Introduction
Developmental speech and language disorders are
highly heritable, with complex multifactorial inheritance in the
majority of cases (1). A
heterozygous mutation of the forkhead-box protein P2 (FOXP2)
gene causes a rare Mendelian speech and language disorder, and was
the first speech and language disorder-associated gene to be
identified (2). The FOXP2
gene (OMIM, 605317), located on 7q31, encodes a transcription
factor that contains a polyglutamine (poly-Q) tract and a forkhead
DNA binding domain (2). Previous
studies have reported that point mutations and chromosomal
abnormalities of FOXP2 are associated with speech and
language disorders, which are characterized by difficulties with
the learning and production of sequences of oral movements
(3–7).
Speech sound disorder (SSD) is the prevalent speech
disorder in preschool children, and individuals with SSD have
difficulty producing speech sounds appropriate for their age and
dialect. In our previous study on a Chinese Han population, it was
observed that FOXP2 may confer vulnerability to SSD
(8). Deletion of the heterozygous
triplet codon, CAA in exon 5 of FOXP2 resulted in five
probands presenting with SSD. There are 40 poly-Q tracts encoded by
(CAG)4 CAA (CAG)4 (CAA)2 (CAG)2 (CAA)2 (CAG)3 (CAA)5 (CAG)2 (CAA)2
(CAG)5 CAA (CAG)5 CAA CAG within exon 5. This same heterozygous CAA
deletion (c.520delCAA, p.Q174del) was present in five probands, but
absent in normal control subjects. The entire mutated sequence
(mutation site italicized) is as follows: (CAG)4 CAA (CAG)4 (CAA)2
(CAG)2 (CAA)2 (CAG)3 (CAA)4 (CAG)2 (CAA)2 (CAG)5 CAA (CAG)5
CAA CAG. The amino acid, glutamine is encoded by CAG and CAA
codons, and the c.520delCAA mutation is predicted to result in a
reduction in the length of the poly-Q tract from 40 to 39 residues.
Butland et al (9)
investigated the length of CAG-poly-Q tracts in the entire human
genome in a sample of healthy individuals, and observed that
FOXP2 has the longest mean and maximum poly-Q tract length
with relatively little variance. Previous studies have demonstrated
that the length of CAG-only tracts is correlated with Q-tract
variance (10–12) and interruptions in CAG codons may
stabilize highly repetitive poly-Q tracts (10,13).
Thus, FOXP2 may have increased stability and be less polymorphic
with a shorter CAG repeat that is interrupted by CAA codons.
The expansion of poly-Q-encoding CAG codon repeats
has been identified as pathogenic in numerous genes associated with
neurodegenerative disorders, including the HD-associated huntingtin
gene in Huntington's disease (14). However, the clinical significance
of reductions in poly-Q tracts is unclear. Vernes et al
(15) demonstrated that FOXP2, a
transcription factor expressed in neurons, binds to and markedly
downregulates the expression of contactin-associated protein-like 2
(CNTNAP2). The CNTNAP2 gene, located on 7q35–36.1,
encodes contactin-associated protein 2 (CASPR2), which belongs to
the neurexin ion transporter family (16) and is highly expressed in
language-associated circuits of the brain (17). CNTNAP2 has been implicated
in multiple neurodevelopmental disorders, including autism
(18–23), and may affect early language
acquisition in normal individuals (24). However, to the best of our
knowledge, the potential association between CNTNAP2 and SSD
has not been investigated. In the present study, the
transcriptional regulation of poly-Q reduction mutations of
FOXP2 on CNTNAP2 was investigated.
Materials and methods
Vector construction
The GV287 lentiviral expression system containing
the green fluorescent protein (GFP) coding sequence (Genechem Co.,
Ltd., Shanghai, China) was used to generate FOXP2 expression
vectors. The T-vector-FOXP2 containing wild-type
FOXP2 cDNA (NM_014491.3, isoform I) was purchased from
OriGene Technologies Inc. (Rockville, MD, USA). The coding sequence
of FOXP2 was inserted into the lentiviral vector, GV287 by
homologous recombination. The wild-type FOXP2 lentivirus was
designated as LV-FOXP2-WT.
The poly-Q reduction (c.520delCAA) FOXP2
mutant vector was generated by four rounds of polymerase chain
reaction (PCR) using genomic DNA from an SSD patient that included
a heterozygous CAA deletion, as previously described (8). The primer sequences and vector
construction strategy are presented in Table I and Fig. 1, respectively. Primers 3 and 6 were
used to amplify the poly-Q reduction fragment, and primers 1 and 4
were used to obtain the sequence upstream of FOXP2. A
homologous recombination arm was introduced in primer 1, and
primers 2 (also including a homologous recombination arm) and 5
were used to amplify the downstream sequence. The entire
FOXP2 coding sequence was subsequently amplified using
primers 1 and 2 and the above PCR products (a, b and c) as
template. The mutant form of FOXP2 was inserted into the
lentiviral vector, GV287 by homologous recombination and designated
as LV-FOXP2-MT. The empty lentiviral vector, GV287 (termed LV-NC)
served as a negative control. All constructed vectors were
confirmed by direct sequencing.
 | Table IPrimers used in the present
study. |
Table I
Primers used in the present
study.
