, Tran T,
Paez-Cortez J, Boppidi K, Lu J, Fine A and Ai X: A Shh/miR-206/BDNF
cascade coordinates innervation and formation of airway smooth
muscle. J Neurosci. 31:15407–15415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hufbauer M, Lazić D, Reinartz M, Akgül B,
Pfister H and Weissenborn SJ: Skin tumor formation in human
papillomavirus 8 transgenic mice is associated with a deregulation
of oncogenic miRNAs and their tumor suppressive targets. J Dermatol
Sci. 64:7–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Georgantas RW III, Streicher K, Luo X,
Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW,
Richman L, et al: MicroRNA-206 induces G1 arrest in melanoma by
inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res.
27:275–286. 2014. View Article : Google Scholar
|
|
12
|
Mu Y, Zhou H, Li W, Hu L and Zhang Y:
Evaluation of RNA quality in fixed and unembedded mouse embryos by
different methods. Exp Mol Pathol. 95:206–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yi R, Poy MN, Stoffel M and Fuchs E: A
skin microRNA promotes differentiation by repressing 'stemness'.
Nature. 452:225–229. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hildebrand J, Rütze M, Walz N, Gallinat S,
Wenck H, Deppert W, Grundhoff A and Knott A: A comprehensive
analysis of microRNA expression during human keratinocyte
differentiation in vitro and in vivo. J Invest Dermatol. 131:20–29.
2011. View Article : Google Scholar
|
|
15
|
Wang WX, Wilfred BR, Baldwin DA, Isett RB,
Ren N, Stromberg A and Nelson PT: Focus on RNA isolation: Obtaining
RNA for microRNA (miRNA) expression profiling analyses of neural
tissue. Biochim Biophys Acta. 1779:749–757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang F, Hayashi K, Kaneda M, Lao K and
Surani MA: A sensitive multiplex assay for piRNA expression.
Biochem Biophys Res Commun. 369:1190–1194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Mowla SJ, Farhadi HF, Pareek S, Atwal JK,
Morris SJ, Seidah NG and Murphy RA: Biosynthesis and
post-translational processing of the precursor to brain-derived
neurotrophic factor. J Biol Chem. 276:12660–12666. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee EJ, Baek M, Gusev Y, Brackett DJ,
Nuovo GJ and Schmittgen TD: Systematic evaluation of microRNA
processing patterns in tissues, cell lines and tumors. RNA.
14:35–42. 2008. View Article : Google Scholar :
|
|
20
|
Takada S, Berezikov E, Yamashita Y,
Lagos-Quintana M, Kloosterman WP, Enomoto M, Hatanaka H, Fujiwara
S, Watanabe H, Soda M, et al: Mouse microRNA profiles determined
with a new and sensitive cloning method. Nucleic Acids Res.
34:e1152006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sempere LF, Freemantle S, Pitha-Rowe I,
Moss E, Dmitrovsky E and Ambros V: Expression profiling of
mammalian microRNAs uncovers a subset of brain-expressed microRNAs
with possible roles in murine and human neuronal differentiation.
Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wienholds E and Plasterk RH: MicroRNA
function in animal development. FEBS Lett. 579:5911–5922. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miura P, Amirouche A, Clow C, Bélanger G
and Jasmin BJ: Brain-derived neurotrophic factor expression is
repressed during myogenic differentiation by miR-206. J Neurochem.
120:230–238. 2012. View Article : Google Scholar
|
|
24
|
Park H and Poo MM: Neurotrophin regulation
of neural circuit development and function. Nat Rev Neurosci.
14:7–23. 2013. View
Article : Google Scholar
|
|
25
|
LeMaster AM, Krimm RF, Davis BM, Noel T,
Forbes ME, Johnson JE and Albers KM: Overexpression of
brain-derived neurotrophic factor enhances sensory innervation and
selectively increases neuron number. J Neurosci. 19:5919–5931.
1999.PubMed/NCBI
|
|
26
|
González-Martínez T, Fariñas I, Del Valle
ME, Feito J, Germanà G, Cobo J and Vega JA: BDNF, but not NT-4, is
necessary for normal development of Meissner corpuscles. Neurosci
Lett. 377:12–15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jackman A and Fitzgerald M: Development of
peripheral hindlimb and central spinal cord innervation by
subpopulations of dorsal root ganglion cells in the embryonic rat.
J Comp Neurol. 418:281–298. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peters EM, Botchkarev VA, Müller-Röver S,
Moll I, Rice FL and Paus R: Developmental timing of hair follicle
and dorsal skin innervation in mice. J Comp Neurol. 448:28–52.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Valdés-Sánchez T, Kirstein M,
Pérez-Villalba A, Vega JA and Fariñas I: BDNF is essentially
required for the early postnatal survival of nociceptors. Dev Biol.
339:465–476. 2010. View Article : Google Scholar : PubMed/NCBI
|