| Product | Primer
sequence | bp |
|---|
| a | P1 F:
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGATGCAGGAATCTGCGACAG-3′
P4 R: 5′-CTGCTGAGGAGACAGGACTTG-3′ | 378 |
| b | P3 F:
5′-CAAGTCCTGTCTCCTCAGCAG-3′
P6 R: 5′-GATGAGTCCCTGACGCTGAAG-3′ | 393 |
| c | P5 F:
5′-CTTCAGCGTCAGGGACTCATC-3′
P2 R: 5′-TCCTTGTAGTCCATACCTTCCAGATCTTCAGATAAAGGC-3′ | 1457 |
| d | P1 F:
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGATGCAGGAATCTGCGACAG-3′
P2 R:5′-TCCTTGTAGTCCATACCTTCCAGATCTTCAGATAAAGGC-3′ | 2186 |
Establishment of stably transfected
SH-SY5Y cell lines
The SH-SY5Y human cell-line (Genechem Co., Ltd.) was
used for expression analysis. Cells were plated at a density of
5×104 cells/well on poly-L-lysine-coated 6-well plates
(Sigma-Aldrich, St. Louis, MO, USA) in Dulbecco's modified Eagle's
medium/Ham's F12 nutrient mixture (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) as expansion medium (2
ml/well). Cells were grown at 37°C in the presence of 5%
CO2. SH-SY5Y cells were infected with the appropriate
lentivirus (LV-FOXP2-WT, LV-FOXP2-MT or LV-NC). Three days
following infection, the efficiency of lentiviral expression was
determined by visualizing the GFP reporter using a MicroPublisher
3.3 RTV microscope (Olympus Corporation, Tokyo, Japan). Stably
infected cells were cultured and harvested at 80% confluence for
subsequent RNA and protein extraction. Untreated SH-SY5Y cells
served as a blank control (control group).
RNA extraction and quantitative reverse
transcription (qRT)-PCR analysis
SH-SY5Y cells from each group were cultured in
12-well plates in expansion medium (2 ml/well) for three days, and
total RNA was extracted using Invitrogen TRIzol reagent (Thermo
Fisher Scientific, Inc.). The first strand cDNA was synthesized
using Axygen M-MLV-Reverse Transcriptase (Corning Life Sciences,
Union City, CA, USA). qRT-PCR was subsequently performed using an
Applied Biosystems ABI 7500 system (Thermo Fisher Scientific, Inc.)
with the SYBR Green Master mix (Takara Bio, Inc., Otsu, Japan).
Primers were synthesized as follows: Forward, 5′-GCG TCA GGG ACT
CAT CTCC-3′ and reverse, 5′-GAG GTC TAG CCC TCC ATG TTTA-3′ for
FOXP2; and forward, 5′-TTT AAT GGC CCG TTT ATGTC-3′ and
reverse, 5′-TTG TTT CGC ACT GGT TTAGG-3′ for CNTNAP2; and
forward, 5′-TGA CTT CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT
GTT GCT GTA GCC AAA-3′ for GAPDH (Genechem Co., Ltd.). The cycling
parameters were as follows: 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec, 60°C for 1 min, and 72°C for 30 sec.
Dissociation curves were generated to ensure that a single and
specific product was amplified. Cycle quantification values
(Cq) were analyzed by Applied Biosystems SDS1.4 software
(Thermo Fisher Scientific, Inc.) and relative quantification of the
FOXP2 and CNTNAP2 expression levels was determined
using the comparative Cq method with the GAPDH
transcript serving as an internal control.
Western blot analysis
Total protein (300 µg) was isolated from
infected SH-SY5Y cells and the protein concentration was determined
using a BCA Protein assay kit (GE Healthcare Life Sciences, Logan,
UT, USA). Equal quantities of protein (40 µg) were denatured
and separated by 10% SDS-PAGE (VE-180; Tanon Science &
Technology Co., Ltd., Shanghai, China) at 80 V for ~2 h. Gels were
transferred to polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA) at 300 mA for 150 min, membranes were blocked
with 5% milk in Tris-buffered saline Tween-20 (TBST; Genechem Co.,
Ltd.) buffer and incubated with primary antibodies, rabbit
monoclonal anti-FOXP2 (1:1,000; cat. no. 5337; Cell Signaling
Technology, Inc., Danvers, MA, USA), rabbit polyclonal anti-CASPR2
(1:1,000; cat. no. 3731; Cell Signaling Technology, Inc.) or mouse
monoclonal anti-GAPDH (1:10,000; cat. no. KC-5G4; KangChen Bio-Tech
Co. Ltd., Shanghai, China), overnight at 4°C. Following incubation
with goat polyclonal anti-rabbit and anti-mouse immunoglobulin
G-horseradish peroxidase secondary antibodies (1:2,000; sc-2004 and
sc-2005; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
detection was assessed with an ECL detection kit (Thermo Fisher
Scientific, Inc.). Densitometry was conducted using Image J 1.47
software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All analyses were performed using SPSS version 17.0
(SPSS, Inc., Chicago, IL, USA). All data are represented as the
mean ± standard deviation. Post hoc analysis of variance,
Student-Newman-Keuls (SNK) test and an independent sample Student's
t-test were used to investigate differences in CNTNAP2
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
Construction of the FOXP2 poly-Q
reduction mutant
Exon 5 of FOXP2 is a highly repetitive
sequence, which facilitated generation of the poly-Q reduction
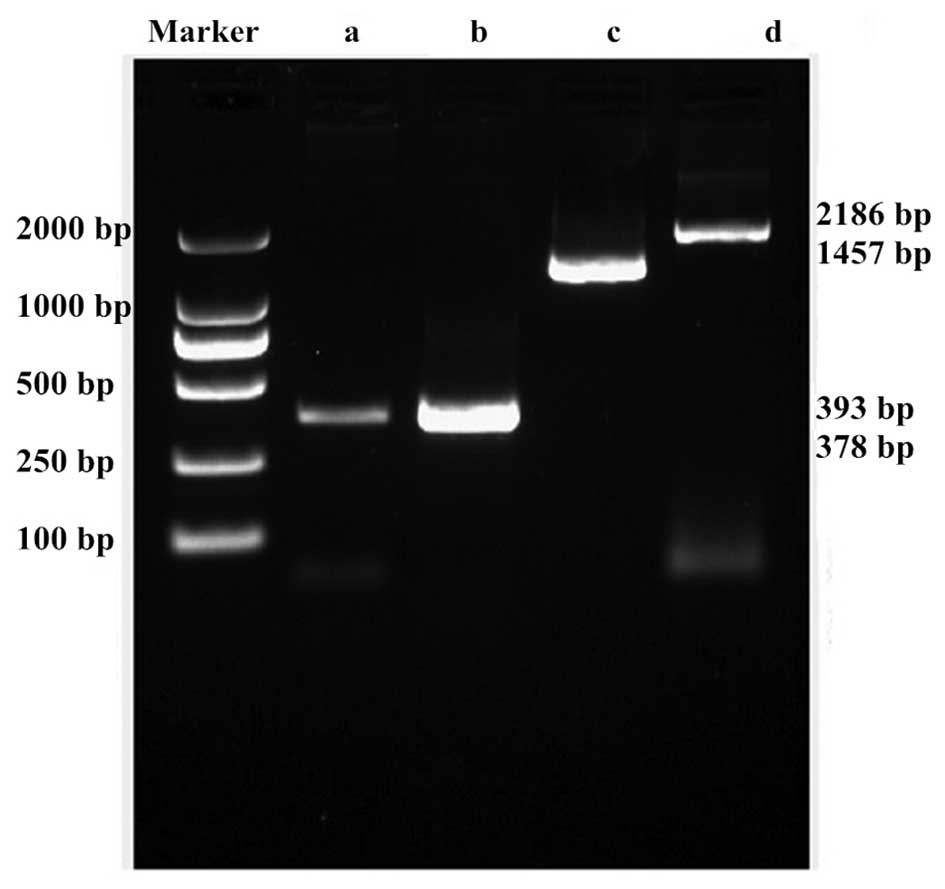
mutant (c.520delCAA) by multiple rounds of PCR (Fig. 1). The amplified products were
visualized by agarose gel electrophoresis and ethidium bromide
staining (Fig. 2), and the PCR
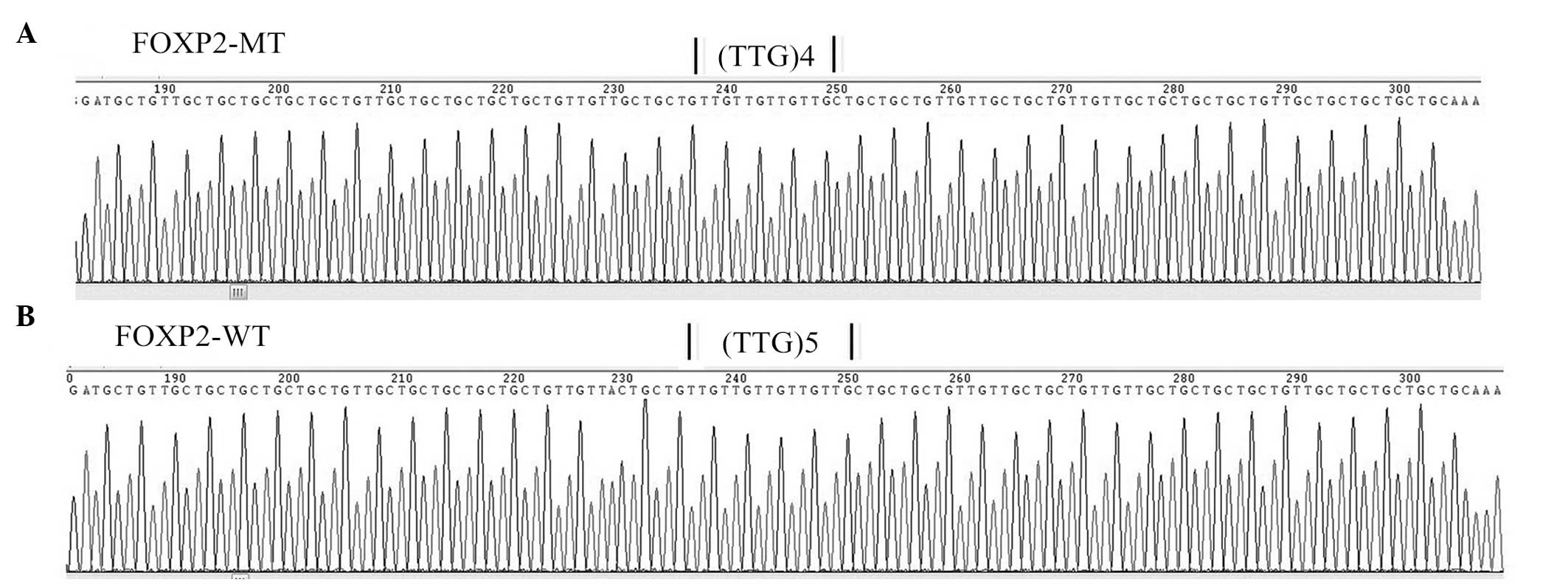
products were confirmed by sequencing. Reverse sequencing of CAACAG
repeats with one CAA deletion confirmed the poly-Q reduction in the
mutant, compared with the wild-type sequence (Fig. 3). The coding sequences of wild-type
and mutant FOXP2 were integrated into the GV287 lentiviral
expression system by homologous recombination to achieve stable
expression of FOXP2.
SH-SY5Y cells were stably infected with
LV-FOXP2-WT and LV-FOXP2-MT
The human SH-SY5Y cell line is a commonly used
cellular model for neuronal function. SH-SY5Y cells were infected
with the LV-FOXP2-WT, LV-FOXP2-MT or LV-NC constructs, and
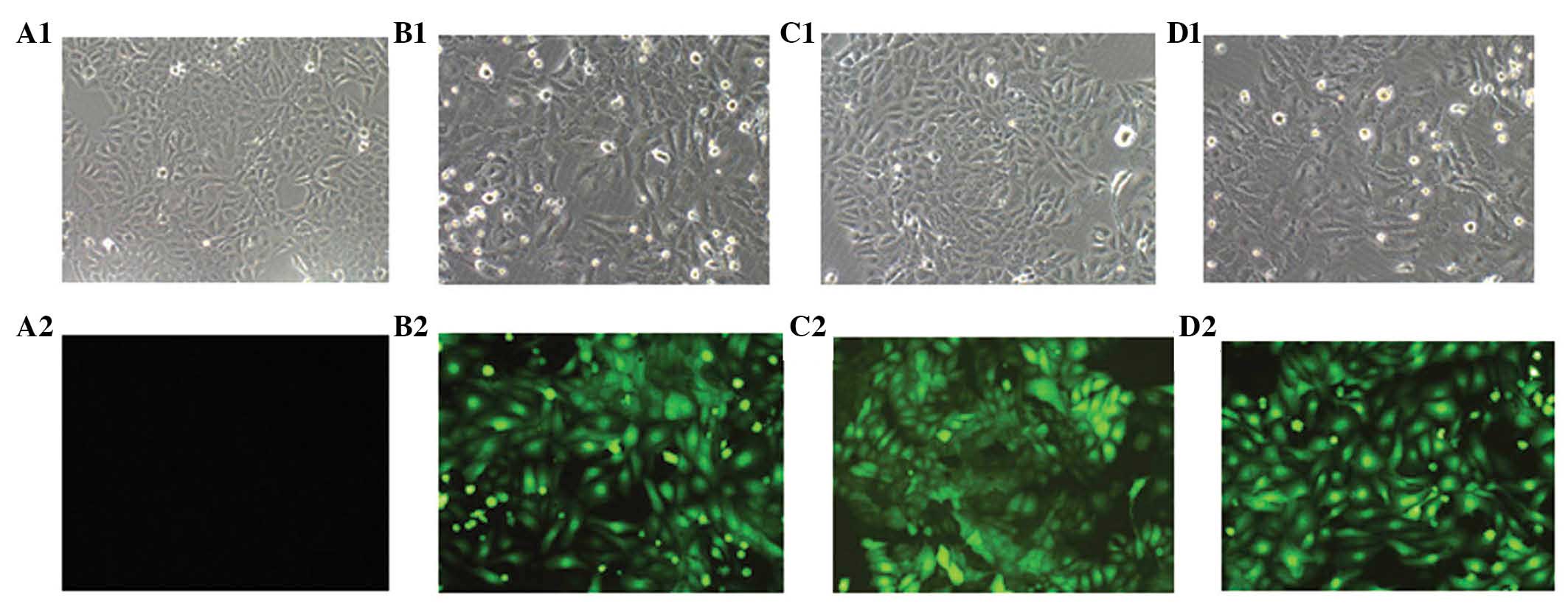
expression of GFP was visualized by fluorescence microscopy to
determine the efficiency of infection. As presented in Fig. 4, a high intensity of enhanced GFP
fluorescence was apparent in the cells infected with all three
constructs, however, this was not observed in the untreated cells
(control group). A fluorescence ratio of >80% was considered to
indicate stably transfected cells.
Wild-type FOXP2 and the poly-Q reduction
mutant were highly expressed in infected SH-SY5Y cells
FOXP2 expression levels were determined by
qRT-PCR, and FOXP2 mRNA expression levels were significantly
higher in SH-SY5Y cells harboring LV-FOXP2-WT and LV-FOXP2-MT
constructs (P<0.05) (Fig. 5A).
In addition, FOXP2 protein levels were higher in infected cells, as
determined by western blotting (Fig.
5B). These results indicate that the SH-SY5Y cell model
overexpressed wild-type FOXP2 and the poly-Q reduction mutant.
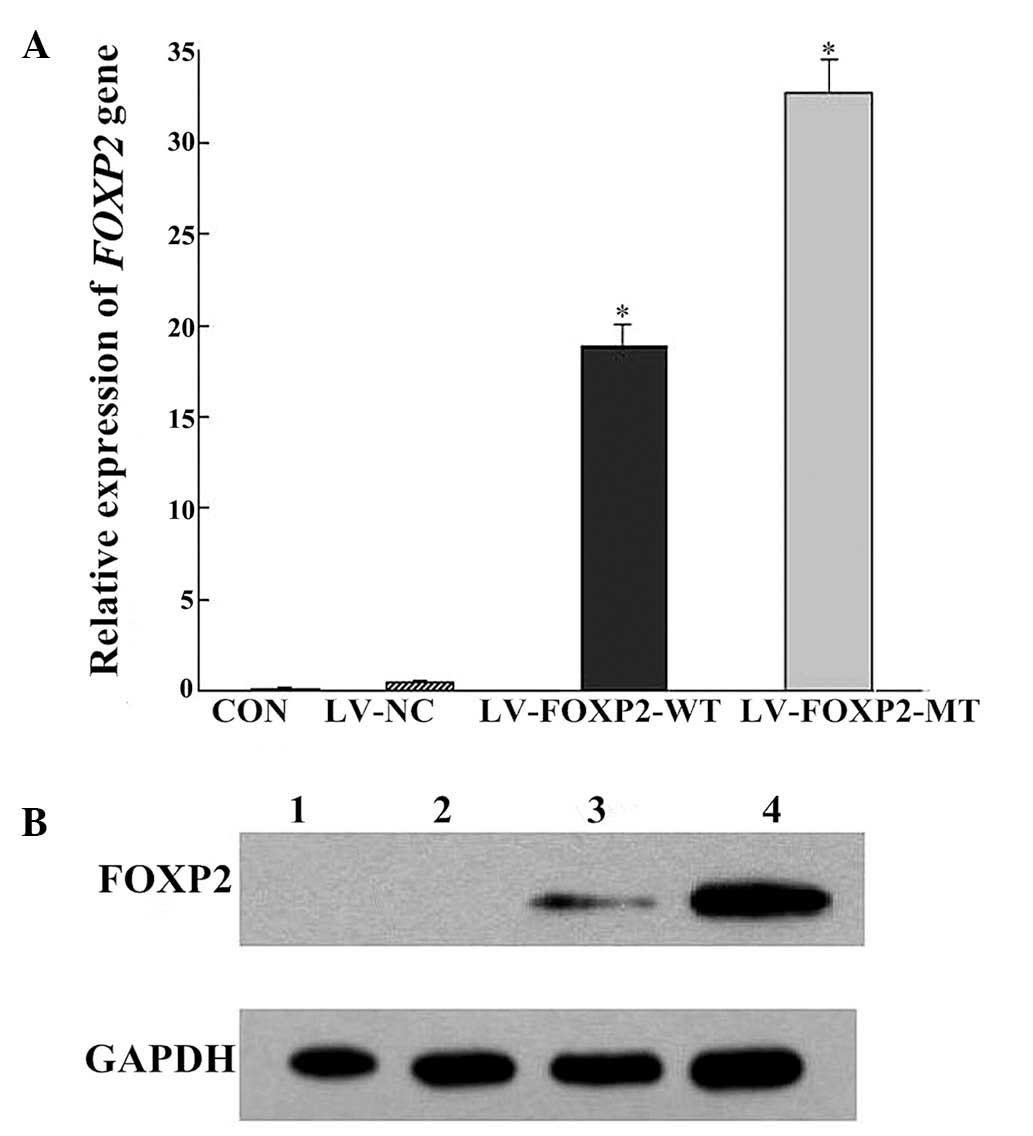 | Figure 5FOXP2 expression in cells transfected
with LV-FOXP2-WT and LV-FOXP2-MT as determined by (A) quantitative
reverse transcription-polymerase chain reaction
(*P<0.05, vs. the LV-NC) and (B) western blotting
(*P<0.05, vs. the LV-NC). Lane 1, CON; lane 2, LV-NC;
lane 3, LV-FOXP2-WT; and lane 4, LV-FOXP2-MT. FOXP2, forkhead box
protein P2; MT, mutant; WT, wild-type; LV, lentivirus; NC, negative
control; CON, control. |
The FOXP2 poly-Q reduction mutant
upregulated the expression of CNTNAP2
In order to investigate the role of poly-Q reduction
on CNTNAP2 expression levels, CNTNAP2 mRNA expression
levels in the stably infected cell lines were determined by
qRT-PCR. CNTNAP2 mRNA levels were observed to be 0.105-fold
lower in cells infected with LV-FOXP2-WT than those infected with
LV-NC, indicating inhibition of CNTNAP2 expression by
wild-type FOXP2 (Fig. 6A). In
addition, CNTNAP2 mRNA expression levels were 1.788-fold
higher in cells infected with LV-FOXP2-MT than those infected with
LV-NC, and 17.03-fold higher than those infected with LV-FOXP2-WT.
These results indicate that poly-Q reduction of FOXP2 reversed the
suppressive effect of the wild-type protein and upregulated
transcription of CNTNAP2.
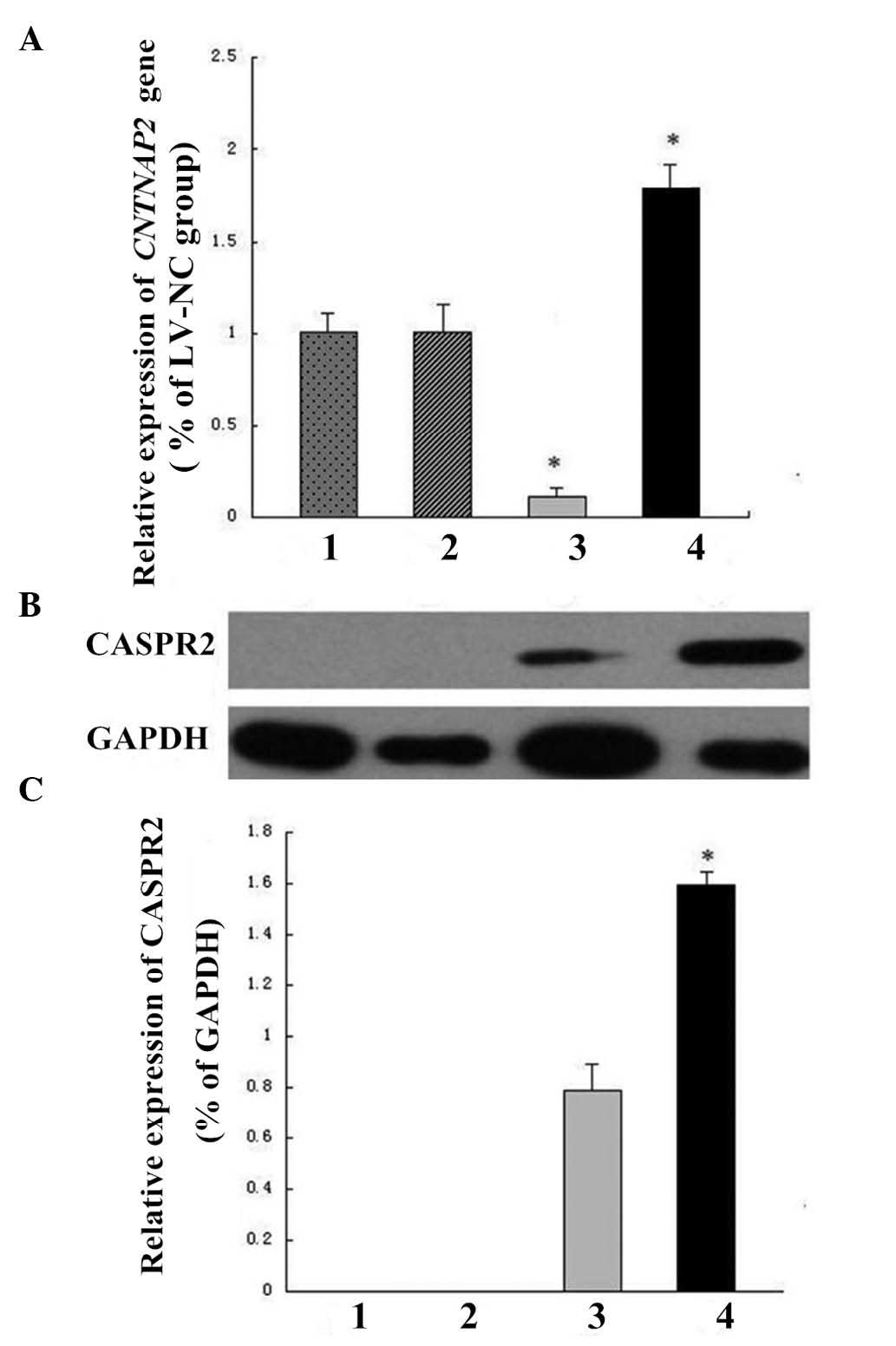 | Figure 6Upregulation of CNTNAP2 expression by
FOXP2-MT as determined by qRT-PCR and western blotting. (A)
CNTNAP2 transcription identified using qRT-PCR.
CNTNAP2 expression level is significantly higher in the
cells transfected with LV-FOXP2-MT than in the cells of the CON and
LV-NC groups; the CNTNAP2 expression level is significantly
lower in cells transfected with LV-FOXP2-WT
(*P<0.01). (B) Analysis of FOXP2 and CASPR2 protein
levels by western blotting. The density of the band corresponding
to CASPR2 in the SH-SY5Y-FOXP2-MT cells is markedly higher than
that in the SH-SY5Y-FOXP2-WT cells. Lane 1, CON; lane 2, LV-NV;
lane 3, LV-FOXP2-WT; and lane4, LV-FOXP2-MT. (C) The level of
CASPR2 expression in the LV-FOXP2-MT group is significantly higher
than that in the LV-FOXP2-WT group (*P<0.01).
CNTNAP2, contactin-associated protein-like 2; LV, lentivirus; NC,
negative control; CASPR2, contactin-associated protein-like 2;
FOXP2, forkhead box protein P2; MT, mutant; qRT-PCR, quantitative
reverse transcription-polymerase chain reaction; CON, control; WT,
wild-type. |
The levels of the CASPR2 protein, encoded by
CNTNAP2, were subsequently determined by western blotting,
and CASPR2 expression was not observed in the untreated SH-SY5Y
cells (control) or cells infected with LV-NC (Fig. 6B). However, low expression levels
of CASPR2 were detected in cells infected with LV-FOXP2-WT, and
significantly higher expression levels of CASPR2 were demonstrated
in cells infected with LV-FOXP2-MT (P<0.01), indicating an
upregulation of CNTNAP2 by the FOXP2 poly-Q reduction mutant
(Fig. 6C).
Discussion
In the present study, the SH-SY5Y human
neuroblastoma cell line was engineered to stably express wild-type
FOXP2 and a poly-Q reduction mutant. Using these cell lines, the
effects of poly-Q reduction on the FOXP2-mediated regulation of
CNTNAP2 was assessed, and the FOXP2 mutant containing a
shorter poly-Q tract was demonstrated to upregulate the expression
of CNTNAP2 at the mRNA and protein levels.
The FOXP2 gene encodes an important nervous
system transcription factor that includes a long poly-Q tract in
exon 5 that varies in length between 34 and 40 glutamine residues
(9). However, the physiological
consequences of mutations in this poly-Q tract are not well
understood. Wassink et al (25) previously detected small internal
deletions in the Q40 tract in two families with autism. MacDermot
et al (3) reported a
heterozygous insertion of the sequence, CAGCAGCAACAA into the
poly-Q region of exon 5 in a proband with developmental verbal
dyspraxia. In our previous study the poly-Q coding region of
FOXP2 in SSD patients from a Chinese Han population was
sequenced, and it was observed that a single CAA codon was deleted,
which decreased the number of glutamines from 40 to 39 (8). The underlying mechanism by which
FOXP2 poly-Q reduction affects SSD requires further
investigation.
Identification of putative target genes that may
interact with or be regulated by the FOXP2 neural transcription
factor is important. The CNTNAP2 gene encodes the CASPR2
protein, which is highly abundant in the frontal and temporal
lobes, in the striatal circuits of the developing human brain, and
in the frontal cortex of the adult brain (17). These regions are involved in speech
and language learning. Vernes et al (15) reported that FOXP2 markedly
downregulates the expression of CNTNAP2. The effects of
FOXP2 poly-Q reduction on the expression of CNTNAP2 in
SH-SY5Y cells, which have particularly low endogenous FOXP2
expression levels (26), was also
investigated in the current study. The cells were stably infected
with wild-type and mutant FOXP2 expression vectors using a
lentiviral expression system, and GFP fluorescence was used to
confirm a successful infection and compare the expression levels.
Expression of wild-type and mutant FOXP2 was indicated at the mRNA
and protein levels. However, the expression of mutant FOXP2 was
significantly higher than that of the wild-type protein, for
reasons that remain to be elucidated, however, post-transcriptional
regulation, such as mRNA degradation may contribute to this
variance. The expression level of CNTNAP2 was significantly
decreased in cells expressing wild-type FOXP2 compared with the
non-expressing controls, which was consistent with a previous study
(15). Notably, the expression
level of CNTNAP2 was significantly higher in cells
expressing the FOXP2 poly-Q reduction mutant, which reversed the
repressive effect of the wild-type protein, and thus markedly
upregulated the level of CNTNAP2 expression. To the best of
our knowledge, the present study is the first to demonstrate that
this divergence in the regulatory activity of wild-type and mutant
FOXP2 on CNTNAP2 expression, may underlie the pathogenic
mechanism by which poly-Q reduction is associated with SSD.
Furthermore, these results indicate a possible association between
CNTNAP2 and SSD.
The method by which deletion of a single glutamine
from the poly-Q tract of FOXP2 markedly affects the transcription
and expression of CNTNAP2 remains to be elucidated.
According to a previous study (27), the number of repeats in the FOXP2
poly-Q tract is relatively monomorphic in a healthy population, and
in patients presenting with severe speech and language disorders,
although 40 is considered to be a large number of repeats,
therefore, greater variability may be expected. Sequences may
include up to 14 CAA interruptions among the CAG repeats, which
prevent slippage and expansion. Therefore, deletion of a CAA codon
may decrease the stability of poly-Q tracts, which has been
observed to form a hairpin structure in FOXP2 transcripts
(27). Deletion of CAA may alter
the configuration of the hairpin, which may affect the binding of
FOXP2 to the CNTNAP2 promoter region.
In the present study, the results of the qRT-PCR
experiments demonstrate CNTNAP2 was expressed in the CON and
LV-NC cells, however, the CASPR2 protein was not observed in these
cells in the western blotting experiments. By contrast, the CASPR2
protein was detected in SH-SY5Y cells transfected with wild-type
FOXP2 and the poly-Q mutant. The reason for this discrepancy
has not yet been elucidated, and it has not, to the best of our
knowledge, been reported whether CASPR2 is expressed in SH-SY5Y
cells. CNTNAP2 is a target of FOXP2, by which it is directly
regulated (15). CASPR2 expression
may be switched off in the absence of FOXP2, however when present,
FOXP2 initiates transcription of CNTNAP2 and subsequent
accumulation of CASPR2.
In conclusion, the present study demonstrates that
CNTNAP2 expression was significantly upregulated in SH-SY5Y
cells expressing a poly-Q reduction mutation of FOXP2.
CNTNAP2 may, therefore, also be associated with SSD. Further
in vivo investigations using animal models are required to
clarify the exact role of poly-Q reduction of FOXP2 in
SSD.
Acknowledgments
The present study was supported by a grant from the
National Natural Science Foundation of China (grant nos. 81101019
and 81100187).
References
|
1
|
Fisher SE, Lai CS and Monaco AP:
Deciphering the genetic basis of speech and language disorders.
Annu Rev Neurosci. 26:57–80. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai CS, Fisher SE, Hurst JA, Vargha-Khadem
F and Monaco AP: A forkhead-domain gene is mutated in a severe
speech and language disorder. Nature. 413:519–523. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
MacDermot KD, Bonora E, Sykes N and Coupe
AM: Identification of FOXP2 truncation as a novel cause of
developmental speech and language deficits. Am J Hum Genet.
76:1074–1080. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shriberg LD, Ballard KJ, Tomblin JB, Duffy
JR, Odell KH and Williams CA: Speech, prosody, and voice
characteristics of a mother and daughter with a 7;13 translocation
affecting FOXP2. J Speech Lang Hear Res. 49:500–525. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug
J, Nakabayashi K, Finucane B, Hartung D, Innes M, Kerem B, Nowaczyk
MJ, et al: Absence of a paternally inherited FOXP2 gene in
developmental verbal dyspraxia. Am J Hum Genet. 79:965–972. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeesman S, Nowaczyk MJ, Teshima I, Roberts
W, Cardy JO, Brian J, Senman L, Feuk L, Osborne LR and Scherer SW:
Speech and language impairment and oromotor dyspraxia due to
deletion of 7q31 that involves FOXP2. Am J Med Genet A.
140:509–514. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lennon PA, Cooper ML, Peiffer DA,
Gunderson KL, Patel A, Peters S, Cheung SW and Bacino CA: Deletion
of 7q31.1 supports involvement of FOXP2 in language
impairment:clinical report and review. Am J Med Genet A. 143
A:791–798. 2007. View Article : Google Scholar
|
|
8
|
Zhao Y, Ma H, Wang Y, Gao H, Xi C, Hua T,
Zhao Y and Qiu G: Association between FOXP2 gene and speech sound
disorder in Chinese population. Psychiatry Clin Neurosci.
64:565–573. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Butland SL, Devon RS, Huang Y, Mead CL,
Meynert AM, Neal SJ, Lee SS, Wilkinson A, Yang GS, Yuen MM, et al:
CAG-encoded polyglutamine length polymorphism in the human genome.
BMC Genomics. 8:1262007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jodice C, Giovannone B, Calabresi V,
Bellocchi M, Terrenato L and Novelletto A: Population variation
analysis at nine loci containing expressed trinucleotide repeats.
Ann Hum Genet. 61:425–438. 1997. View Article : Google Scholar
|
|
11
|
Wren JD, Forgacs E, Fondon JW III,
Pertsemlidis A, Cheng SY, Gallardo T, Williams RS, Shovet RV, Minna
JD and Garner HR: Repeat polymorphisms within gene regions:
Phenotypic and evolutionary implications. Am J Hum Genet.
67:345–356. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mularoni L, Guigó R and Albà MM: Mutation
patterns of amino acid tandem repeats in the human proteome. Genome
Biol. 7:R332006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sobczak K and Krzyzosiak WJ: Patterns of
CAG repeat interruptions in SCA1 and SCA2 genes in relation to
repeat instability. Hum Mutat. 24:236–247. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
MacDonald M: the Huntington's Disease
Collaborative Research Group: A novel gene containing a
trinucleotide repeat that is expanded and unstable on Huntington's
disease chromosomes. Cell. 72:971–983. 1993. View Article : Google Scholar
|
|
15
|
Vernes SC, Newbury DF, Abrahams BS,
Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE,
Geschwind DH, et al: A functional genetic link between distinct
developmental language disorders. N Engl J Med. 359:2337–2345.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakabayashi K and Scherer SW: The human
contactin-associated protein-like 2 gene (CNTNAP2) spans over 2 Mb
of DNA at chromosome 7q35. Genomics. 73:108–112. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abrahams BS, Tentler D, Perederiy JV,
Oldham MC, Coppola G and Geschwind DH: Genome-wide analyses of
human perisylvian cerebral cortical patterning. Proc Natl Acad Sci
USA. 104:17849–17854. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alarcón M, Abrahams BS, Stone JL, Duvall
JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter
DH, et al: Linkage, association, and gene-expression analyses
identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet.
82:150–159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arking DE, Cutler DJ, Brune CW, Teslovich
TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH Jr, et al: A
common genetic variant in the neurexin superfamily member CNTNAP2
increases familial risk of autism. Am J Hum Genet. 82:160–164.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rossi E, Verri AP, Patricelli MG,
Destefani V, Ricca I, Vetro A, Ciccone R, Giorda R, Toniolo D,
Maraschio P and Zuffardi O: A 12Mb deletion at 7q33-q35 associated
with autism spectrum disorders and primary amenorrhea. Eur J Med
Genet. 51:631–638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bakkaloglu B, O'Roak BJ, Louvi A, Gupta
AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG,
Stillman AA, et al: Molecular cytogenetic analysis and resequencing
of contactin associated protein-like 2 in autism spectrum
disorders. Am J Hum Genet. 82:165–173. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jackman C, Horn ND, Molleston JP and Sokol
DK: Gene associated with seizures, autism and hepatomegaly in an
Amish girl. Pediatr Neurol. 40:310–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Hu Z, He Y, Xiong Z, Long Z, Peng Y,
Bu F, Ling J, Xun G, Mo X, et al: Association analysis of CNTNAP2
polymorphisms with autism in the Chinese Han population. Psychiatr
Genet. 20:113–7. 2010.PubMed/NCBI
|
|
24
|
Whitehouse AJ, Bishop DV, Ang QW, Pennell
CE and Fisher SE: CNTNAP2 variants affect early language
development in the general population. Genes Brain Behav.
10:451–456. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wassink TH, Piven J, Vieland VJ, Pietila
J, Goedken RJ, Folstein SE and Sheffield VC: Evaluation of FOXP2 as
an autism susceptibility gene. Am J Med Genet. 114:566–569. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Konopka G, Bomar JM, Winden K, Coppola G,
Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA and Geschwind
DH: Human-specific transcriptional regulation of CNS development
genes by FOXP2. Nature. 462:213–217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sobczak K and Krzyzosiak WJ: CAG repeats
containing CAA interruptions form branched hairpin structures in
spinocerebellar ataxia type 2 transcripts. J Biol Chem.
280:3898–3910. 2005. View Article : Google Scholar
|